Open Access Article
Open Access ArticleInfluence of ions to modulate hydrazone and oxime reaction kinetics to obtain dynamically cross-linked hyaluronic acid hydrogels†
Shujiang
Wang‡
ab,
Ganesh N.
Nawale‡
 b,
Oommen P.
Oommen
b,
Oommen P.
Oommen
 c,
Jöns
Hilborn
b and
Oommen P.
Varghese
c,
Jöns
Hilborn
b and
Oommen P.
Varghese
 *b
*b
aMaisonneuve-Rosemont Hospital Research Centre & Dept. of Ophthalmology, University of Montreal, Montreal, Canada
bTranslational Chemical Biology Laboratory, Department of Chemistry, Ångström Laboratory, Uppsala University, 751 21, Uppsala, Sweden
cBioengineering and Nanomedicine Lab, Faculty of Medicine and Health Technologies and BioMediTech Institute, Tampere University, Korkeakoulunkatu 3, Tampere-33720, Finland. E-mail: oommen.varghese@kemi.uu.se
First published on 4th July 2019
Abstract
Dynamic covalent chemistry forming hydrazone and oxime linkages is attractive due to its simplicity, selectivity and compatibility under aqueous conditions. However, the low reaction rate at physiological pH hampers its use in biomedical applications. Herein, we present different monovalent and bivalent aqueous salt solutions as bio-friendly, non-toxic catalysts which can drive the hydrazone and oxime reactions with excellent efficacy at physiological pH. Direct comparison of hydrazone and oxime reactions using a small molecule model, without any salt catalysis, indicated that oxime formation is 6-times faster than hydrazone formation. Addition of different salts (NaCl, NaBr, KCl, LiCl, LiClO4, Na2SO4, MgCl2 and CaCl2) accelerated the pseudo-first-order reaction kinetics by ∼1.2–4.9-fold for acylhydrazone formation and by ∼1.5–6.9-fold for oxime formation, in a concentration-dependent manner. We further explored the potential of such catalysts to develop acylhydrazone and oxime cross-linked hyaluronic acid (HA) hydrogels with different physicochemical properties without changing the degree of chemical modification. Analogous to the small molecule model system, the addition of monovalent and divalent salts as catalysts significantly reduced the gelling time. The gelling time for the acylhydrazone cross-linked HA-hydrogel (1.6 wt%) could be reduced from 300 min to 1.2 min by adding 100 mM CaCl2, while that for the oxime cross-linked HA-hydrogel (1.2 wt%) could be reduced from 68 min to 1.1 min by adding 50 mM CaCl2. This difference in the gelling time also resulted in hydrogels with differential swelling properties as measured after 24 h. Our results are the first to demonstrate the use of salts, for catalyzing hydrogel formation under physiologically relevant conditions.
Introduction
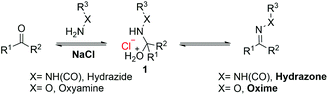
Hydrogels are commonly composed of highly hydrated polymer structures with tunable biophysical and biochemical properties allowing many biomedical applications.1–3 Hydrogels developed from biopolymers are generally less toxic and could be used to prepare biomaterials that mimic the native extracellular matrix (ECM) produced by cells.4 When such materials can be obtained by employing mild reaction conditions without any toxic excipients, they can be used to deliver living cells and sensitive biomolecules in vivo for biomedical applications.5 There are different methods to develop hydrogels including chemically modified polymers having reactive functional groups to obtain covalent cross-links, physical entanglement or electrostatic interactions between oppositely charged polymers or materials.6–9 Among these, dynamic covalent chemistry, such as hydrazone and oxime formation reactions, has gained much attention due to its simplicity, selectivity and efficiency