Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Optimization of interstrand interactions enables burn detection with a collagen-mimetic peptide†
Jesús M.
Dones
 ab,
I. Caglar
Tanrikulu
ab,
I. Caglar
Tanrikulu
 ac,
Jenu V.
Chacko
ac,
Jenu V.
Chacko
 d,
Alexandra B.
Schroeder
e,
Trish T.
Hoang
c,
Angela L. F.
Gibson
d,
Alexandra B.
Schroeder
e,
Trish T.
Hoang
c,
Angela L. F.
Gibson
 *f,
Kevin W.
Eliceiri
*f,
Kevin W.
Eliceiri
 *de and
Ronald T.
Raines
*de and
Ronald T.
Raines
 *abc
*abc
aDepartment of Chemistry, Massachusetts Institute of Technology, 77 Massachusetts Avenue, Cambridge, MA 02139, USA. E-mail: rtraines@mit.edu
bDepartment of Chemistry, University of Wisconsin – Madison, 1101 University Avenue, Madison, WI 53706, USA
cDepartment of Biochemistry, University of Wisconsin – Madison, 433 Babcock Drive, Madison, WI 53706, USA
dDepartment of Biomedical Engineering, University of Wisconsin – Madison, 1550 Engineering Drive, Madison, WI 53706, USA
eDepartment of Medical Physics, University of Wisconsin School of Medicine and Public Health, 1111 Highland Avenue, Madison, WI 53705, USA. E-mail: eliceiri@wisc.edu
fDepartment of Surgery, University of Wisconsin School of Medicine and Public Health, 600 Highland Avenue, Madison, WI 53792, USA. E-mail: gibson@surgery.wisc.edu
First published on 30th October 2019
Abstract
Collagen is an abundant component of the extracellular matrix and connective tissues. Some collagen-mimetic peptides (CMPs) that do not form homotrimers can anneal to damaged tissue. Here, through a computational screen, we identify (flpHypGly)7 as an optimal monomeric CMP for heterotrimer formation. We find that (flpHypGly)7 forms stable triple helices with (ProProGly)7 but not with itself. The nonnatural amino acid HflpOH, which is (2S,4S)-4-fluoroproline, is not toxic to human fibroblasts or keratinocytes. Conjugation of (flpHypGly)7 to a fluorescent dye enables the facile detection of burned collagenous tissue with high specificity. The ubiquity of collagen and the prevalence of injuries and diseases that disrupt endogenous collagen suggests widespread utility for this approach.
Introduction
Collagen—the basic building block of the extracellular matrix (ECM)—is the most abundant protein in vertebrates. Comprising a third of the total protein and three quarters of the dry weight of skin in humans, collagen is an essential component of all connective tissues.1 Collagen also plays a crucial role in maintaining the biological and structural integrity of the ECM. Its continual remodeling impacts and controls cell differentiation and proliferation,2 and atypical organization of the collagen matrix is a biomarker for various diseases, including fibrosis, connective tissue disorders, and cancer.3–7Natural collagen is comprised of three left-handed, polyproline II-type (PPII) helices aligned in a parallel manner and coiled with each other to form a right-handed triple helix.1,8 Each of the peptide strands is constituted by a continuous repeat of Xaa-Yaa-Gly units. Even though the Xaa and Yaa positions can accommodate any amino acid, (2S)-proline (Pro; 28% abundance) and (2S,4R)-4-hydroxyproline (Hyp; 38% abundance) are the most prevalent, respectively. As a result, ProHypGly is the most common triplet in collagen (10.5%).9 These natural preferences at the Xaa and Yaa positions promote the conformational stability of the collagen triple helix.1
Collagen stability is governed by stereoelectronic effects that preorganize its prevalent pyrrolidine rings.1 A Cγ-endo and Cγ-exo pyrrolidine-ring pucker is preferred at the Xaa and Yaa position, respectively, in stable triple-helical structures. Substituents at the 4-position of a proline residue can enforce the preferred pyrrolidine-ring pucker via a gauche10 or steric11 effect, lowering the entropic cost for forming a triple helix and increasing its conformational stability (Table 1).1 For example, the stereospecific installation of a fluoro group generates a gauche effect that leads to a Cγ-endo pucker in (2S,4S)-4-fluoroproline (flp) and supports the Cγ-exo pucker in (2S,4R)-4-fluoroproline (Flp) to a greater extent than does the 4R-hydroxy group of Hyp.12–14 The impact of such substitutions are apparent in the thermostability of collagen-mimetic peptides (CMPs). The value of Tm, which is the temperature at the midpoint of the thermal transition between a triple helix and its component strands, tracks the strength of the inductive effect induced by the substitutions.15 For example, replacing Hyp with Flp at the Yaa position in (ProYaaGly)7 leads to ΔTm = 9 °C (Table 1).
