Open Access Article
Open Access ArticleTotal chemical synthesis of murine ISG15 and an activity-based probe with physiological binding properties†
Bo-Tao
Xin‡
 a,
Jin
Gan‡
a,
Daniel J.
Fernandez
b,
Klaus-Peter
Knobeloch
b,
Paul P.
Geurink
a,
Jin
Gan‡
a,
Daniel J.
Fernandez
b,
Klaus-Peter
Knobeloch
b,
Paul P.
Geurink
 a and
Huib
Ovaa
a and
Huib
Ovaa
 *a
*a
aOncode Institute and Department of Cell and Chemical Biology, Leiden University Medical Center (LUMC), 2333 ZC Lei-den, The Netherlands. E-mail: H.Ovaa@lumc.nl
bInstitute of Neuropathology, Faculty of Medicine, University of Freiburg, Freiburg, Germany, Breisacherstraße 64, 79106 Freiburg, Germany
First published on 31st October 2019
Abstract
The linear synthesis of the N-terminal domain of mISG15 has been developed which enables the synthesis of full-length mISG15 and the activity-based probe Rho-mISG15-PA via native chemical ligation. Pilot experiments showed that the synthetic proteins were properly folded and recognized by endogenous enzymes. Our synthesis strategy allows the generation of other mISG15-based probes and reagents that can accelerate the research in this field.
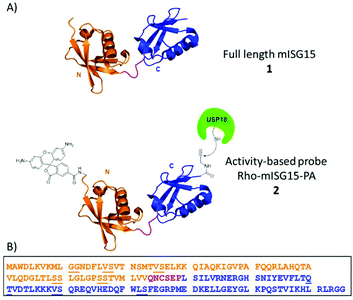
The innate immune system serves as the first line of defence against viral and bacterial infections in mammalian cells. Activation of this system initiates the release of Type-I interferons (IFN-α and IFN-β), which eventually leads to the transcription of more than 300 interferon-stimulated genes (ISGs). These ISGs encode for many different proteins such as cytokines, chemokines, transcription factors and enzymes that regulate the host immune response. One of the most strongly induced proteins is ubiquitin (Ub)-like modifier ISG15 (interferon stimulated gene of ∼15 kDa).1,2 This 157 amino acid containing protein is translated as a 17.8 kDa pro-protein and subsequently processed into its mature form. ISG15 contains two characteristic Ub-like β-grasp domains connected via a short, unstructured hinge region, such that it forms a linear dimer of Ub-like modules (Fig. 1A).3
Similar to Ub, ISG15 is conjugated post-translationally to the ε-amine of a target protein lysine residue by means of an enzymatic cascade in which the Ube1L (E1 activating enzyme),4 UbCH8 (E2 conjugating enzyme)5,6 and several E3 ligases (for example mouse HERC6 and human HERC5)7–10 are involved. This process is called ISGylation.11
ISG15 is deconjugated by specific proteases in a process termed deISGylation, comparable to deubiquitination by deubiquitinating enzymes (DUBs). In fact, the first (and to date only) human deISGylase was initially found to bind Ub and hence named Ub binding protein 43 (UBP43), or Ub specific protease 18 (USP18), although it does not display DUB activity.12 USP18 can process pro-ISG15. However, mice lacking the catalytic activity of USP18 do not show compromise, demonstrating that either other proteases with ISG15 selectivity exist and/or USP18 is not the isopeptidase for processing ISG15.13 Indeed, several DUBs have been reported to be cross-reactive with ISG15 (e.g. USP2, 14 and 21).14,15 However the observation that USP18C61A shows higher levels on ISGylation suggests that USP18 represents the major deISGylase for substrates within the context of the whole organism.16
Most Ub-like proteins are found in all eukaryotic species but ISG15 has evolved with the IFN system and is therefore only found in vertebrates.17 ISG15 plays a critical role in the antiviral response. For example the overexpression of ISG15 inhibits HIV replication in HIV infected cells.18 ISG15 knock-down in mice led to a higher susceptibility to several viral infections and ISG15 silencing increased viral growth in different cell types.19 Interestingly, the antiviral activity of ISG15 either requires its conjugation as shown by the fact that also Ube1L-deficient mice display a higher susceptibility to viral infection.2 Alternatively some viruses are counteracted in a conjugation independent manner. Notably, the transcription of ISG15 as well as the E1/E2/E3 enzymes and USP18 are highly regulated by Type-I IFN.20
The antiviral properties of ISG15 are further reflected by the finding that several viruses have developed immune evasion strategies that directly target the ISG15 pathway. For example the NS1B protein of influenza-B binds ISG15, thereby blocking the interaction with Ube1L and inhibiting ISGylation.4 The deadly human pathogen Crimean Congo hemorrhagic fever virus (CCHFV) targets ISG15 conjugates by expressing a deISGylating enzyme.21
Besides its function as a protein modifier, extracellular ISG15 has been reported to stimulate the IFN-γ secretion through binding to the LFA-1 cell surface receptor.22
Despite its immunological importance, the ISG15 system has remained relatively unexplored compared to other Ub-(like) proteins and one of the main reasons for that is the minimal availability of convenient (chemical) tools to interfere with the system. Successful chemical synthesis of ubiquitin allowed the development of ubiquitin-based tools, such as assay reagents, inhibitors and probes which tremendously boosted the research in the ubiquitin field.23,24 Along this line, we reasoned that the development of a method to synthesize full-lengthISG15 would open new avenues for the preparation of ISG15-based research tools which will likely accelerate the research in this field as well.
