Open Access Article
Open Access ArticleLupeol inhibits the proliferation and migration of MDA-MB-231 breast cancer cells via a novel crosstalk mechanism between autophagy and the EMT
Xin
Zhang
ab,
Zhanwang
Gao
ab,
Kehan
Chen
a,
Qingyuan
Zhuo
a,
Meixian
Chen
a,
Jiansong
Wang
ac,
Xiaoping
Lai
*a and
Lingli
Wang
 *ab
*ab
aSchool of Pharmaceutical Sciences, Guangzhou University of Chinese Medicine, no. 232, Waihuandong Road, Guangzhou Higher Education Mega Center, Guangzhou 510006, People's Republic of China. E-mail: wlingli@gzucm.edu.cn
bDongguan Institute of Guangzhou University of Chinese Medicine, no. 4, Libin Road, Songshan Lake Science and Technology Industrial Park, Dongguan, 523000, People's Republic of China
cBaiyunshan Pharmaceutical General Factory, Guangzhou Baiyunshan Pharmaceutical Holdings Co., Ltd., Guangzhou, 510515, PR China
First published on 21st April 2022
Abstract
Triple-negative breast cancer is the most aggressive type of breast cancer, with a poor prognosis, while effective treatment options are limited. In this study, the anti-tumor effect of lupeol, a natural triterpenoid, toward breast cancer cells and the underlying mechanisms were examined. We firstly predict the primary pathways of lupeol inhibited to TNBC by a network pharmacology approach, which indicated that lupeol may inhibit TNBC via multiple signaling pathways. In addition, experimental data showed that lupeol exhibited outstanding anti-proliferative and anti-metastatic abilities in vitro and in vivo. Additional intrinsic mechanism studies revealed that lupeol might induce autophagy by inhibiting the Akt-mTOR pathway, and activating an autophagy inhibited epithelial–mesenchymal transition (EMT). This study demonstrated that lupeol could inhibit TNBC cells by inducing autophagy, suggesting lupeol as a potential treatment alternative or as a dietary supplement for TNBC, as well as offering novel insights into the anti-cancer effect of lupeol.
Introduction
Triple-negative breast cancer (TNBC), representing 10% to 20% of all types of breast cancer (BC), is the most aggressive type of BC, with high mortality rates.1 Compared with other subtypes of breast cancer, TNBC lacks the expressions of estrogen receptors, progesterone (PR), and human epidermal growth factor receptor 2 (HER2), which causes limited treatment-effectiveness and poor patient prognosis.2 Even though there have been outstanding advancements within the healthcare industry in recent years, alternative treatment strategies for TNBC are absent to date. Clinically, radiation therapy and chemotherapy remain the mainstay of TNBC treatment.3 Moreover, TNBC shows high metastasis and recurrence rates in TNBC patients, which causes pain and suffering to individuals and brings a huge burden to families and society. Therefore, it is of profound significance to explore the underlying mechanism of TNBC and search for novel therapeutics.Autophagy, one of the main mechanisms of programmed cell death, is the major pathway for the degradation of cytoplasmic components.4 Recently, it was reported that autophagy plays a fundamental role in regulating tumor development in multiple cancers, which is considered a “double-edged sword” in cancer progression.5–7 On the one hand, appropriate autophagy protects tumor cells from chemotherapy-induced death; on the other hand, excessive autophagy causes autophagic death.8 In TNBC, more and more evidence indicates that autophagy is able to become an appealing strategy for the treatment of TNBC.9 First, autophagy can suppress the proliferation of TNBC cells. It is revealed that the expression of autophagic protein ATG7 in TNBC is lower than in other subtypes of breast cancer, and a high level of ATG7 is accompanied by a better prognosis.10 Second, autophagy inhibits the epithelial–mesenchymal transition (EMT).11 EMT plays an essential role in TNBC cell invasion through adherens junctions and the loss of substrate polarity, which contributes to its recurrence and incurable nature.12 Previous studies have shown that inducing autophagy to degrade EMT transcriptional repressors, such as Snail13 and Twist1,14 can suppress the EMT in breast cancer cells. Moreover, more and more evidence indicated that there is a complex relationship between autophagy-correlated and EMT-correlated signaling pathways. For example, the EMT reveals the viability of potential metastasis of cancer cells from autophagy in renal cell carcinoma, which suggests that the combination of autophagy inhibitors with a current strategy may be more effective. To sum up, increasing evidence highlights that autophagy seems to function as a tumor suppressor and inhibits tumorigenesis during TNBC development. TNBC shows highly aggressive metastasis, and it is difficult to control clinically by anti-proliferation alone. Therefore, it is meaningful to focus on its proliferation and its metastasis by elucidating the underlying biological mechanism that connects the two.
Lupeol, as a natural pentacyclic triterpene, is present in various vegetables, fruits, and traditional Chinese medicines, such as Atractylodes macrocephala Koidz and Astragalus membranaceus.15 It has been studied that lupeol possesses a variety of pharmacological functions applied, for instance, in antioxidant, antiviral, antihypertensive, anti-inflammatory, and anti-tumor agents.16 With extensive studies, an increasing number of molecular mechanisms of lupeol has been discovered due to its significant anti-tumor function in recent years, which include inducing apoptosis through Akt/mTOR, EGFR/STAT3, and Wnt pathways, and inhibiting migration and invasion of cancer cells through RhoA-Rock1, MAPK/ERK and so on.17 In breast cancer, lupeol can induce apoptotic effects and inhibit the proliferation of MCF-7 cells and MCF-10A cells. However, the studies of lupeol for TNBC treatment are few, while precise mechanisms in TNBC are not known clearly enough as well.
