Open Access Article
Open Access ArticleNew geldanamycin derivatives with anti Hsp properties by mutasynthesis†
Jekaterina
Hermane‡
a,
Simone
Eichner‡
a,
Lena
Mancuso
a,
Benjamin
Schröder‡
a,
Florenz
Sasse
 b,
Carsten
Zeilinger
c and
Andreas
Kirschning
b,
Carsten
Zeilinger
c and
Andreas
Kirschning
 *a
*a
aInstitute of Organic Chemistry, Leibniz Universität Hannover, Schneiderberg 1B, 30167 Hannover, Germany. E-mail: andreas.kirschning@oci.uni-hannover.de
bHelmholtz-Center for Infection Research, Department of Microbial Communication, Inhoffenstr. 7, 38124 Braunschweig, Germany
cLeibniz Universität Hannover, Institute of Biophysics and Center of Biomolecular Drug Research (BMWZ), Schneiderberg 38, D-30167 Hannover, Germany
First published on 7th May 2019
Abstract
Mutasynthetic supplementation of the AHBA blocked mutant strain of S. hygroscopicus, the geldanamycin producer, with 21 aromatic and heteroaromatic amino acids provided new nonquinoid geldanamycin derivatives. Large scale (5 L) fermentation provided four new derivatives in sufficient quantity for full structural characterisation. Among these, the first thiophene derivative of reblastatin showed strong antiproliferative activity towards several human cancer cell lines. Additionally, inhibitory effects on human heat shock protein Hsp90α and bacterial heat shock protein from H. pylori HpHtpG were observed, revealing strong displacement properties for labelled ATP and demonstrating that the ATP-binding site of Hsps is the target site for the new geldanamycin derivatives.
Introduction
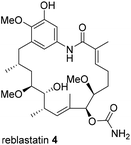
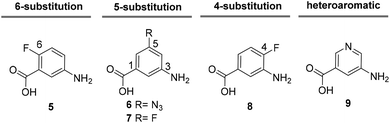
Besides classical concepts of natural product synthesis, namely total- and semisynthesis, strategies that rely on the manipulation of microbial secondary metabolites not after, but in the course of their biosynthesis, have broadened the synthetic portfolio of chemists and biotechnologists. One strategy employs mutant organisms blocked in the biosynthesis of endogenous key precursors on a genetic level. According to Rinehart,1 mutational biosynthesis or mutasynthesis2 aims to produce genetic mutants of an organism in which the formation of key biosynthetic building blocks required for the assembly of natural metabolites are blocked.3 In turn, administration of structurally modified building blocks, so-called mutasynthons, to the blocked mutant may result in their incorporation and lead to the synthesis of novel metabolites, which can be isolated and evaluated for their therapeutic potential.4 In principle, mutasynthesis allows access to compound libraries of pharmacologically relevant and complex novel products. While it is simple to produce gene knock-outs that are useful in the creation of blocked deletion mutants, it is critical that the modified organism performs enzymatic catalysis with high levels of efficiency, selectivity and a broad substrate tolerance when presented with non-natural precursors.Ansamycin antibiotics, specifically the ansamitocins, rifamycin and geldanamycin, have been a prime target to create natural product libraries through mutasynthesis.5 Geldanamycin (3; hydroquinone form in Scheme 1), produced by Streptomyces hygroscopicus, is a potential antitumor drug6 that binds to the N-terminal ATP-binding domain of heat shock protein 90 (Hsp90) and inhibits its ATP-dependent chaperone activity.7
 | ||
| Scheme 1 Key features of geldanamycin biosynthesis.8,9 | ||
Heat shock proteins are essential for survival because they regulate a diverse set of cellular processes and consequently, geldanamycin has been a lead structure for at least two clinical candidates against cancer.
However, activity of the benzoquinone form of geldanamycin first depends on reductive activation to hydroquinone 3 by the enzyme NAD(P)H/quinone oxidoreductase 1 (NQO1).7,10–12 Thus, it is the hydroquinone itself that is regarded to be the motif, that binds to the ATP binding domain. The benzoquinone-based geldanamycins are far from being ideal antitumor drugs, because they show hepatotoxic side effects, in part, because of their Michael-acceptor properties toward natural thiols such as glutathione.12,13
As mentioned previously, mutasynthesis has been successfully employed to access new geldanamycin analogues, particularly by the groups of Lee, Hong,16,17 by Menzella13 and through our own efforts.9,18 All published mutasyntheses of new phenolic derivatives of geldanamycin, which we call reblastatin derivatives in this work as they commonly lack the double bond at C4–C5 or a quinone/hydroquinone moiety, rely on the disruption of genes coding for AHBA formation and feeding of unnatural 3-aminobenzoic acid derivatives. In our preliminary studies, we demonstrated that aromatic mutasynthons 5–9 are successfully processed to new reblastatin derivatives (Fig. 1). These results showed that modifications at positions 4–6 (AHBA numbering),19 as well as heteroaromatic mutasynthons, are in principle tolerated by the biosynthetic assembly line, although the scope is still unknown.