So far, intein chemistry has been applied to prepare full-length ISG15-based probes, equipped with a reactive moiety and reporter group,14,25 however this method is bound to the limitations of protein expression and therefore lacks the convenient “at will attachment” of any desired chemical entity at any site, a feature that is available in a fully synthetic approach.26–28 We here report the first chemical synthesis of full-length mISG15 and an activity-based probe derivative thereof, rhodamine-ISG15-propargylamide (Rho-ISG15-PA), (1 and 2 in Fig. 1) using solid-phase peptide chemistry and native-chemical ligation, and show that the synthetic proteins are properly recognized by USP18 and Ube1L.
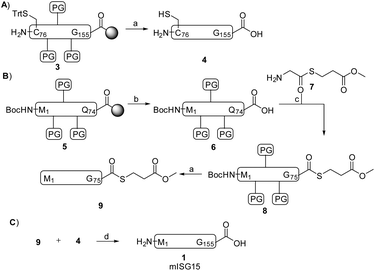
ISG15 can be considered as a linear dimer of two ubiquitin-like modules (Fig. 1, ISG15 has 155 AA versus ubiquitin 76 AA), and we therefore decided to cut the protein into two parts at Cys76 and first synthesize the N-terminal domain (M1-Q74) and C-terminal domain (C76-G155) separately via a linear SPPS approach. In the final step, the full-length products could then be constructed via assembling these two parts together through native chemical ligation (NCL). The mutation of N75G was introduced at the NCL site because we reasoned that this mutation, that is located in the unstructured hinge region, would greatly increase the conversion rate in NCL, yet have a minimal impact on the structure and activity of mISG15.
The C-terminal domain C76-G155 (peptide 3, Scheme 1) was synthesized by SPPS using a similar method reported previously by our group for the preparation of Ac-mISG1580-154-PA.25 Standard Fmoc-based SPPS conditions were used (4 eq. Fmoc protected AA, 4 eq. PyBOP, 8 eq. DIPEA and double couplings). After treatment of the peptide with a cleavage mixture (TFA/H2O/phenol/TIPS) followed by HPLC purification, 38 mg of peptide 4 was obtained with an overall yield of 16%.
The SPPS of the N-terminal domain (M1-Q74) turned out to be more challenging. We explored various dipeptide “aggregation breakers” such as pseudoprolines29 and dimethoxybenzyl (DMB)30 at different positions in the sequence and eventually identified five sites that required these building blocks. The optimized synthesis strategy is shown in Fig. 1B. N-Terminal domain M1-Q74, peptide 5 (Scheme 1) was prepared on a mild-acid-labile chlorotrityl resin and the last coupling involved a Boc-protected amino acid. After mild acidic cleavage from the resin by HFIP, side chain protected peptide 6 was obtained and the H-Gly-thioester (7) was selectively incorporated to give building block 8 using Sakakibara's method to elongate protected peptides,31 which could prevent the epimerization of the stereocenter in Q74. After global deprotection and HPLC purification, the N-terminal domain (9) was ready (overall yield: 20 mg, 10%). Native chemical ligation of the C-terminal domain (4) and the N-terminal domain (9) was carried out in buffer containing 6 M Gnd·HCl, 0.15 M sodium phosphate and 0.25 mM MPAA (pH 7.5) at 37 °C overnight. The reaction mixture was analysed by LC-MS (Fig. S1†), which showed the formation of product (1) and the hydrolysed product of peptide 9. After purification by HPLC and gel-filtration, pure full-length mISG15 (1) was obtained (1.2 mg, 0.070 μmol, 5.3%). The purity of mISG15 was confirmed by SDS-PAGE (Fig. 2A) and MS (Fig. 2B) analysis.