| Xaa (R1,R2) | Yaa (R1,R2) |
T
m![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) b (°C) b (°C) |
|||||||
|---|---|---|---|---|---|---|---|---|---|
| Flp (F,H) | Mop (OMe,H) | Hyp (OH,H) | Mep (H,CH3) | Clp (Cl,H) | Pro (H,H) | (XaaProGly)7 | (XaaProGly)10 | ||
| a Values of IE were calculated with molecular mechanics. Values <2.8 kcal mol−1 are depicted in gray typeface (see note §). b Values of Tm for the denaturation of the indicated triple helices were determined previously by circular dichroism spectroscopy.1 ND, not determined; NH, no helix. | |||||||||

|
flp (H,F) | 2.8 | 3.4 | 3.0 | 1.1 | 5.6 | 1.3 | 33 | 58 |
| mep (CH3,H) | −0.4 | −0.3 | −0.1 | −0.6 | 2.4 | −0.4 | 13 | ND | |
| Pro (H,H) | −0.1 | −0.6 | 0.3 | −0.2 | 1.2 | 0.0 | NH | 31–41 | |
T
m
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) b(°C) b(°C) |
|||||||||
| (ProYaaGly)7 | 45 | ND | 36 | 29 | 23 | NH | |||
| (ProYaaGly)10 | 91 | 70 | 61–69 | ND | 52 | 31–41 | |||
Not all Xaa and Yaa pairings are compatible. The register of the strands in a triple helix is staggered such that each cross-section contains an Xaa, Yaa, and Gly residue.1 Even though (flpProGly)7 and (ProFlpGly)7 form highly stable triple helices (Table 1),16 (flpFlpGly)7 is a monomer in solution due to unfavorable steric interactions between neighboring flp and Flp residues (Fig. 1).17 Still, mixtures of (flpFlpGly)7 and (ProProGly)7 produce stable triple helices because of the preorganization of (flpFlpGly)7 and the minimal steric clashes between flp/Flp and neighboring Pro residues.17
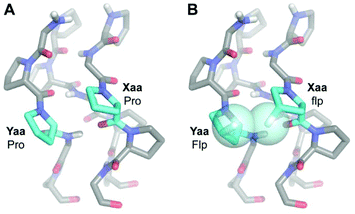 | ||
| Fig. 1 Molecular models of neighboring Xaa and Yaa positions on strands of a collagen triple helix. (A) When Xaa and Yaa are Pro, there is not a steric clash between the neighboring side chains (cyan). (B) When flp and Flp are placed similarly, clashing fluoro groups (cyan spheres) deter strand-association.17 | ||
Because (flpFlpGly)7 and (ProProGly)7 form only heterotrimeric triple helices,‡ they can anneal readily to exposed strands on denatured collagen (Fig. 2). Such annealing could allow for the development of diagonistic or therapeutic agents conjugated to a monomeric CMP.19–22 Previously, we have used CMP-conjugates to deliver fluorescent dyes,23 a sunscreen,24 and growth factors25 to damaged collagen. Although (flpFlpGly)7 seemed to outperform (ProProGly)7 as an annealing strand,23 its exceptional propensity was hindered by its poor solubility, which can complicate formulation and dosing, and diminish bioavailability.26 The highly soluble (ProHypGly)7 peptide can be successful in this context, but its homotrimerization necessitates a denaturation step prior to application,27,28 complicating its use. Another approach invokes protecting groups to deter triple-helix formation,29,30 but their removal introduces operational complexity.
 | ||
| Fig. 2 Representation of a collagen-mimetic peptide with a pendant moiety (XCMP) annealing selectively to a damaged site within the collagen triple helix but not forming a homotrimeric triple helix. | ||
Here, we conduct a computational survey of proline analogs to identify (XaaYaaGly)7 variants that are monomeric in solution, but have high ability to anneal with damaged collagen. We confirm the association characteristics of our design in vitro and demonstrate its specificity for damaged collagen by identifying a burned region on a rat-tail tendon ex vivo.