 | ||
| Fig. 1 Aromatic mutasynthons 5–9 successfully transformed into new reblastatin derivatives in preparative amounts. | ||
Herein, we provide a full evaluation of substrate tolerance for an AHBA blocked mutant strain of S. hygroscopicus using a wide variety of aromatic and heteroaromatic amino acids as mutasynthons, and subsequently report the preparation of new reblastatin derivatives by mutasynthesis. Antiproliferative properties of the new metabolites, which were available in sufficient amounts, are compared with their inhibitory effects on the ATP binding domain in human and bacterial heat shock proteins using a microarray competition assay.20
Results and discussion
Mutasynthetic experiments (preliminary screening)
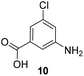
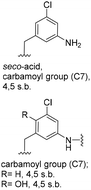
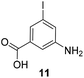
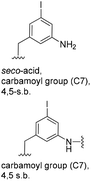
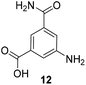
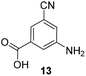
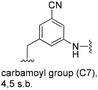
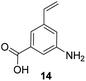
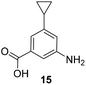
5-Substituted 3-aminobenzoic acids 6 and 7 were found to be the most promising candidates for accessing new reblastatin derivatives using mutasynthesis.18a We extended the scope of substrates to 5-substituted-3-aminobenzoic acids 10–15 (Table 1)21 that included halo-substitution (10 and 11) as well as C-substitution (12–15) Typically, the free acids can be administered as they are activated to the corresponding adenylates in Streptomyces hygroscopicus. Next, these are attacked by phosphopantheteine-thiol to form the PKS-bound thioesters. Preliminary experiments were performed on a small scale (0.038 mmol mutasynthon, 30 mL fermentation broth, 7 d). This set of mutasynthetic experiments revealed that carbamoylation always takes place, and in selected cases hydroxylation at C17 as tailoring modifications can occur. In our hands, desaturation at C4,C5 is a rarely observed transformation when feeding unnatural aminobenzoic acids and for the new examples reported here, it was not observed at all. Amide 12 was not processed as judged by UPLC-HRMS analysis but the functionally related nitrile 13 provided one macrolactam metabolite after feeding to the AHBA(−) mutant strain of S. hygroscopicus. Other C-substituents such as the vinyl group present in benzoic acid 14 or the cyclopropyl group as in 15 were also converted into the amidinated macrolactams. In several cases, we detected the presence of seco acids (for mutasynthons 10, 11 and 15) and in one case also the “seco amide” (for mutasynthon 15). The “seco amide” is the amide derivative of the corresponding seco acid, which cannot serve as substrate for macrolactamisation anymore.| Mutasynthon21 | Formula | Proposed structurea,b |
|---|---|---|
| a Analysis by UPLC-HRMS. b s.b. = single bond. | ||
|
C29H44N2O7Cl[M + H]+: calc.: 555.2837, found: 555.2834 |
|
| C28H42N2O7Cl[M + H]+, calc.: 537.2731, found: 537.2748 | ||
| C28H41N2O6ClNa[M + Na]+: calc.: 575.2500, found: 575.2505 | ||
|
C28H44N2O7I[M + H]+: calc.: 647.2193, found: 647.2184 |
|
| C28H42N2O6I[M + H]+: calc.: 629.2088, found: 629.2072 | ||
|
— | Not processed |
|
C29H42N3O6[M + H]+: calc.: 528.3074, found: 528.3070 |
|
|
C30H44N2O6Na[M + Na]+: calc.: 551.3097, found: 551.3105 |
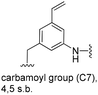
|
|
C31H49N2O7[M + H]+: calc.: 561.3540, found: 561.3527 |
|
| C31H50N3O6[M + H]+: calc.: 560.3700, found: 560.3710 | ||
| C31H46N2O6Na[M + Na]+: calc.: 565.3254, found: 565.3254 | ||
| C31H46N2O7Na[M + Na]+: calc.: 581.3203, found: 581.3208 | ||
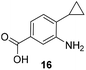
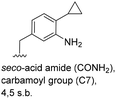
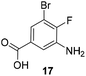
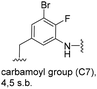
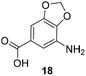
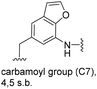
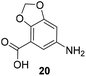
The second series of experiments utilised diversely substituted aminobenzoic acids 16–20 in which the third and in some cases a fourth substituent are located at positions 4 and 5 or 5 and 6, respectively (Table 2). With the successful biotransformation of 4-fluoro-3-aminobenzoic acid 8, precedence for the acceptance of 4-substituted mutasynthons was already at hand.18b 3-Amino-4-cyclopropylbenzoic acid 16 was processed to the carbamoylated seco acid (amide form) so that macrolactamisation did not occur, which is likely due to steric hindrance in the ortho-position of the aniline group. 4,5-Disubstituted aminobenzoic acid 18 behaved in the same manner, whereas the 4-fluoro-5-bromo derivative 17 yielded the expected amidinated macrolactam. Obviously, a small fluorine substituent at the 4 position as is also the case for benzoic acid 8 does not hamper macrolactamisation by the amide synthase (GdmF; Scheme 1).