and the requirement of mild reaction conditions.9–13 However, these conjugation strategies are associated with a low reaction rate at physiological pH.14 To overcome this problem, several synthetic catalysts have been developed to enhance the rates of hydrazone and oxime formation reactions.15–18 The most commonly used catalysts include aniline19 and aniline analogues18,20 which demonstrate good catalytic efficiency at acidic pH with aldehyde substrates, while the catalytic efficiency declines dramatically at physiological pH, especially with the less reactive keto substrates.15 Other nucleophilic catalysts were also developed to improve coupling reactions at physiological pH.15–17,20,21 The reaction mechanism for hydrazone and oxime formation at neutral pH involves the formation of a charged intermediate 1, followed by a dehydration step of the intermediate 1 which is the rate-limiting step (Scheme 1).We have recently demonstrated that carboxylates can promote the protonation of the hydroxyl oxygen atom in 1 at neutral pH by forming a cyclic intermediate.22 We also discovered that simple salts such as sodium chloride (NaCl) could accelerate oxime formation at neutral pH with both the aldehyde and less reactive keto substrates by stabilizing the rate limiting transition state, thereby promoting the dehydration step.23 It is important to note that so far the salt based catalytic system has only been applied to oxime-based conjugation reactions; however, the effect of salts on the formation of hydrazone at neutral pH remains unclear. We hereby report the effects of different salts on the reaction kinetics of acylhydrazone and oxime formation reactions. We also demonstrate the influence of salts on the cross-linking efficiency of biomacromolecules to form hydrogels. We explored the gelling kinetics in the presence of different salts, without changing the degree of chemical modification on the polymers.
Results and discussion
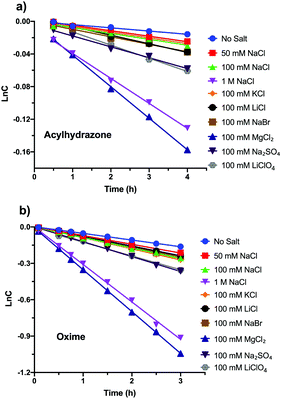
To evaluate the effects of various salts on the reaction kinetics of acylhydrazone and oxime formation reactions, we first performed model reactions with a small molecule model, namely, 4-nitrobenzaldehyde as an electrophile and adipic dihydrazide or (aminooxy)methane as a nucleophile (Scheme S1, ESI†). The pseudo-first-order reaction rates were analyzed in 10 mM phosphate buffer (PB, pH 7.4) containing 10% DMF by employing different salt concentrations (Fig. 1). Interestingly, an aqueous solution containing NaCl indeed accelerated both the acylhydrazone and oxime formation reactions in a salt concentration-dependent manner, which corroborates with our recent observation of oxime reaction catalysis with saline.23In the case of acylhydrazone, when 50 mM NaCl was used as a catalyst, the reaction rates were increased by ∼1.2-fold as compared to that of the uncatalyzed reaction (Fig. 2a). Gratifyingly, when the NaCl concentration was further increased to 100 mM and 1 M, we observed an ∼1.4-fold and ∼3.9-fold increase in the reaction rate, compared to that of the uncatalyzed reaction (Fig. 2a). When similar salt concentrations (50 mM, 100 mM and 1 M NaCl) were used in the oxime formation reaction, we observed ∼1.5-fold, ∼1.6-fold and ∼5.8-fold increments in the rate as compared to those of the uncatalyzed reactions (Fig. 2b). Though the reaction kinetics indicated a similar enhancement of reaction rates for both acylhydrazone and oxime reactions, it should be noted that the oxime formation reaction was ∼6.2-times faster than the acylhydrazone formation reaction without any salt (kobs = 0.054 for oxime and kobs = 0.0087 for acylhydrazone).