Based on previous works, we aim to predict the effective targets of lupeol and the potential mechanism of anti-BC through a network pharmacology approach and verify the exact mechanisms of lupeol treated TNBC, to lay the foundations for future drug development and clinical applications of lupeol.
Methods and materials
Analysis of targets related to TNBC
The differential gene of TNBC was analyzed by obtaining the data of transcriptome profiling from TCGA (https://portal.gdc.cancer.gov/). Differential genes with P < 0.0001 and log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2FC > 2 were considered clinically significant. The targets of lupeol were predicted from SwissTargetPrediction (https://www.swisstargetprediction.ch) and TCMSP (https://lsp.nwu.edu.cn/tcmspsearch.php). Then, the target of lupeol related to TNBC was validated using the differential gene of TNBC. These validation results were further analyzed by PPI, GO, and KEGG pathways.
2FC > 2 were considered clinically significant. The targets of lupeol were predicted from SwissTargetPrediction (https://www.swisstargetprediction.ch) and TCMSP (https://lsp.nwu.edu.cn/tcmspsearch.php). Then, the target of lupeol related to TNBC was validated using the differential gene of TNBC. These validation results were further analyzed by PPI, GO, and KEGG pathways.
PPI network construction and clustering analysis
Target proteins were introduced by STRING database version 11.0 (https://string-db.org/). The organism was set to Homo sapiens, and the minimum required interaction score was set to 0.4 to construct a protein–protein interaction network (PPI). The PPI network consists of nodes and edges, representing target protein and protein–protein interactions, respectively. The blue line stands for “gene co-evolution” evidence, while the red line for “gene fusion” and the yellow line for “text mining”. Then the core targets were identified through network analysis using Cytoscape software (v.3.7.1).GO and KEGG pathway analysis
As a statistical analysis software, R studio is a combination of statistical analysis and graphic display and is being exploited in GO and KEGG enrichment analyses. The enriched terms with P < 0.01 were considered significant. The top 30 enrichment results were used to visualize and analyze pathways by R studio.Molecular complex detection (MCODE), which is a Cytoscape plugin, was used to analyze cluster modules in the PPI network. The MCODE criteria for selection were as follows: degree cutoff = 2, node score cutoff = 0.2, k-core = 2, and max depth = 100.
Cell culture and compounds
MDA-MB-231 was purchased from the Cell Bank of the Chinese Academy of Sciences and cultured in DMEM with high glucose (Gibco, USA) and 10% of FBS at 37 °C in 5% CO2. Lupeol, SC79 (an activator of AKT), and 3-MA (autophagy inhibitor) were obtained from MedChemExpress (MCE, China), dissolved in DMSO, and diluted with DMEM to the desired concentration with a final DMSO concentration that did not exceed 0.5%.Cell viability assay and colony formation assay
MDA-MB-231 cells (3000 cells per well) were seeded into a 96-well plate, and the cytotoxicity of lupeol was estimated using the CCK8 assay (Biosharp, China). Briefly, the cells were cultured in a 37 °C/5%CO2 incubator for 24 h and then treated with lupeol at 0, 1.5625, 3.125, 6.25, 12.5, 25, 50, and 100 μM for 24 h and 48 h, respectively. For CCK8 detection, 10 μl of CCK8 reagent was added 4 h before analysis. Absorbance was recorded at 450 nm with a microplate reader.MDA-MB-231 cells (300–500 cells per well) were seeded in 6-well plates and then treated with different concentrations of lupeol and cultured for 2 weeks, while media were changed every 3 days. At the end of incubation, the cells were washed twice with PBS, fixed with paraformaldehyde for 30 min and stained with 0.1% crystal violet solution (Solarbio, Beijing, China) for 30 min, and dried overnight. The intensity of crystal violet staining was quantified by Image J (version 2.0.0).
Wound healing assay
MDA-MB-231 cells were cultured in 6-well plates. A straight scratch was made while the cells were grown to almost 90% density. Then, the cells were treated with various lupeol concentrations for 24 h. Cell migration activity was photographed at 0, 12, and 24 h, and the quantified values were compared using Image J (version 2.0.0).Immunofluorescence (IF) analysis
MDA-MB-231 cells were spread on a glass slide in 6-well plates and incubated with different concentrations of lupeol for 24 h. Next, the cells were subjected to 4% paraformaldehyde fixation for 15 min, washed with cold PBS 3 times, and blocked with 5% BSA solution for 1 h. Then, the cells were immunostained with 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 LC3 antibody for 2 h followed by incubation with 1
500 LC3 antibody for 2 h followed by incubation with 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 secondary antibodies for 1 h. Finally, the nucleus was counterstained with DAPI (G1012, Servicebio, China) and immunofluorescence was imaged with a NIKON ECLIPSE C1 fluorescence microscope (Tokyo, Japan).
200 secondary antibodies for 1 h. Finally, the nucleus was counterstained with DAPI (G1012, Servicebio, China) and immunofluorescence was imaged with a NIKON ECLIPSE C1 fluorescence microscope (Tokyo, Japan).