Results and discussion
(flpHypGly)7 is an optimal CMP
An optimal CMP is a highly soluble monomer under physiological conditions, but forms strong triple helices with natural collagen strands. To search for such (XaaYaaGly)7 variants, we defined two sets of proline analogs that are compatible with the steric and electronic requirements at the Xaa and Yaa positions (Table 1).1 The resulting 18 combinatorial Xaa⋯Yaa pairs represent interactions between Xaa and Yaa residues on the neighboring strands of an (XaaYaaGly)7 homotrimer. To determine the extent of steric repulsion in each homotrimer, we applied molecular mechanics calculations to models derived from a high-resolution structure of a (ProProGly)10 triple helix (PDB entry 1kf6).31 Briefly, Xaa and Yaa residues were positioned on neighboring positions on the triple helix. Following energy minimization, the interaction between the Xaa⋯Yaa pair was calculated as the energy difference between the homotrimeric helix and the free strands.32,33 Three known examples of monomeric CMPs—(ProProGly)7, (flpFlpGly)7, and (clpClpGly)7—served as experimental benchmarks for data analysis.17,34Interaction energies (IE) revealed that four combinations—Xaa flp⋯Flp, flp⋯Mop, flp⋯Hyp, and flp⋯Clp—engender unfavorable interactions (Table 1).§ Among these four, a flp⋯Flp pair of Xaa⋯Yaa residues (IE = 2.8 kcal mol−1) merits special consideration. flp in the Xaa position and Flp in the Yaa position are individually the most stabilizing 4-substituted proline analogs. Yet, (flpFlpGly)7 does not form a stable triple helix, suggesting that Xaa⋯Yaa clashes of IE ≥ 2.8 kcal mol−1 are likely to produce monomeric peptides. Peptides that satisfied this criterion include (flpMopGly)7, (flpHypGly)7, and (flpClpGly)7 (IE = 3.4, 3.0, and 5.6 kcal mol−1, respectively). The higher solubility bestowed by Hyp (hydroxy group) compared to Mop (methoxy group) or Clp (chloro group) led to explore (flpHypGly)7 experimentally.¶ Notably, a stereospecific H→F substitution is the only difference between a flp-Hyp-Gly unit and the isologous Pro-Hyp-Gly unit of natural collagen. Moreover, deleterious O)(F steric clashes engendered by (flpHypGly)7 will not preclude annealing to strands of human collagen, which lack flp entirely and have Hyp in only 38% of its Xaa-Yaa-Gly units.9 Finally, the HflpOH monomer is readily accessible from natural HHypOH by an SN2 reaction.35 We note that CMPs containing flp-Hyp-Gly units have been examined previously,36,37 though in contexts distinct from those described herein. For example, (flpHypGly)9 was found to form a stable triple helix at ambient temperature, complicating its use in biological contexts.37 Hence, we examined a shorter peptide: (flpHypGly)7.