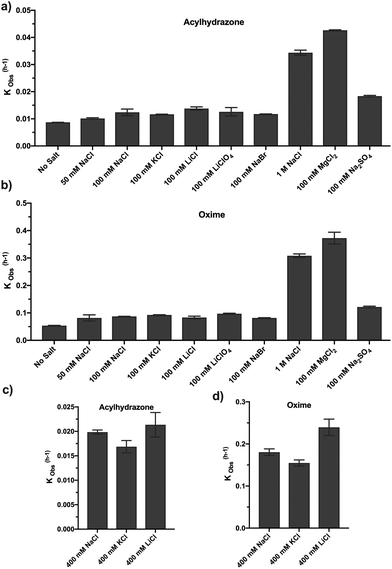
Since both the cation and anion participate in the stabilization of the rate-limiting step, overcoming the α-effect induced by nucleophilic reagents23,24 such as hydrazide and aminooxy groups, we investigated the effect of other ions on the reaction rate. All the monovalent salts tested (NaCl, NaBr, KCl, LiCl, and LiClO4) indicated similar levels of increase in the reaction rate (∼1.4–1.8-fold) with both hydrazide and aminooxy substrates when the salt concentration was kept at 100 mM. Interestingly, when 100 mM Na2SO4 and MgCl2 were used in the reaction, a ∼2.1-fold and ∼4.9-fold increase in the reaction rate was observed, respectively, for acylhydrazone formation (Fig. 2a). A similar pattern of rate increase (2.3-fold and ∼7-fold) was observed for oxime formation using the same catalysts (Fig. 2b). We also attempted to perform experiments with CaCl2 as a catalyst; however, at a 100 mM concentration of CaCl2, we observed precipitation of the salt. Taken together, these results indicate that different ions in the reaction mixture catalyse the oxime and acylhydrazone formation reactions at physiological pH. We believe that this effect is due to the increased ionic strength in the reaction medium which increases the local concentration of the organic molecules (‘salting-out effect’) and a decrease in the energy of the transition state, similar to our previous observation.23 In order to evaluate the salt effect, we performed reactions with a salt known to have a salt-out (LiCl) or salt-in (KCl) effect based on the Hofmeister series with a higher salt concentration (400 mM).25 These experiments indicated that for both the hydrazone and oxime-formation reactions, LiCl accelerated the reaction rate, suggesting a salt-out effect, whereas KCl reduced the rate as compared to the NaCl catalyzed reaction (Fig. 2c and d). In order to determine whether our results are not due to an ‘indirect’ field effect, we compared the reaction rates using LiCl and LiClO4 having different anions. These experiments indicated a similar increase in the reaction rate, suggesting that the observed reaction rate is not due to the field effect but due to the salt-out effect (Fig. S1a and S1b in the ESI†). It is noteworthy that the rate of the salt catalyzed reaction could be further enhanced by performing such reactions at a higher temperature (Fig. S1c in the ESI†).
To exemplify the utility of these catalysts for bioconjugation, we explored the effects of salts to catalyze hydrazone and oxime cross-linking reactions to prepare hydrogels. For this purpose, we chemically modified hyaluronic acid (HA), a natural extracellular matrix component, with aldehyde and hydrazide/aminooxy modifications. HA is known to play a significant role in cell proliferation and migration26 and in maintaining the stem cell niche within our body.27 Therefore hydrogels using HA components have been extensively used for several biomedical applications, such as cell delivery,28 and drug delivery29 and in the field of regenerative medicine.30 We have previously developed a hydrazone-cross-linked HA hydrogel29 and used it to deliver recombinant human bone morphogenetic protein-2 (rhBMP-2) to regenerate bone in vivo.8,31 We have also utilized such materials to deliver small molecule drugs32 and large nucleic acid drugs28,33,34 and for tuning the differential release of proteins.35 One of the key aspects of hydrogel preparation is the reaction kinetics of gel formation. We have recently shown that by modulating the thiol pKa, in a thiol-functionalized HA, it is possible to control the cross-linking density and hydrogel swelling properties of the disulfide cross-linked HA hydrogel.36 This prompted us to investigate the effect of gelling kinetics on the properties of hydrazone and oxime cross-linked hydrogels.