Western blot analysis
Total protein of cells and tumor tissues was extracted using RIPA lysate (P0013B, Beyotime, China) and quantified according to the BCA kit. Equal amounts of proteins were subjected to SDS-PAGE and transferred to PVDF membranes. Then, the membranes were probed with different primary antibodies: LC3 (RRID: AB-2844592), mTOR (RRID: AB2835169), ATG7 (RRID: AB2838097), N-cadherin (RRID: AB2835344), E-cadherin (RRID: AB-2833315), Beclin-1 (RRID: AB-2837614), and P62 (RRID:). All antibodies were purchased from Affinity Biosciences (China). Finally, proteins were incubated followed by rabbit-goat secondary antibody (RRID: AB-2839429), developed with ECL advance reagent (Affinity Biosciences, LOT#1927B02, China) and visualized with a Bioworld ComplexTM2000 developer (Tokyo, Japan), while β-actin (RRID: AB-2839420), GAPDH (RRID: AB-2839421) and tubulin beta (RRID: AB-2827688) were used as loading controls and Image J software (version 2.0.0) was used to analyze the gray value.Real-time PCR
Total RNA of cells was extracted using Trizol reagent. Quantitative real-time PCR for LC3, Beclin-1, and P62 mRNA expressions was carried out. Primer sequences are listed in Table 1. The relative expression levels of the respective mRNAs were quantified by the 2−ΔΔCt method with GAPDH as an endogenous reference.| Genes | Primer sequence (5′-3′) |
|---|---|
| LC3 | F: AGTGGAAGATGTCCGGCTCAT; R: GCTGCTTCTCACCCTTGTATCG |
| Beclin-1 | F: AGCTCAGTACCAGCGGGAGT; R: TGGAAGGTGGCATTGAAGAC |
| P62 | F: CACCGCACCGTGAAGGCCTACCTTC; R: CACCGCGCTACACAAGTCGTAGTCT |
| β-Actin | F: GCCAACCGTAAAAGATG; R: CCAGGATAGAGCCACCAAT |
Mouse tumor models
All animal experiments were approved by the Ethics Committee of Guangzhou University of Chinese Medicine (Permit number is ZYD-2021-226, Guangzhou, China). Female BALB/c nude mice were purchased from Zhuhai BesTest Bio-Tech Co., Ltd (Zhuhai, China).For a subcutaneous xenograft tumor model, 15 female BALB/c nude mice (4 weeks old) were injected with MDA-MB-231 cells (1–2 × 106 cells) subcutaneously. When the tumor size reached 100 mm3 (about 5 days later), the mice were randomly divided into three groups (5 mice per group): control group (corn oil), low dose group (20 mg per kg per 2 days), and high dose group (40 mg per kg per 2 days), the whole experiment lasting for 21 days. During the treatment, the size of the tumor and the weight of mice were recorded every 2 days. At the end of the experiments, all mice were sacrificed via cervical dislocation. Then, the tumors were isolated, weighed, and imaged, with part of the tumor tissues preserved in liquid nitrogen for western blot analysis, and part fixed with formaldehyde for further histopathological analysis.
For a pulmonary metastasis model, 18 female BALB/c nude mice (4 weeks old) were injected with MDA-MB-231 cells (1 × 105 cells) through the tail vein. About 6 days later, the same grouping and administration treatments as those of the experiments described above were applied. 21 days later, the lungs were isolated and the numbers of metastatic nodules were counted. Then, part of the lung tissues was preserved in liquid nitrogen for western blot analysis, and part fixed with formaldehyde for further histopathological analysis.
Immunohistochemistry (IHC) analysis
Sections of tumor were submerged in EDTA antigenic retrieval reagent (pH 8.0) and microwaved for antigenic retrieval. Then, the slides were incubated at 37 °C for 40 min with LC3 antibody (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500), Atg5 antibody (1
500), Atg5 antibody (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500), MMP-2 antibody (1
500), MMP-2 antibody (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400), and E-cadherin antibody (1
400), and E-cadherin antibody (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 400), respectively, and HRP polymer conjugated secondary antibody was applied for 30 min and then developed with ECL Advance reagent using a microscope.
400), respectively, and HRP polymer conjugated secondary antibody was applied for 30 min and then developed with ECL Advance reagent using a microscope.