(flpHypGly)7 is structured only as a heterotrimer
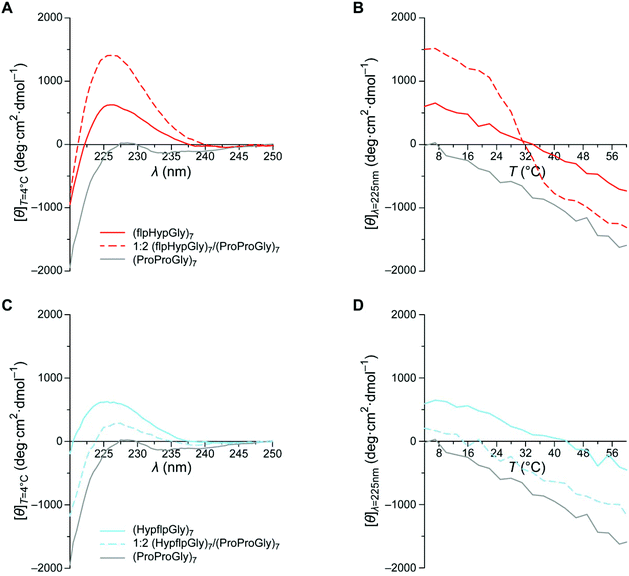
The (flpHypGly)7 peptide and its variants were synthesized by microwave-assisted solid-phase peptide synthesis and purified by reversed-phase HPLC. Samples prepared in 50 mM HOAc were heated to 65 °C, cooled to 4 °C, and equilibrated at 4 °C for ≥24 h prior to analysis. The structure of peptides and their mixtures was assessed with circular dichroism (CD) spectroscopy, following the characteristic CD signature for the collagen triple helix and PPII helix at ∼225 nm.CD spectra for (flpHypGly)7 and (ProProGly)7 reveal shallow peaks near 225 nm (Fig. 3A). These samples exhibit no cooperative denaturation with increasing temperature, indicating the absence of triple helices in solution (Fig. 3B). This behavior is typical for PPII structures that do not associate into triple helices.36–38 In marked contrast, a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 (flpHypGly)7/(ProProGly)7 solution exhibits a strong collagen signature in its CD spectrum (Fig. 3A), as well as cooperative denaturation with Tm = 28 °C (Fig. 3B and S2A†). A 2
2 (flpHypGly)7/(ProProGly)7 solution exhibits a strong collagen signature in its CD spectrum (Fig. 3A), as well as cooperative denaturation with Tm = 28 °C (Fig. 3B and S2A†). A 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (flpHypGly)7/(ProProGly)7 solution exhibits similar thermostability (Fig. S1A, S1B, and S2B†). The results of these mixing experiments indicate that (flpHypGly)7 can form a stable triple helix with (ProProGly)7 strands, but not with itself.
1 (flpHypGly)7/(ProProGly)7 solution exhibits similar thermostability (Fig. S1A, S1B, and S2B†). The results of these mixing experiments indicate that (flpHypGly)7 can form a stable triple helix with (ProProGly)7 strands, but not with itself.
(HypflpGly)7 does not form a triple helix
Next, we sought to use a (flpHypGly)7–dye conjugate in a physiological context, but we needed a negative control. A free (that is, unconjugated) dye could serve as a negative control for such experiments. The disparity between the physicochemical attributes of a CMP–dye conjugate and the dye alone could, however, compromise data-interpretation.To mimic the attributes of the (flpHypGly)7 peptide, we synthesized (HypflpGly)7, which is a compositional isomer (CI). Swapping the flp and Hyp residues generates a sequence that violates the conformational restrictions of the Xaa and Yaa positions (Table 1), and should preclude triple-helix formation.12,39 Indeed, the CD spectrum of (HypflpGly)7 mimics closely that of (flpHypGly)7 (Fig. 3C), and does not exhibit the cooperative transition of triple-helix denaturation (Fig. 3D). Further, the CD spectrum of a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 or 2
2 or 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (HypflpGly)7/(ProProGly)7 mixture is a linear combination of the individual (ProProGly)7 and (HypflpGly)7 spectra and exhibits no evidence of a triple helix (Fig. 3C and S1C†). These spectra, together with the absence of a cooperative transition (Fig. 3D and S1D†), provide strong evidence that (HypflpGly)7 does not interact with (ProProGly)7. Because (HypflpGly)7 has the same composition as does (flpHypGly)7 but lacks its annealing properties, (HypflpGly)7 serves as an ideal negative control for our physiological experiments.
1 (HypflpGly)7/(ProProGly)7 mixture is a linear combination of the individual (ProProGly)7 and (HypflpGly)7 spectra and exhibits no evidence of a triple helix (Fig. 3C and S1C†). These spectra, together with the absence of a cooperative transition (Fig. 3D and S1D†), provide strong evidence that (HypflpGly)7 does not interact with (ProProGly)7. Because (HypflpGly)7 has the same composition as does (flpHypGly)7 but lacks its annealing properties, (HypflpGly)7 serves as an ideal negative control for our physiological experiments.