To develop hydrazone cross-linked HA hydrogels, we first prepared aldehyde and hydrazide derivatives of HA using our previously optimised method.29,37 It is noteworthy that the hydrazide unit contained a urea-type of linkage which allowed the delocalisation of charges, resulting in improved stability of the hydrazone linkages. The degree of modification was observed to be 6% for hydrazide modification and 12% for aldehyde modification. To develop oxime cross-linked HA-hydrogels, we synthesized a novel aminooxy HA employing a carbodiimide coupling reaction with HA at pH 4.7 with a 10-fold excess of the diaminooxy linker (with respect to the carbodiimide, Scheme S3, ESI†) with a dual modification.37 The diaminooxy linker was prepared following a reported method (Scheme S2, ESI†).38 The aminooxy modified HA derivative was purified by dialysis and characterized by 1H NMR analysis (Fig. S1, ESI†). The degree of aminooxy substitution was determined by UV-vis spectrophotometry by following a modified trinitrobenzene sulfonic acid (TNBS) assay.39 These experiments indicated a 7% modification of HA-aminooxy residues with respect to the disaccharide repeat unit. The chemically modified polymers were used to prepare HA hydrogels in PB (10 mM, pH 7.4) with different concentrations of salts by mixing an equal volume ratio (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) of HA-hydrazide/aminooxy and HA-aldehyde derivatives at 16 mg mL−1 concentrations (Scheme S4, ESI†) to obtain gels with 1.6 wt%. The influence of the aqueous salt solution on the cross-linking reaction kinetics was determined by rheological measurements employing oscillatory time sweeps on the reaction mixture at a 1 Hz frequency and a controlled gap distance of 1 mm. The storage modulus (G′) and loss modulus (G′′) were monitored against time to determine the gel point (the time when G′ = G′′).
1) of HA-hydrazide/aminooxy and HA-aldehyde derivatives at 16 mg mL−1 concentrations (Scheme S4, ESI†) to obtain gels with 1.6 wt%. The influence of the aqueous salt solution on the cross-linking reaction kinetics was determined by rheological measurements employing oscillatory time sweeps on the reaction mixture at a 1 Hz frequency and a controlled gap distance of 1 mm. The storage modulus (G′) and loss modulus (G′′) were monitored against time to determine the gel point (the time when G′ = G′′).
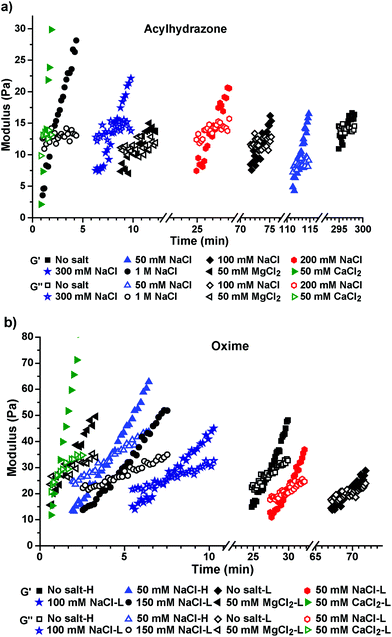
The acylhydrazone cross-linked HA hydrogels exhibited a slower reaction rate with a gelation time of ∼5 h under uncatalyzed reaction conditions (Fig. 3a and 4a). Interestingly, the gelation time decreased to ∼2 h in the presence of 50 mM NaCl, which further decreased to ∼71.6 min and ∼26.2 min with 100 mM and 200 mM NaCl, respectively. When the NaCl concentration was further increased to 300 mM and 1 M, the gelation time was dramatically decreased to ∼8.3 min and ∼2.3 min, respectively (Fig. 3a and 4a).