Statistics
All data were expressed as the mean ± standard deviation of at least three independent experiments. Statistical differences among the groups were analyzed with SPSS (version 20.0) and a one-way analysis of variance was utilized for statistical analysis. Significant differences were labeled as follows: *, P < 0.05; **, P < 0.01; ***, P < 0.001.Results
Lupeol suppresses proliferation in vitro and in vivo
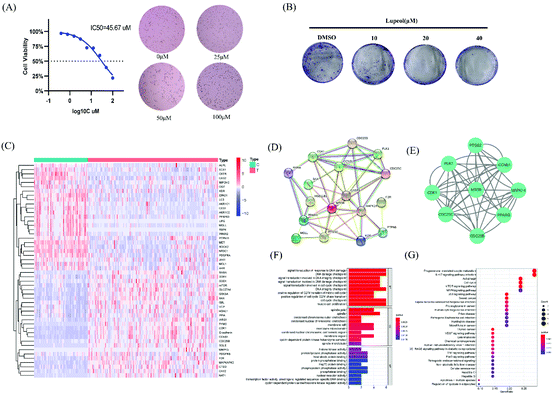
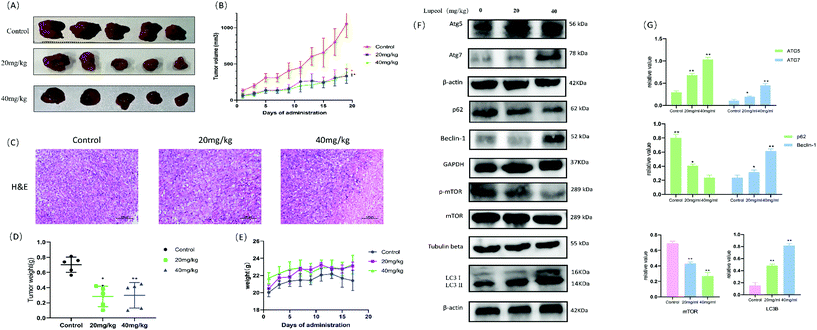
To investigate the potential anti-proliferative effect of lupeol, MDA-MB-231 cells were assessed by the CCK8 assay and treated at a certain concentration gradient of lupeol. As shown in Fig. 1A, the proliferation of MDA-MB-231 was significantly inhibited by lupeol with concentration-dependent cytotoxicity (IC50 = 45.67 μM). Consistently, a colony formation assay was also performed to verify the anti-proliferation effect of lupeol. As shown in Fig. 1B, the colony formation abilities of the cells at lupeol concentrations of 20 and 40 μM was shown to be significantly inhibited compared with the control group (P < 0.05).Furthermore, to determine whether lupeol inhibited the growth of MDA-MB-231 both in vitro and in vivo, MDA-MB-231 tumor-bearing mice were treated with lupeol at 20 mg kg−1 and 40 mg kg−1. As shown in Fig. 3A and B, treatment with lupeol significantly inhibited MDA-MB-231 tumor growth and weights (P < 0.05) in a dose-dependent manner compared with the control group, while the bodyweight of mice did not differ significantly among the groups.
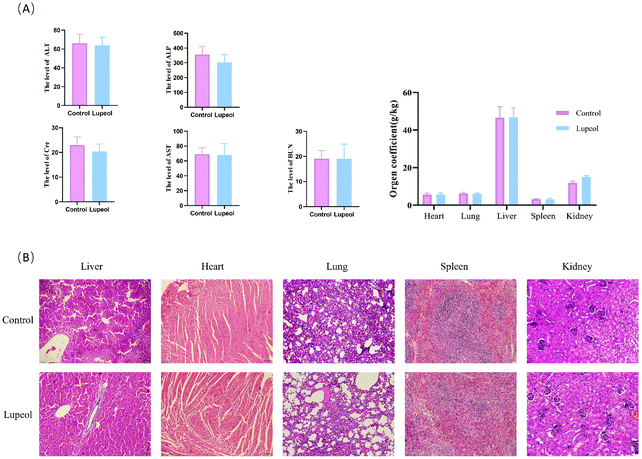 | ||
| Fig. 3 Evaluation of the safety of lupeol in vivo. (A) Effects of lupeol on the serum biochemical markers and (B) H&E staining of the major organs after the treatment with lupeol. | ||
In addition, a safety assessment of lupeol was performed. The results are presented as follows (see Fig. 4A and B). Serum ALT, AST, Cre, Bun, and ALP activities were not significantly different in the three groups, and H&E staining of the major tissues (heart, liver, spleen, lungs, and kidneys) also showed that lupeol had no apparent toxicity in vivo, while the weight of every organ was no different among the groups as well. Collectively, these results suggested that lupeol had obviously and significantly inhibited proliferation of TNBC in vivo and in vitro.
Analysis of lupeol related to clinical breast cancer targets
First, 201 clinical samples derived from the University of Pittsburgh were selected, of which there were 143 TNBC tissues and 59 normal breast tissues. Transcriptome profiling analysis and clustering of the 201 samples were performed. Of these, 38 gene targets were significantly downregulated and 56 gene targets were significantly upregulated in clinical breast cancer tissues, which were plotted as a heatmap with 50 significant differential genes (Fig. 1C).Then, 93 gene targets of lupeol were collected from TCMSP and SwissTargetPrediction. Of these targets, 17 differential genes are related to TNBC targets. By constructing a PPI network of these 17 common gene targets using the STRING database (Fig. 1D) and analyzing by degree, betweenness, and closeness in the CytoNCA of Cytoscape, 9 core gene targets were identified, which are mTOR, PTGS2, MAPK14, CCNB1, CDC25B, CDC25C, PPARG, CDK1, and PLK1 (Fig. 1E).
Next, 22 gene targets for this functional profiling were analyzed. Signal transduction in response to DNA damage, the DNA damage checkpoint, the positive regulation of the G2/M transition of the mitotic cell cycle, and the positive regulation of cell cycle G2/M phase transition were enriched by GO-BP analysis (Fig. 1F). KEGG pathway enrichment demonstrated that progesterone-mediated oocyte maturation, the IL-17 signaling pathway, autophagy, cell cycle, mTOR signal pathway, and MAPK signaling pathway were significantly enriched (Fig. 1G).