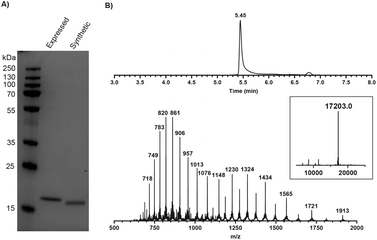 | ||
Fig. 2 Validation of synthetic mISG15 by (A) SDS-PAGE showing synthetic and expressed mISG15 and (B) MS analysis of the synthetic full-length mISG15 (1). Calculated mass: 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 202.4 Da. 202.4 Da. | ||
To verify the correct folding of purified synthetic mISG15, we compared the biochemical function of synthetic and expressed mISG15. We treated synthetic and expressed mISG15 with UBE1L (human ISG15 E1), which initiates the mISG15 conjugation cycle.5 Using the procedures previously reported by our group,32 mISG15 can be conjugated to the TAMRA-labelled dipeptide TAMRA-Lys(SH)Gly under the activation of UBE1L. The efficient formation of 5-TAMRA-Lys(mISG15)-Gly-OH proved that synthetic mISG15 was recognized and processed with the same efficiency as expressed mISG15 (Fig. 3). These results were further confirmed by MS analysis and the mass shift corresponded to the addition of the TAMRA-labelled dipeptide (Fig. S2†).
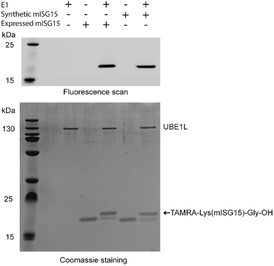 | ||
| Fig. 3 Validation of synthetic mISG15 via the reactivity of both expressed and synthetic mISG15 with UBE1L. | ||
With a successful synthesis approach in hand, we then focused on the generation of mISG15-based tools to profile and visualize enzymes involved in the ISGylation. For this purpose, we first designed an activity-based probe (Fig. 1, Rho-mISG15-PA) with an alkyne at the C-terminus as a warhead and a rhodamine label at the N-terminus as a fluorescent tag. The synthesis of Rho-mISG15-PA was carried out in the same fashion as the preparation of mISG15 (shown in Scheme S1 and Fig. S3†). In brief, the C-terminal domain, C76-G154 was prepared by SPPS with an N-terminal Boc protecting group and the protected peptide was cleaved from the trityl resin. After coupling propargyl amine to the C-terminal carboxylic acid, the protein was deprotected and purified by HPLC. The N-terminal domain, M1-Q74 was also prepared by SPPS and the N-terminus was coupled to DiBoc-Rho-COOH.26 The thioester was prepared as described above and after NCL and purifications, the Rho-mISG15-PA probe (2) was obtained (0.21 mg, 0.012 μmol, 1.9% yield).
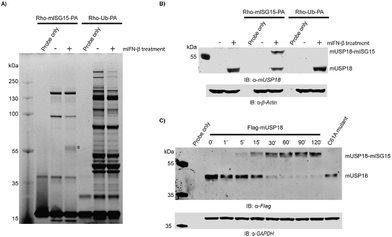
In order to investigate the structural integrity and activity of this fully synthetic ISG15 probe we performed a labelling experiment in lysates from mouse NMuMG cells. Since USP18 expression is highly regulated by Type-I IFN, the cell were treated with or without IFN-β for 24 hours prior to cell lysis. Both lysates were treated with probe 2 and we used the pan-DUB probe Rho-Ub-PA33 as a control. The samples were resolved by SDS-PAGE and all probe-labelled proteins were visualized by in-gel fluorescence imaging (Fig. 4A and S5A†). A clear band around 55 kDa can be seen in the IFN-β treated sample that is labelled with the ISG15 probe, and this band is absent in the non IFN-β treated sample. In Ub probe samples, no new band was observed in the IFN-β treated sample when compared to the non IFN-β treated sample. This band was further verified to be ISG15-probe-labelled USP18 (43 kDa USP18 + 17 kDa probe) by western blot using anti-mUSP18 antibody (Fig. 4B and Fig. S6†). Our results confirmed the previous conclusion that USP18 conveniently reacts with ISG15-PA but lacks any reactivity towards Ub-PA.12 A few other bands appear in both IFN-β and non-treated ISG15 probe samples, which most likely correspond to ISG15 cross-reactive DUBs.14
To further confirm that our probe could label mUSP18, HEK293T cells transiently overexpressing Flag-mUSP18 wildtype or Flag-mUSP18 C61A (active site mutant) were lysed and incubated with the Rho-mISG15-PA probe. After 30 min, almost full labelling was achieved and there was no labelling in the sample with the C61A active site mutant (Fig. 4C and S5B†). These results show that our probe can label mUSP18 via the active site cysteine. Furthermore, our results indicate that the asparagine 75 to glycine substitution in the hinge region hardly affected the structural properties as well as the activity of the protein.