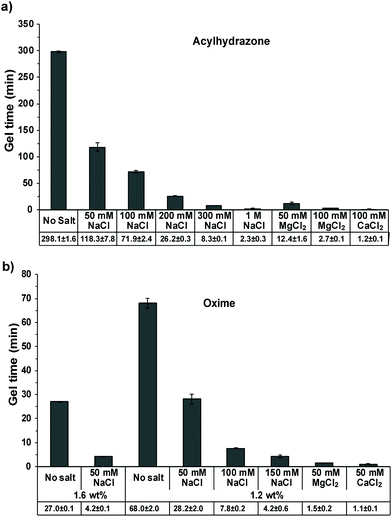 | ||
| Fig. 4 The gel time of (a) acylhydrazone cross-linked HA-hydrogel with an HA concentration of 1.6 wt% and (b) oxime cross-linked HA-hydrogel with an HA concentration of 1.6 wt% or 1.2 wt%. | ||
When 100 mM MgCl2 and 100 mM CaCl2 were used as catalysts, the gelation time reduced to ∼2.7 min and ∼1.2 min, respectively, for the acylhydrazone-cross-linked hydrogel (Fig. 3 and 4). In the oxime-cross-linked hydrogel formation reaction (1.2 wt%), 50 mM MgCl2 and 50 mM CaCl2 reduced the gelation time to ∼1.5 min and ∼1.1 –min, respectively. We then proceeded to evaluate the hydrogel rheological properties after 48 h of mixing the gel components. For this purpose, we prepared HA hydrogels with different salts as discussed above. After 24 h, these hydrogels were suspended in 10 mM PBS for an additional 24 h to remove excess salts, and then the rigidity of the hydrogels was analyzed by performing rheological measurements using oscillatory frequency sweep. The observed rheological data (Table 1) therefore represent a relative measure of the cross-linking densities that were obtained using different salt concentrations during the cross-linking reaction.
| Hydrogel | Salt | G′ (Pa) | G′′ (Pa) |
|---|---|---|---|
| Acylhydrazone | No salt | 416.5 ± 8.2 | 13.8 ± 1.1 |
| 50 mM NaCl | 435.8 ± 13.0 | 13.7 ± 2.1 | |
| 50 mM MgCl2 | 1145.0 ± 25.1 | 17.6 ± 0.2 | |
| 150 mM NaCl | 1240.0 ± 18.0 | 17.7 ± 0.8 | |
| Oxime | No salt | 2101.5 ± 17.5 | 34.2 ± 6.4 |
| 50 mM NaCl | 3171.0 ± 156.0 | 39.6 ± 5.4 | |
| 50 mM MgCl2 | 2067.0 ± 13.0 | 22.2 ± 5.6 |
As shown in Table 1, for acylhydrazone cross-linked hydrogels, the rigidity increased with an increase in the salt concentration. For oxime hydrogels, however, the addition of 50 mM NaCl increased the storage modulus but not when 50 mM MgCl2 was used. We believe this is due to the extremely high reaction rate when MgCl2 was used as a catalyst resulting in inhomogeneous hydrogel formation due to inefficient mixing of the gel precursors. These results indicate that optimal reaction kinetics for oxime formation is needed for appropriate gel formation. Too fast reactions will not permit mixing to allow reactive functional groups to meet each other, while slow reactions would require a longer time until complete conversion.
In order to validate the influence of hydrogel gelling kinetics on the swelling behaviour of the biomaterials, we performed experiments with 1.6 wt% HA-hydrogels having acylhydrazone and oxime cross-linkages. As anticipated, without any salts, the acylhydrazone cross-linked hydrogels exhibited excessive swelling (∼75%) within three days while the oxime cross-linked gels demonstrated only ∼20% swelling (Fig. 5).
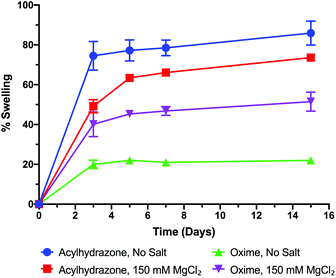 | ||
| Fig. 5 The effect of reaction kinetics on the swelling of acylhydrazone and oxime cross-linked HA-hydrogels. HA solid content is 1.6 wt%. | ||
Addition of MgCl2, which is expected to reduce the reaction time significantly, reduced the swelling behaviour of the acylhydrazone cross-linked materials (∼50% in three days). This result corroborates with the increase in the storage modulus as a result of an increase in the gelling kinetics. In the case of the oxime reaction, on the other hand, we observed an increase in swelling (∼40% in three days) upon the addition of MgCl2 (Fig. 5). We believe that this unusual behavior with MgCl2 is due to the limited possibility to achieve homogeneous mixing of the gel components due to the too high reaction rates and too short gelation time. It is interesting to note that the initial swelling ratio after the first five days was maintained even after 15 days of incubation, indicating that the dynamic cross-linked gels once formed remain stable at physiological pH in the presence of salts in the buffer (Fig. 5).