Through integrated analysis of the above results and based on previous works, we speculated that the mTOR signaling pathway and autophagy may be the primary pathways in lupeol inhibited TNBC. The Akt-mTOR pathway plays an important role in the development of TNBC.18,19 In addition, mTOR is an essential inhibitor of autophagy, which controls the initiation signals of autophagy.20 Therefore, we have carried out further experiments to verify the conjecture.
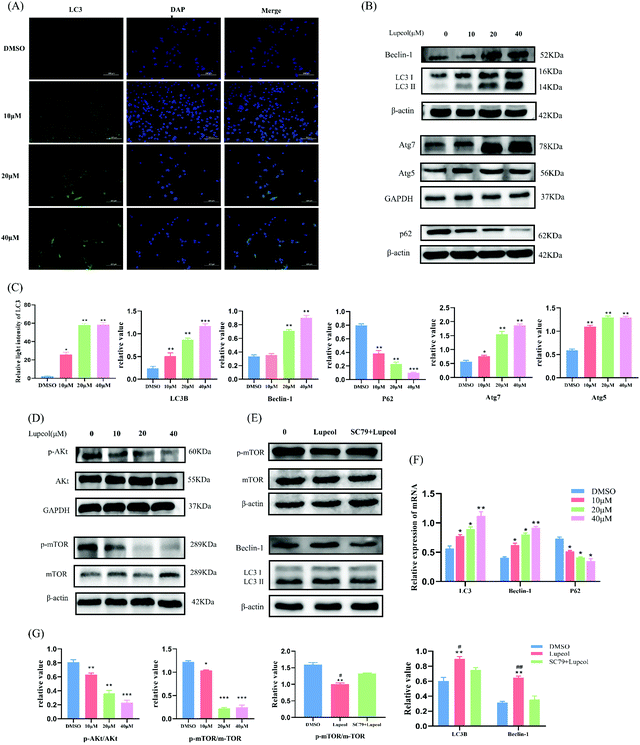
Lupeol induces autophagic cell death by inhibiting the Akt-mTOR pathway
To verify the results of the bioinformatics analysis and investigate the reason behind the growth inhibition induced by lupeol in MDA-MB-231 cells, western blot and IF were utilized to examine autophagosomes and, as a result, we found that the expressions of LC-3II, Beclin1, Atg7, and Atg5 were increased with increasing concentrations of lupeol (P < 0.05), whereas the expression of P62 was decreased (P < 0.05) (Fig. 2A–C), while the same trend held for mRNA expression levels (Fig. 2F). It is known that LC3 is localized on autophagosomes and the increasing ratio of LC3II/LC3I is the specific marker of autophagic formation;21 also, P62 is a substrate of autophagy and it can be degraded in autolysosomes, which means that enhanced autophagic formation causes the decrease of P62.22 Beclin1 is a key initiator of autophagy,23 so the upregulation of Beclin-1 suggests that lupeol upregulation of autophagy is initiated by the upstream signal. Therefore, the present results suggest that lupeol induces autophagosome formation in MDA-MB-231 cells.Moreover, western blot results also showed that both p-Akt and p-mTOR decreased as a result of treatment with lupeol for 24 h in MDA-MB-231 cells, while the total amount of AKT and mTOR proteins are unchanged (Fig. 2D). To further explore the regulatory relationship of AKT–mTOR pathways and lupeol-induced autophagy, MDA-MB-231 cells were further treated with lupeol and SC79 (an activator of AKT). After treatment with SC79, the expression of p-mTOR was increased, whereas the expressions of LC3 and Beclin1 were decreased, suggesting that the AKT–mTOR pathway is involved in lupeol-induced autophagy in MDA-MB-231 cells (Fig. 2E).
Lupeol suppresses migration in vitro and in vivo
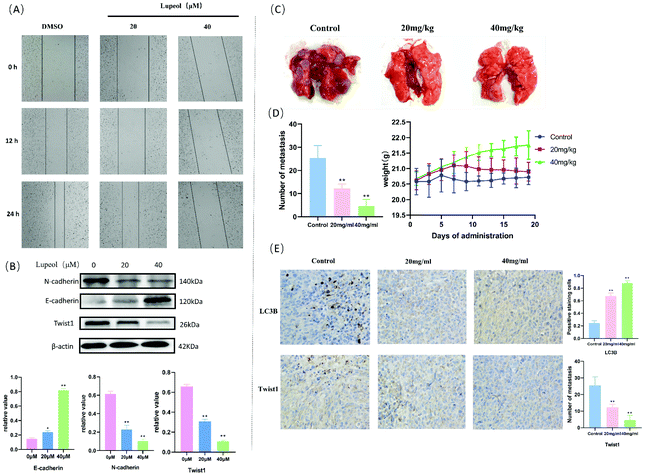
To evaluate the anti-migration ability of lupeol in MDA-MB-231 cells, wound healing assays were used and the results are shown in Fig. 5A, which indicates that lupeol could significantly inhibit the mobilities of MDA-MB-231 cells in a concentration-dependent manner. According to the western blot analysis (Fig. 5B), inhibition of migration led to up-regulation of E-cadherin and down-regulation of N-cadherin and Twist1 in MDA-MB-231 cells in response to lupeol treatment (P < 0.05). Twist1 is a basic helix–loop–helix protein that induces the EMT in early embryonic morphogenesis, cancer development, and cancer metastasis.24Then, we established a lung metastasis model of MDA-MB-231 cells in mice to reveal the efficacy of lupeol in vivo. As shown in Fig. 5, the numbers of tumor nodules in the lungs of 20 mg kg−1 (12.20 ± 1.924, P < 0.05) and 40 mg kg−1 (4.60 ± 2.88, P < 0.05) treatment groups were significantly decreased compared with the control group (25.40 ± 5.32), while there were no significant differences in the weight of each group (P > 0.05) (Fig. 5C and D). Moreover, we observed that the protein levels of Twist1 decreased gradually with the administration of increasing lupeol concentrations compared with LC3B in the immunohistochemistry assay (Fig. 5E). The above results implied that lupeol shows effective inhibition of MDA-MB-231 pulmonary metastatic tumors.