HflpOH is not cytotoxic
The proteolysis of (flpHypGly)7 would generate three amino acids: HflpOH, HHypOH, and HGlyOH. Two of these amino acids, HHypOH and HGlyOH, are prevalent in human collagen; HflpOH, though, is nonnatural. As 1/5 of FDA-approved drugs contain fluorine,40 we expected HflpOH to be well-tolerated in humans. Nevertheless, HflpOH itself is not a component of any approved drug. Accordingly, we tested the tolerance of two relevant cell types for this amino acid: human fibroblasts and primary human epidermal keratinocytes. Fibroblasts are the most common cells in human connective tissue, and maintain its health and integrity. Keratinocytes constitute 90% of the cells in the epidermis, which is the barrier against environmental damage. We found that HflpOH is not detectably toxic to either fibroblasts or keratinocytes at concentrations up to 1 mM (Fig. 4).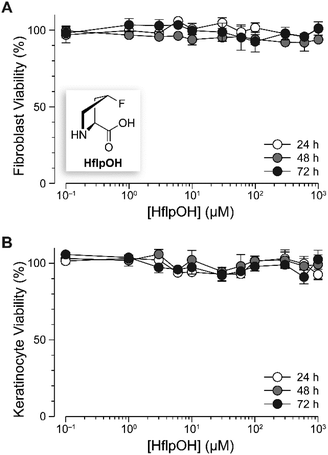 | ||
| Fig. 4 Graph showing the effect of HflpOH on the viability of human fibroblasts (A) and primary human epidermal keratinocytes (B). Data were obtained with a tetrazolium dye-based assay for metabolic activity.44 Values are the mean ± SEM (n = 3, biological replicates). | ||
(flpHypGly)7 anneals to damaged collagen ex vivo
To evaluate the binding of a (flpHypGly)7–dye conjugate to fibrillar collagen, we used the rat-tail tendon as a testbed, as it is comprised primarily of fibrillar collagen.41 Type I collagen from rat-tail tendons has been studied extensively, and is accessible by well-developed procedures.42 For the treatment of rat-tail tendon, we prepared variants of (flpHypGly)7 and (HypflpGly)7 that are conjugated via their N-terminal amino group to a cyanine 5 (Cy5) dye, obtaining Cy5CMP and Cy5CI, respectively (Fig. 5A). We choose Cy5 because of the far-red wavelength of its fluorescence excitation (λmax = 652 nm) and emission (λem = 672 nm), which allows for detection in tissue, as well as its high brightness (ε × Φ = 0.18).43 To test whether (flpHypGly)7 can distinguish damaged collagen from healthy collagen, we used a 350 °C soldering iron to burn a rat-tail tendon through glass (Fig. 5B). We then treated the tendon with Cy5CMP and Cy5CI, incubated for 1 h at 37 °C, and washed extensively with PBS. | ||
| Fig. 5 Multiphoton microscopy images of the annealing of fluorescently labeled CMP and its compositional isomer to collagen fibers from rat tails. (A) Amino acid sequences of the Cy5CMP and Cy5CI peptides. (B) Method for probing for damaged collagen in a rat-tail tendon. (C) Superimposed images of a damaged rat-tail tendon using second harmonic generation (SHG) in the green channel and the fluorescence from Cy5CMP in the red channel. (D) Superimposed images of the SHG channel of a damaged rat-tail tendon and the fluorescence from the compositional isomer, Cy5CI. (E) Bar graph of the mean fluorescence intensity in the indicated regions of interest (ROIs) as determined with ImageJ software.48 | ||
Second harmonic generation (SHG) microscopy is a nonlinear optical method that is used extensively to identify and visualize fibrillar collagens in tissues.45–47 Using a multiphoton laser-scanning microscope, data can be acquired from both SHG and Cy5 channels, revealing the distinction between damaged and healthy collagen (SHG) and its detection with our probe (Cy5). The fibrils exposed to 350 °C show little or no signal through the SHG channel, confirming the absence of intact collagen structure in the burn area (Fig. S3†). Upon treatment with Cy5CMP, the regions that fail to show a strong SHG signal exhibit a strong Cy5 fluorescence (Fig. 5C), indicating the association Cy5CMP with damaged collagen. Importantly, the same intensity is not observed in intact regions. In contrast, the distinction between damaged and healthy collagen is not apparent with the negative control, Cy5CI (Fig. 5D).
Quantification of Cy5 fluorescence reveals that the localization of Cy5CMP on burned collagen is greater by 25-fold (i.e., 148/6) than its fluorescence on intact collagen (Fig. 5E). Similarly, the labeling of these regions by Cy5CMP is greater by 5.3-fold (i.e., 148/28) that is that by the Cy5CI peptide. These results confirm annealing by (flpHypGly)7 to regions of collagen damage. Finally, we note that Cy5CMP is highly soluble in water, showing no evidence of precipitation despite its pendant hydrophobic dye.