Conclusions
In summary, we present a simple and efficient strategy to modulate hydrazone and oxime formation kinetics at physiological pH by utilizing simple salts as catalysts. Increasing the NaCl concentration or using divalent salts such as MgCl2 or CaCl2 can significantly accelerate the reaction rates for such reactions. The increase in the reaction rates was demonstrated in both small molecules and multifunctional biomacromolecules and utilized for designing acylhydrazone and oxime-based hydrogels under physiologically relevant conditions. This simple strategy to modulate hydrazone and oxime reaction kinetics will provide a great advantage for developing biomaterials or for bioconjugation applications.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We acknowledge financial support from the Swedish Strategic Research ‘StemTherapy’ (Dnr 2009-1035) and the Swedish Foundation for Strategic Research (SSF, SBE13-0028).Notes and references
- A. S. Hoffman, Adv. Drug Delivery Rev., 2012, 64, 18–23 CrossRef.
- J. Kopecek, Biomaterials, 2007, 28, 5185–5192 CrossRef CAS.
- D. Seliktar, Science, 2012, 336, 1124–1128 CrossRef CAS.
- M. W. Tibbitt and K. S. Anseth, Biotechnol. Bioeng., 2009, 103, 655–663 CrossRef CAS.
- J. A. Burdick and G. D. Prestwich, Adv. Mater., 2011, 23, H41–H56 CrossRef CAS.
- A. Sigen, Q. Xu, P. McMichael, Y. Gao, X. Li, X. Wang, U. Greiser, D. Zhou and W. Wang, Chem. Commun., 2018, 54, 1081–1084 RSC.
- T. R. Hoare and D. S. Kohane, Polymer, 2008, 49, 1993–2007 CrossRef CAS.
- H. J. Yan, T. Casalini, G. Hulsart-Billström, S. Wang, O. P. Oommen, M. Salvalaglio, S. Larsson, J. Hilborn and O. P. Varghese, Biomaterials, 2018, 161, 190–202 CrossRef CAS.
- A. E. G. Baker, R. Y. Tam and M. S. Shoichet, Biomacromolecules, 2017, 18, 4373–4384 CrossRef CAS.
- O. P. Oommen, S. J. Wang, M. Kisiel, M. Sloff, J. Hilborn and O. P. Varghese, Adv. Funct. Mater., 2013, 23, 1273–1280 CrossRef CAS.
- K. Jeet and R. T. Raines, Angew. Chem., Int. Ed., 2008, 47, 7523–7526 CrossRef.
- M. Rashidian, M. M. Mahmoodi, R. Shah, J. K. Dozier, C. R. Wagner and M. D. Distefano, Bioconjugate Chem., 2013, 24, 333–342 CrossRef CAS.
- G. N. Grover, J. Lam, T. H. Nguyen, T. Segura and H. D. Maynard, Biomacromolecules, 2012, 13, 3013–3017 CrossRef CAS.
- E. H. Cordes and W. P. Jencks, J. Am. Chem. Soc., 1962, 84, 832–837 CrossRef CAS.
- D. K. Kölmel and E. T. Kool, Chem. Rev., 2017, 117, 10358–10376 CrossRef.
- D. Larsen, A. M. Kietrys, S. A. Clark, H. S. Park, A. Ekebergh and E. T. Kool, Chem. Sci., 2018, 9, 5252–5259 RSC.
- M. Rashidian, M. M. Mahmoodi, R. Shah, J. K. Dozier, C. R. Wagner and M. D. Distefano, Bioconjugate Chem., 2013, 24, 333–342 CrossRef CAS.
- M. Wendeler, L. Grinberg, X. Wang, P. E. Dawson and M. Baca, Bioconjugate Chem., 2014, 25, 93–101 CrossRef CAS.