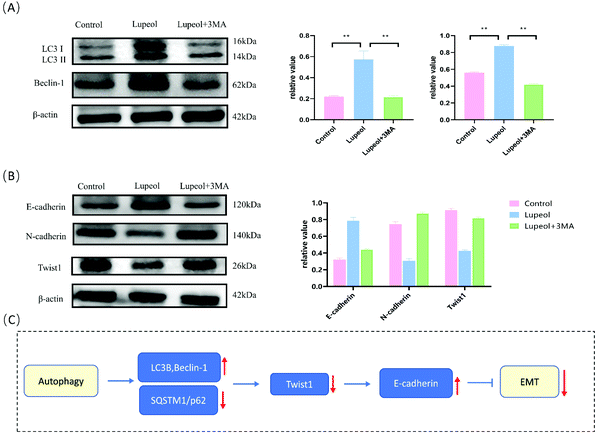
Lupeol reverses the EMT by inducing autophagy
To further investigate whether autophagy directly contributes to the EMT in the present study, the effect of adding autophagy inhibitor 3MA was examined. As shown in Fig. 6A and B, the expression levels of lupeol-induced autophagy proteins and E-cadherin were significantly decreased after lupeol treatment with 3MA, while the expression of Twist1 and N-cadherin was increased. In tumor development, N-cadherin and E-cadherin belong to the classical cadherin family, and the switch of E-cadherin to N-cadherin is the key event in the EMT, while Twist1 represses the promoter of CDH1 that encodes for E-cadherin.25 In previous studies, it is found that enhancing the formation of autophagosomes can inhibit the expression of Twist1, which thereby suppresses N-cadherin and induces E-cadherin, finally inhibiting the EMT (Fig. 6C).26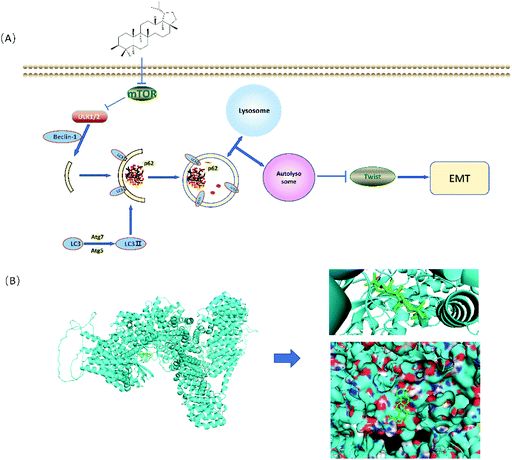 | ||
| Fig. 6 (A) Schematic for the regulation of lupeol induced autophagy and inhibited EMT and (B) molecular docking analysis of lupeol and mTOR. | ||
Discussion
Breast cancer is one of the most common cancers nowadays, but early diagnosis, and surgery combined with targeting therapy directed at a specific subtype of breast cancer, has achieved curative results clinically. However, due to the lack of molecular targets for drugs, and with its highly aggressive prognosis, treatment of TNBC remains a colossal challenge and there is an urgent need to develop novel therapeutic strategies against TNBC driven by this demanding situation. Indeed, lupeol has been proposed to perform anti-tumor functions and might possess a potential utilization to inhibit TNBC, which led us to investigate the exact action and immanent mechanism of lupeol treatment of TNBC.27In the present study, the underlying mechanisms of lupeol against TNBC were firstly predicted through network pharmacology, which is based on the analysis of network models and system biology. The TCGA database was utilized to describe the prognosis of different cancers, and we observed the differences between TNBC and other breast cancers, which inspired us to further investigate the correlation between TNBC and lupeol, and finally discover a possible mechanism for lupeol treated TNBC. Our findings pointed towards progesterone-mediated oocyte maturation, IL-17 signaling pathway, autophagy, cell cycle, mTOR signal pathways, and MAPK signaling pathway being the primary mechanisms of lupeol against TNBC (Fig. 1). Owing to the vital role autophagy plays in the process of TNBC,28 we thereby focused on the role of autophagy in lupeol treatment against TNBC.