Conclusions
In conclusion, we have reported the first preparation of the 155 amino acid protein mISG15 through SPPS chemistry and native chemical ligation. Many efforts have been made to optimize the synthesis of the N-terminal domain through SPPS and five critical “aggregation breaker” positions were identified. The successful synthesis of the N-terminal domain enabled us to obtain full-length mISG15 and the Rho-mISG15-PA probe, which were properly recognized and efficiently processed by UBE1L and USP18, respectively. With our established method for the preparation of the N-terminal domain and full length mISG15, it is now possible to design and synthesize various mISG15-based reagents or probes to investigate the complicated biological function of mISG15. For example, our synthesis route allows for the incorporation of photo-crosslinking amino acids or the introduction of other amino acid mutations anywhere in the sequence to study the proteins that could interact with mISG15. As most of the ISG15 function in particular with respect to ISG15deconjugation by USP18 is attributed to its C-terminal domain, this will especially become valuable to elucidate the (so far understudied) specific role(s) of the N-terminal domain within ISG15. We expect that the strategy reported here to synthesize full-length mISG15 will lead to an expansion of the current limited ISG15 toolbox which will concomitantly result in a better understanding of ISG15 biology. In addition, the acquired chemical knowledge will aid in the chemical preparation of the even more challenging human ISG15.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank Dris el Atmioui, Cami Talavera Ormeño and Paul Hekking (Department of Cell and Chemical Biology, LUMC, Leiden) for SPPS assistance. This work was supported by a VICI grant and a grant from N.W.O. Gravitation Programme Institute for Chemical Immunology (ICI) to H. O.Notes and references
- D. Zhang and D.-E. Zhang, J. Interferon Cytokine Res., 2011, 31, 119–130 CrossRef CAS PubMed.
- A. G. van der Veen and H. L. Ploegh, Annu. Rev. Biochem., 2012, 81, 323–357 CrossRef CAS PubMed.
- J. Narasimhan, M. Wang, Z. Fu, J. M. Klein, A. L. Haas and J.-J. P. Kim, J. Biol. Chem., 2005, 280, 27356–27365 CrossRef CAS PubMed.
- W. Yuan and R. M. Krug, EMBO J., 2001, 20, 362–371 CrossRef CAS PubMed.
- K. I. Kim, N. V. Giannakopoulos, H. W. Virgin and D. E. Zhang, Mol. Cell. Biol., 2004, 24, 9592–9600 CrossRef CAS PubMed.
- C. Zhao, S. L. Beaudenon, M. L. Kelley, M. B. Waddell, W. Yuan, B. A. Schulman, J. M. Huibregtse and R. M. Krug, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 7578–7582 CrossRef CAS PubMed.
- D. Oudshoorn, S. van Boheemen, M. T. Sanchez-Aparicio, R. Rajsbaum, A. Garcia-Sastre and G. A. Versteeg, PLoS One, 2012, 7, e29870 CrossRef CAS PubMed.
- L. Ketscher, A. Basters, M. Prinz and K. P. Knobeloch, Biochem. Biophys. Res. Commun., 2012, 417, 135–140 CrossRef CAS PubMed.
- J. J. Wong, Y. F. Pung, N. S. Sze and K. C. Chin, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 10735–10740 CrossRef CAS PubMed.
- A. Dastur, S. Beaudenon, M. Kelley, R. M. Krug and J. M. Huibregtse, J. Biol. Chem., 2006, 281, 4334–4338 CrossRef CAS PubMed.
- Y. J. Jeon, H. M. Yoo and C. H. Chung, Biochim. Biophys. Acta, 2010, 1802, 485–496 CrossRef CAS PubMed.
- M. P. Malakhov, O. A. Malakhova, K. I. Kim, K. J. Ritchie and D. E. Zhang, J. Biol. Chem., 2002, 277, 9976–9981 CrossRef CAS PubMed.