Conclusions
An optimized CMP can act as a vehicle for the detection of mammalian collagen that has suffered burn damage. Burn injuries affect more than 7 million individuals worldwide and result in significant morbidity and mortality.49 Current methods to diagnose burns face challenges, such as the disparity between the clinical identification of burn depth and the histopathologic determination of cellular and ECM damage.50 Further, only 70% of visual assessments of full-thickness burn injuries are accurate.51 Our approach could enable the reliable detection of thermally damaged collagen in a clinical setting. The ubiquity of collagen as well as the disruptive effects of fibrosis, connective-tissue disorders, and cancer on collagen in the ECM suggest its utility in other regimens as well.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by grant R56 AR044276 (NIH), the Laboratory for Optical and Computational Instrumentation, and the Morgridge Institute for Research. We thank Dr Ellen Arena for useful input on the manuscript and image analysis.Notes and references
- M. D. Shoulders and R. T. Raines, Annu. Rev. Biochem., 2009, 78, 929–958 CrossRef CAS PubMed.
- P. Lu, K. Takai, V. M. Weaver and Z. Werb, Cold Spring Harbor Perspect. Biol., 2011, 3, a005058 Search PubMed.
- M. C. Skala, J. M. Squirrell, K. M. Vrotsos, J. C. Eickhoff, A. Gendron-Fitzpatrick, K. W. Eliceiri and N. Ramanujam, Cancer Res., 2005, 65, 1180–1186 CrossRef CAS PubMed.
- S. J. Lin, S. H. Jee, C. J. Kuo, R. J. Wu, W. C. Lin, J. S. Chen, Y. H. Liao, C. J. Hsu, T. F. Tsai, Y. F. Chen and C. Y. Dong, Opt. Lett., 2006, 31, 2756–2758 CrossRef PubMed.
- N. D. Kirkpatrick, M. A. Brewer and U. Utzinger, Cancer Epidemiol. Biomarkers Prev., 2007, 16, 2048–2057 CrossRef CAS PubMed.
- E. Dimitrow, M. Ziemer, M. J. Koehler, J. Norgauer, K. Konig, P. Elsner and M. Kaatz, J. Invest. Dermatol., 2009, 129, 1752–1758 CrossRef CAS PubMed.
- M. W. Conklin, J. C. Eickhoff, K. M. Riching, C. A. Pehlke, K. W. Eliceiri, P. P. Provenzano, A. Friedl and P. J. Keely, Am. J. Pathol., 2011, 178, 1221–1232 CrossRef PubMed.
- K. Okuyama, C. Hongo, R. Fukushima, G. Wu, H. Narita, K. Noguchi, Y. Tanaka and N. Nishino, Biopolymers, 2004, 76, 367–377 CrossRef CAS PubMed.
- J. A. M. Ramshaw, N. K. Shah and B. Brodsky, J. Struct. Biol., 1998, 122, 86–91 CrossRef CAS PubMed.
- E. S. Eberhardt, N. Panasik Jr. and R. T. Raines, J. Am. Chem. Soc., 1996, 118, 12261–12266 CrossRef CAS PubMed.
- M. D. Shoulders, J. A. Hodges and R. T. Raines, J. Am. Chem. Soc., 2006, 128, 8112–8113 CrossRef CAS PubMed.
- L. E. Bretscher, C. L. Jenkins, K. M. Taylor, M. L. DeRider and R. T. Raines, J. Am. Chem. Soc., 2001, 123, 777–778 CrossRef CAS PubMed.
- M. L. DeRider, S. J. Wilkens, M. J. Waddell, L. E. Bretscher, F. Weinhold, R. T. Raines and J. L. Markley, J. Am. Chem. Soc., 2002, 124, 2497–2505 CrossRef CAS PubMed.
- R. W. Newberry and R. T. Raines, Top. Heterocycl. Chem., 2017, 48, 1–26 CAS.
- M. D. Shoulders and R. T. Raines, Adv. Exp. Med. Biol., 2009, 611, 251–252 CrossRef CAS PubMed.