In the early stage of autophagy, Beclin1 and class III PI3K assemble as a complex and are activated by the ULK1 complex to induce the formation of an autophagic vesicle. Subsequently, ATG are proteins involved in the lipidation of LC3II, which is stably localized on the autophagosome membrane and used as a specific marker to detect autophagosome formation.29 Our results showed that lupeol promoted the expressions of Beclin1 ATG5, ATG7, and LC3II, which indicated that lupeol induced the formation of autophagosomes (Fig. 2D). To explore the causes leading to this phenomenon, Akt–mTOR pathways were detected, and as a result, p-Akt and p-mTOR were inhibited after treating with lupeol in vitro and in vivo (Fig. 2F). It is known that Akt–mTOR signaling is a major negative regulatory pathway of autophagy because the mTORC1 complex can bind directly to ULK1,30 which disrupts ULK1 complex formation and thus suppresses autophagy (Fig. 7A). In particular, we also performed a molecular docking study to investigate the relationship between lupeol and mTOR. Interestingly, there is a great possibility of protein–protein interactions between lupeol and mTOR (Fig. 7B), which needs further confirmation through co-immunoprecipitation. To summarize, the above findings show that lupeol induced autophagy through Akt–mTOR to suppress TNBC.
Acquisition of TNBC invasive properties and the difficulty of its prognosis are driven by the EMT, while Twist1 is considered a primary regulator of the EMT.26 Twist1 is a helix–loop–helix protein and works in early embryonic morphogenesis, cancer development, and cancer metastasis. It is reported that the inhibition of autophagy can promote the expression of Twist1 protein through P62 accumulation; here, instead, it is whether enhanced autophagy or P62 inhibition causes the decrease of Twist1.31 During this study, lupeol suppressed the expression of Twist1 and N-cadherin and finally reduced the metastases in vitro and in vivo (Fig. 5). This phenomenon disappeared in the presence of autophagy inhibitors (Fig. 6), which suggested that lupeol suppressed the metastases of MDA-MB-231 cells through Twist1 for the first time. However, the potential application of lupeol in the clinical treatment of TNBC prognosis still needs further exploration.
Conclusions
Taken together, our results demonstrate that lupeol can suppress the proliferation of TNBC by inducing autophagy via the PI3K/mTOR pathway, and inhibit the metastasis of TNBC by reducing the EMT through autophagy-mediated Twist1. Our study reveals the mechanism of lupeol inhibited TNBC and validates that lupeol-induced autophagy could be a promising therapeutic medicine for TNBC, as well as providing new insights into the treatment and prognosis of TNBC.Author contributions
All the authors have made contributions to the manuscript and approved the version to be submitted. X. P. L. and L. L. W.: the co-corresponding authors, responsible for designing the experiments and providing the funding support. X. Z.: the first author, responsible for writing the article and preparing figures. Z. W. G., K. H. C., Q. Y. Z., and M. X. C.: responsible for performing the experiments and analyzing the data.Conflicts of interest
All authors declared that no conflicts of interest exist.Acknowledgements
This work was supported by the 2021 Dongguan Social Development Technology Key Project (no. 20211800905502).References
- H. Sung, J. Ferlay, R. L. Siegel, M. Laversanne, I. Soerjomataram and A. Jemal, et al., Global Cancer Statistics 2020: GLOBOCAN Estimates of Incidence and Mortality Worldwide for 36 Cancers in 185 Countries, CA Cancer J. Clin., 2021, 71, 209–249 CrossRef PubMed.
- J. L. da Silva, N. C. Cardoso Nunes, P. Izetti, G. G. de Mesquita and A. C. de Melo, Triple negative breast cancer: A thorough review of biomarkers, Crit. Rev. Oncol. Hematol., 2020, 145, 102855 CrossRef PubMed.
- S. Izadi, A. Nikkhoo, M. Hojjat-Farsangi, A. Namdar, G. Azizi and H. Mohammadi, et al., CDK1 in Breast Cancer: Implications for Theranostic Potential, Anticancer Agents Med. Chem., 2020, 20, 758–767 CrossRef CAS PubMed.
- D. J. Klionsky, G. Petroni, R. K. Amaravadi, E. H. Baehrecke, A. Ballabio and P. Boya, et al., Autophagy in major human diseases, EMBO J., 2021, 40, e108863 CrossRef CAS PubMed.
- Y. Wang, N. Wu and N. Jiang, Autophagy provides a conceptual therapeutic framework for bone metastasis from prostate cancer, Cell Death Dis., 2021, 12, 909 CrossRef PubMed.
- Y. Zhen, R. Zhao, M. Wang, X. Jiang, F. Gao and L. Fu, et al., Flubendazole elicits anti-cancer effects via targeting EVA1A-modulated autophagy and apoptosis in Triple-negative Breast Cancer, Theranostics, 2020, 10, 8080–8097 CrossRef CAS PubMed.
- Y. Li, X. Feng, Y. Zhang, Y. Wang, X. Yu and R. Jia, et al., Dietary flavone from the Tetrastigma hemsleyanum vine triggers human lung adenocarcinoma apoptosis via autophagy, Food Funct., 2020, 11, 9776–9788 RSC.
- W. Cao, J. Li, K. Yang and D. Cao, An overview of autophagy: Mechanism, regulation and research progress, Bull. Cancer, 2021, 108, 304–322 CrossRef PubMed.
- Y. S. Abd El-Aziz, J. Gillson, P. J. Jansson and S. Sahni, Autophagy: A promising target for triple negative breast cancers, Pharmacol. Res., 2022, 175, 106006 CrossRef CAS PubMed.
- M. Li, J. Liu, S. Li, Y. Feng, F. Yi and L. Wang, et al., Autophagy-related 7 modulates tumor progression in triple-negative breast cancer, Lab. Invest., 2019, 99, 1266–1274 CrossRef PubMed.