- J. L. Potter, J. Narasimhan, L. Mende-Mueller and A. L. Haas, J. Biol. Chem., 1999, 274, 25061–25068 CrossRef CAS PubMed.
- A. Catic, E. Fiebiger, G. A. Korbel, D. Blom, P. J. Galardy and H. L. Ploegh, PLoS One, 2007, 2, e679 CrossRef PubMed.
- Y. Ye, M. Akutsu, F. Reyes-Turcu, R. I. Enchev, K. D. Wilkinson and D. Komander, EMBO Rep., 2011, 12, 350–357 CrossRef CAS PubMed.
- L. Ketscher, R. Hannss, D. J. Morales, A. Basters, S. Guerra, T. Goldmann, A. Hausmann, M. Prinz, R. Naumann, A. Pekosz, O. Utermohlen, D. J. Lenschow and K. P. Knobeloch, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 1577–1582 CrossRef CAS PubMed.
- J. B. Andersen and B. A. Hassel, Cytokine Growth Factor Rev., 2006, 17, 411–421 CrossRef CAS PubMed.
- B. Skaug and Z. J. Chen, Cell, 2010, 143, 187–190 CrossRef CAS PubMed.
- D. J. Lenschow, Viruses, 2010, 2, 2154–2168 CrossRef CAS PubMed.
- A. J. Sadler and B. R. Williams, Nat. Rev. Immunol., 2008, 8, 559–568 CrossRef CAS PubMed.
- N. Frias-Staheli, N. V. Giannakopoulos, M. Kikkert, S. L. Taylor, A. Bridgen, J. Paragas, J. A. Richt, R. R. Rowland, C. S. Schmaljohn, D. J. Lenschow, E. J. Snijder, A. Garcia-Sastre and H. W. T. Virgin, Cell Host Microbe, 2007, 2, 404–416 CrossRef CAS PubMed.
- C. D. Swaim, A. F. Scott, L. A. Canadeo and J. M. Huibregtse, Mol. Cell, 2017, 68, 581–590 CrossRef CAS PubMed.
- R. Ekkebus, D. Flierman, P. P. Geurink and H. Ovaa, Curr. Opin. Chem. Biol., 2014, 23, 63–70 CrossRef CAS PubMed.
- K. F. Witting, M. P. C. Mulder and H. Ovaa, J. Mol. Biol., 2017, 429, 3388–3394 CrossRef CAS PubMed.
- A. Basters, P. P. Geurink, A. Rocker, K. F. Witting, R. Tadayon, S. Hess, M. S. Semrau, P. Storici, H. Ovaa, K. P. Knobeloch and G. Fritz, Nat. Struct. Mol. Biol., 2017, 24, 270–278 CrossRef CAS PubMed.
- P. P. Geurink, B. D. M. van Tol, D. van Dalen, P. J. G. Brundel, T. E. T. Mevissen, J. N. Pruneda, P. R. Elliott, G. B. A. van Tilburg, D. Komander and H. Ovaa, ChemBioChem, 2016, 17, 816–820 CrossRef CAS PubMed.
- G. J. van der Heden van Noort, R. Kooij, P. R. Elliott, D. Komander and H. Ovaa, Org. Lett., 2017, 19, 6490–6493 CrossRef CAS PubMed.
- F. El Oualid, R. Merkx, R. Ekkebus, D. S. Hameed, J. J. Smit, A. de Jong, H. Hilkmann, T. K. Sixma and H. Ovaa, Angew. Chem., Int. Ed., 2010, 49, 10149–10153 CrossRef CAS PubMed.
- T. Haack and M. Mutter, Tetrahedron Lett., 1992, 33, 1589–1592 CrossRef CAS.
- J. Blaakmeer, T. Tijsse-Klasen and G. I. Tesser, Int. J. Pept. Protein Res., 1991, 37, 556–564 CrossRef CAS PubMed.
- S. Sakakibara, Biopolymers, 1995, 37, 17–28 CrossRef CAS PubMed.
- P. P. Geurink, F. El Oualid, A. Jonker, D. S. Hameed and H. Ovaa, ChemBioChem, 2012, 13, 293–297 CrossRef CAS PubMed.
- A. de Jong, K. Witting, R. Kooij, D. Flierman and H. Ovaa, Angew. Chem., Int. Ed., 2017, 56, 12967–12970 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: MS and LC-MS spectra, synthesis procedures and biochemical experimental procedures (PDF). See DOI: 10.1039/c9ob02127b |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2019 |