- J. A. Hodges and R. T. Raines, J. Am. Chem. Soc., 2003, 125, 9262–9263 CrossRef CAS PubMed.
- J. A. Hodges and R. T. Raines, J. Am. Chem. Soc., 2005, 127, 15923–15932 CrossRef CAS PubMed.
- M. D. Shoulders, K. A. Satyshur, K. T. Forest and R. T. Raines, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 559–564 CrossRef CAS PubMed.
- C. Siebler, R. S. Erdmann and H. Wennemers, Chimia, 2013, 67, 891–895 CrossRef CAS PubMed.
- S. Chattopadhyay and R. T. Raines, Biopolymers, 2014, 101, 821–833 CrossRef CAS PubMed.
- H. Wahyudi, A. A. Reynolds, Y. Li, S. C. Owen and S. M. Yu, J. Controlled Release, 2016, 240, 323–331 CrossRef CAS PubMed.
- A. B. Shekhter, A. L. Fayzullin, M. N. Vukolova, T. G. Rudenko, V. D. Osipycheva and P. F. Litvitsky, Curr. Med. Chem., 2019, 26, 506–516 CrossRef CAS PubMed.
- S. Chattopadhyay, C. J. Murphy, J. F. McAnulty and R. T. Raines, Org. Biomol. Chem., 2012, 10, 5892–5897 RSC.
- A. J. Ellison and R. T. Raines, Org. Biomol. Chem., 2018, 16, 7139–7142 RSC.
- S. Chattopadhyay, K. M. Guthrie, L. Teixeira, C. J. Murphy, R. R. Dubielzig, J. F. McAnulty and R. T. Raines, J. Tissue Eng. Regener. Med., 2016, 10, 1012–1020 CrossRef CAS PubMed.
- E. H. Kerns and L. Di, Drug Like Properties: Concept, Structure, Design and Methods, from ADME to Toxicity Optimization, Elsevier, Boston, MA, 2008 Search PubMed.
- A. Y. Wang, X. Mo, C. S. Chen and S. M. Yu, J. Am. Chem. Soc., 2005, 127, 4130–4131 CrossRef CAS PubMed.
- J. Hwang, Y. Huang, T. J. Burwell, N. C. Peterson, J. Connor, S. J. Weiss, S. M. Yu and Y. Li, ACS Nano, 2017, 11, 9825–9835 CrossRef CAS PubMed.
- Y. Li, C. A. Foss, D. D. Summerfield, J. J. Doyle, C. M. Torok, H. C. Dietz, M. G. Pomper and S. M. Yu, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 14767–14772 CrossRef CAS PubMed.
- Y. Li, C. A. Foss, M. G. Pomper and S. M. Yu, J. Visualized Exp., 2014, e51052 Search PubMed.
- R. Berisio, L. Vitagliano, L. Mazzarella and A. Zagari, Protein Sci., 2002, 11, 262–270 CrossRef CAS PubMed.
- S. L. Mayo, B. D. Olafson and W. A. Goddard, J. Phys. Chem., 1990, 94, 8897–8909 CrossRef CAS.
- A. D. MacKerell Jr., D. Bashford, M. Bellott, R. L. Dunbrack Jr., J. D. Evanseck, M. J. Field, S. Fischer, J. Gao, H. Guo, S. Ha, D. Joseph-McCarthy, L. Kuchnir, K. Kuczera, F. T. Lau, C. Mattos, S. Michnick, T. Ngo, D. T. Nguyen, B. Prodhom, W. E. Reiher, B. Roux, M. Schlenkrich, J. C. Smith, R. Stote, J. Straub, M. Watanabe, J. Wiórkiewicz-Kuczera, D. Yin and M. Karplus, J. Phys. Chem., 1998, B102, 3586–3616 CrossRef PubMed.
- M. D. Shoulders, I. A. Guzei and R. T. Raines, Biopolymers, 2008, 89, 443–454 CrossRef CAS PubMed.
- M. S. Chorghade, D. K. Mohapatra, G. Sahoo, M. K. Gurjar, M. V. Mandlecha, N. Bhoite, S. Moghe and R. T. Raines, J. Fluorine Chem., 2008, 129, 781–784 CrossRef CAS PubMed.
- D. Barth, A. G. Milbradt, C. Renner and L. Moroder, ChemBioChem, 2004, 5, 79–86 CrossRef CAS PubMed.