- H. T. Chen, H. Liu, M. J. Mao, Y. Tan, X. Q. Mo and X. J. Meng, et al., Crosstalk between autophagy and epithelial-mesenchymal transition and its application in cancer therapy, Mol. Cancer, 2019, 18, 101 CrossRef PubMed.
- S. González-Martínez, B. Pérez-Mies, D. Pizarro, T. Caniego-Casas, J. Cortés and J. Palacios, Epithelial Mesenchymal Transition and Immune Response in Metaplastic Breast Carcinoma, Int. J. Mol. Sci., 2021, 22, 121 Search PubMed.
- J. Zou, Y. Liu, B. Li, Z. Zheng, X. Ke and Y. Hao, et al., Autophagy attenuates endothelial-to-mesenchymal transition by promoting Snail degradation in human cardiac microvascular endothelial cells, Biosci. Rep., 2017, 37, BSR20171049 CrossRef CAS PubMed.
- Y. X. Shi, Z. W. Sun, D. L. Jia and H. B. Wang, Autophagy deficiency promotes lung metastasis of prostate cancer via stabilization of TWIST1, Clin. Transl. Oncol., 2022 DOI:10.1007/s12094-022-02786-.
- B. Zhu, Q. L. Zhang, J. W. Hua, W. L. Cheng and L. P. Qin, The traditional uses, phytochemistry, and pharmacology of Atractylodes macrocephala Koidz.: A review, J. Ethnopharmacol., 2018, 226, 143–167 CrossRef CAS PubMed.
- F. S. Tsai, L. W. Lin and C. R. Wu, Lupeol and Its Role in Chronic Diseases, Adv. Exp. Med. Biol., 2016, 929, 145–175 CrossRef CAS PubMed.
- K. Liu, X. Zhang, L. Xie, M. Deng, H. Chen and J. Song, et al., Lupeol and its derivatives as anticancer and anti-inflammatory agents: Molecular mechanisms and therapeutic efficacy, Pharmacol. Res., 2021, 164, 105373 CrossRef CAS PubMed.
- M. A. Khan, V. K. Jain, M. Rizwanullah, J. Ahmad and K. Jain, PI3K/AKT/mTOR pathway inhibitors in triple-negative breast cancer: a review on drug discovery and future challenges, Drug Discovery Today, 2019, 24, 2181–2191 CrossRef CAS PubMed.
- R. Costa, H. S. Han and W. J. Gradishar, Targeting the PI3K/AKT/mTOR pathway in triple-negative breast cancer: a review, Breast Cancer Res. Treat., 2018, 169, 397–406 CrossRef CAS PubMed.
- Y. C. Kim and K. L. Guan, mTOR: a pharmacologic target for autophagy regulation, J. Clin. Invest., 2015, 125, 25–32 CrossRef PubMed.
- L. Galluzzi and D. R. Green, Autophagy-Independent Functions of the Autophagy Machinery, Cell, 2019, 177, 1682–1699 CrossRef CAS PubMed.
- T. Lamark, S. Svenning and T. Johansen, Regulation of selective autophagy: the p62/SQSTM1 paradigm, Essays Biochem., 2017, 61, 609–624 CrossRef PubMed.
- H. D. Xu and Z. H. Qin, Beclin 1, Bcl-2 and Autophagy, Adv. Exp. Med. Biol., 2019, 1206, 109–126 CrossRef CAS PubMed.
- T. Liu, X. Zhao, X. Zheng, Y. Zheng, X. Dong and N. Zhao, et al., The EMT transcription factor, Twist1, as a novel therapeutic target for pulmonary sarcomatoid carcinomas, Int. J. Oncol., 2020, 56, 750–760 CAS.
- S. Elzamly, N. Badri, O. Padilla, A. K. Dwivedi, L. A. Alvarado and M. Hamilton, et al., Epithelial-Mesenchymal Transition Markers in Breast Cancer and Pathological Responseafter Neoadjuvant Chemotherapy, Breast Cancer, 2018, 12, 1178223418788074 Search PubMed.
- L. Qiang and Y. Y. He, Autophagy deficiency stabilizes TWIST1 to promote epithelial-mesenchymal transition, Autophagy, 2014, 10, 1864–1865 CrossRef CAS PubMed.
- M. Wang, H. X. Cui, C. Sun, G. Li, H. L. Wang and C. H. Xia, et al., Effect of lupeol on migration and invasion of human breast cancer MDA-MB-231 cells and its mechanism, Yaoxue Xuebao, 2016, 51, 558–562 Search PubMed.
- Y. Han, S. Fan, T. Qin, J. Yang, Y. Sun and Y. Lu, et al., Role of autophagy in breast cancer and breast cancer stem cells (Review), Int. J. Oncol., 2018, 52, 1057–1070 CAS.
- X. Li, S. He and B. Ma, Autophagy and autophagy-related proteins in cancer, Mol. Cancer, 2020, 19, 12 CrossRef CAS PubMed.
- M. Zachari and I. G. Ganley, The mammalian ULK1 complex and autophagy initiation, Essays Biochem., 2017, 61, 585–596 CrossRef PubMed.
- H. Feng, X. Zhao, Q. Guo, Y. Feng, M. Ma and W. Guo, et al., Autophagy resists EMT process to maintain retinal pigment epithelium homeostasis, Int. J. Biol. Sci., 2019, 15, 507–521 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2022 |