- A. Dirksen, S. Dirksen, T. M. Hackeng and P. E. Dawson, J. Am. Chem. Soc., 2006, 128, 15602–15603 CrossRef CAS.
- P. Crisalli and E. T. Kool, J. Org. Chem., 2013, 78, 1184–1189 CrossRef CAS.
- D. Larsen, M. Pittelkow, S. Karmakar and E. T. Kool, Org. Lett., 2015, 17, 274–277 CrossRef CAS.
- S. Wang, D. Gurav, O. P. Oommen and O. P. Varghese, Chem. – Eur. J., 2015, 21, 5980–5985 CrossRef CAS.
- S. Wang, G. N. Nawale, S. Kadekar, O. P. Oommen, N. K. Jena, S. Chakraborty, J. Hilborn and O. P. Varghese, Sci. Rep., 2018, 8, 2193 CrossRef.
- E. G. Sander and W. P. Jencks, J. Am. Chem. Soc., 1968, 90(22), 6154–6162 CrossRef CAS.
- V. Mazzini and V. S. J. Craig, Chem. Sci., 2017, 8, 7052–7065 RSC.
- T. Murakami, S. Otsuki, Y. Okamoto, K. Nakagawa, H. Wakama, N. Okuno and M. Neo, Connect. Tissue Res., 2018, 1–11, DOI:10.1080/03008207.2018.1465053.
- M. A. Solis, Y.-H. Chen, T. Y. Wong, V. Z. Bittencourt, Y.-C. Lin and L. L. H. Huang, Biochem. Res. Int., 2012, 2012, 346972–346972 Search PubMed.
- M. Paidikondala, G. N. Nawale and O. P. Varghese, Biomacromolecules, 2019, 20, 1317–1324 CrossRef CAS.
- O. P. Oommen, S. Wang, M. Kisiel, M. Sloff, J. Hilborn and O. P. Varghese, Adv. Funct. Mater., 2013, 23, 1273–1280 CrossRef CAS.
- J. Patterson, M. M. Martino and J. A. Hubbell, Mater. Today, 2010, 13, 14–22 CrossRef CAS.
- M. Kisiel, M. Ventura, O. P. Oommen, A. George, X. F. Walboomers, J. Hilborn and O. P. Varghese, J. Controlled Release, 2012, 162, 646–653 CrossRef CAS.
- O. P. Oommen, J. Garousi, M. Sloff and O. P. Varghese, Macromol. Biosci., 2014, 14, 327–333 CrossRef CAS.
- H. Yan, O. P. Oommen, D. Yu, J. Hilborn, H. Qian and O. P. Varghese, Adv. Funct. Mater., 2015, 25, 3907–3915 CrossRef CAS.
- M. Paidikondala, V. K. Rangasami, G. N. Nawale, T. Casalini, G. Perale, S. Kadekar, G. Mohanty, T. Salminen, O. P. Oommen and O. P. Varghese, Angew. Chem., Int. Ed., 2019, 58, 2815–2819 CrossRef CAS.
- S. Wang, O. P. Oommen, H. Yan and O. P. Varghese, Biomacromolecules, 2013, 14, 2427–2432 CrossRef CAS.
- D. Bermejo-Velasco, A. Azémar, O. P. Oommen, J. Hilborn and O. P. Varghese, Biomacromolecules, 2019, 20(3), 1412–1420 CrossRef CAS.
- O. P. Varghese, M. Kisiel, E. Martínez-Sanz, D. A. Ossipov and J. Hilborn, Macromol. Rapid Commun., 2010, 31, 1175–1180 CrossRef CAS.
- M. Wendeler, L. Grinberg, X. Wang, P. E. Dawson and M. Baca, Bioconjugate Chem., 2014, 25, 93–101 CrossRef CAS PubMed.
- W. A. Bubnis and C. M. Ofner 3rd, Anal. Biochem., 1992, 207, 129–133 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9py00862d |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2019 |