- L. L. Bennink, Y. Li, B. Kim, I. J. Shin, B. H. San, M. Zangari, D. Yoon and S. M. Yu, Biomaterials, 2018, 183, 67–76 CrossRef CAS PubMed.
- J. L. Lopes, A. J. Miles, L. Whitmore and B. A. Wallace, Protein Sci., 2014, 23, 1765–1772 CrossRef CAS PubMed.
- K. Inouye, Y. Kobayashi, Y. Kyogoku, Y. Kishida, S. Sakakibara and D. J. Prockop, Arch. Biochem. Biophys., 1982, 219, 198–203 CrossRef CAS PubMed.
- H. Zhang, Y. Li and Z.-X. Jiang, J. Biomol. Res. Ther., 2012, 1, e107 Search PubMed.
- S. Roth and I. Freund, J. Chem. Phys., 1979, 70, 1637–1643 CrossRef CAS.
- N. Rajan, J. Habermehl, M. F. Cote, C. J. Doillon and D. Mantovani, Nat. Protoc., 2006, 1, 2753–2758 CrossRef CAS PubMed.
- A. Waggoner and S. Kenneth, Methods Enzymol., 1995, 246, 362–373 CAS.
- A. H. Cory, T. C. Owen, J. A. Barltrop and J. G. Cory, Cancer Commun., 1991, 3, 207–212 CrossRef CAS.
- A. Keikhosravi, J. S. Bredfeldt, A. K. Sagar and K. W. Eliceiri, Methods Cell Biol., 2014, 123, 531–546 Search PubMed.
- R. J. Tran, K. L. Sly and J. C. Conboy, Annu. Rev. Anal. Chem., 2017, 10, 387–414 CrossRef CAS PubMed.
- L. Mostaço-Guidolin, N. L. Rosin and T. L. Hackett, Int. J. Mol. Sci., 2017, 18, E1772 CrossRef PubMed.
- C. T. Rueden, J. Schindelin, M. C. Hiner, B. E. DeZonia, A. E. Walter, E. T. Arena and K. W. Eliceiri, BMC Bioinf., 2017, 18, 529 CrossRef PubMed.
- M. M. Rybarczyk, J. M. Schafer, C. M. Elm, S. Sarvepalli, P. l. Vaswani, K. S. Balhara, L. C. Carlson and G. A. Jacquet, Afr. J. Emerg. Med., 2017, 7, 30–37 CrossRef.
- A. S. Karim, A. Yan, E. Ocotl, D. D. Bennett, Z. Wang, C. Kendziorski and A. L. F. Gibson, Wound Repair Regen., 2018, 27, 150–161 CrossRef.
- J. E. Thatcher, J. J. Squiers, S. C. Kanick, D. R. King, Y. Lu, Y. Wang, R. Mohan, E. W. Sellke and J. M. DiMaio, Adv. Wound Care, 2016, 5, 360–378 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Experimental procedures and additional analytical data. See DOI: 10.1039/c9ob01839e |
| ‡ Neither (flpFlpGly)7 nor (ProProGly)7 forms a stable homotrimeric triple helix, though for different reasons. (flpFlpGly)7 strands are highly preorganized for triple-helix formation, but experience strong interstrand F)(F clashes in a triple helix.17 (ProProGly)7 strands do not engender steric clashes upon association, but are not especially well-organized for triple-helix formation.18 |
| § We also tested clp (R1 = H, R2 = Cl) as a candidate at the Xaa position. Its use, however, produces extremely unfavorable interactions, even when its neighbor is Pro (IE = 13.8 kcal mol−1). This incompatibility is consistent with the low thermostability of triple helices with clpProGly units,34 and disqualifies the use of clp at the Xaa position of an annealing strand. |
¶ Log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P values are HHypOH, −2.64; HMopOH, −2.02; HProOH, −1.72; HFlpOH, −1.71; and HClpOH, −1.40; as calculated with software from Molinspiration (Slovenský Grob, Slovak Republic). P values are HHypOH, −2.64; HMopOH, −2.02; HProOH, −1.72; HFlpOH, −1.71; and HClpOH, −1.40; as calculated with software from Molinspiration (Slovenský Grob, Slovak Republic). |
| This journal is © The Royal Society of Chemistry 2019 |