Recognition of mixed-sequence DNA targets using spermine-modified Invader probes†
Abstract
Double-stranded oligodeoxyribonucleotides with +1 interstrand zipper arrangements of 2′-O-(pyren-1-yl)methyl-RNA monomers are additionally activated for highly specific recognition of mixed-sequence DNA targets upon incorporation of non-nucleotidic spermine bulges.
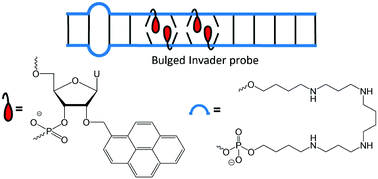
- This article is part of the themed collection: Chemical Biology in OBC


 Please wait while we load your content...
Please wait while we load your content...