| Mutasynthon21 | Formula | Proposed structurea,b |
|---|---|---|
| a Analysis by UPLC-HRMS. b s.b. = single bond. | ||
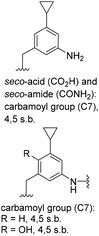
|
C31H50N3O6[M + H]+: calc.: 560.3700, found: 560.3701 |
|
|
C28H40N2O6FBrNa[M + Na]+: calc.: 621.1951, found: 621.2000 |
|
|
C29H46N3O8[M + H]+: calc.: 564.3285, found: 564.3288 |
|
|
C30H42N2O7Na[M + Na]+: calc.: 565.2890, found: 565.2884 |
|
|
— | Not processed |
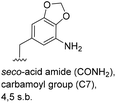
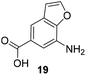
Subtleties concerning the type of substitution at C4 become evident when examining benzoic acid 19, which also gave the corresponding macrolactam ring. Compared to the 2,2-dimethyl-1,3-dioxole ring in 18, the furan moiety is planar and sterically less congested. Finally, substituents located at position 6 as in benzoic acid 20 suppress PKS processing probably due to inefficient loading onto the PKS starter module.
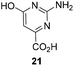
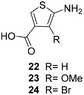
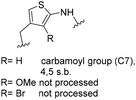
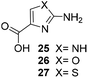
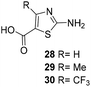
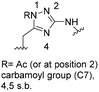
One important result of our previous studies was that, not only are 3-aminobenzoic acid derivatives processed by the PKS of S. hygroscopicus, 5-aminonicotinic acid 9 is also accepted as a PKS substrate. This unprecedented finding initiated a study in our laboratories on the scope and limitations of this new result using a series of six- and five-membered hetarenes 21–34 as mutasynthons (Table 3). The pyrimidine derivative 21, bearing the same substitution pattern as AHBA, failed to serve as a mutasynthon. When exposing a series of five membered hetarenes to the typical fermentation conditions, we found the following general trends. One sulfur atom must be part of the five-membered ring in order to yield mutaproducts, either as macrolactam derivatives or as seco acids. Consequently, thiophene 22, thiazole 28 and thiazine 33 were accepted and processed. However, in the case of the thiazole core the positions of the amino- and carboxyl substituents are crucial, because mutasynthon 27 did not provide new products as judged by HRMS. Therefore, also imidazole 25, oxazole 26 and pyrazine 32 did not serve as mutasynthons. Additional substitution as present in thiophenes 23 and 24 as well as in thiazoles 29 and 30 is also not tolerated by S. hygroscopicus. In view of these findings triazole 31 represents an unusual case, as the sulfur atom is missing in the hetarene ring but UPLC-HRMS revealed formation of a macrocyclic mutaproduct. It has to be noted that the thiophene moiety is a bioisostere for the benzene ring22 and this is also reflected in the present work as 5-aminothiophene-3-carboxylic acid 22 was processed to the corresponding macrolactam by the polyketide synthase.
| Mutasynthon21 | Formula | Proposed structurea,b |
|---|---|---|
| a Analysis by UPLC-HRMS. b s.b. = single bond. | ||
|
— | Not processed |
|
For R = H C26H40N2NaO6S [M + Na]+: calc.: 551.2505, found: 551.2510 |
|
|
— | Not processed |
|
For R = H C25H37N3NaO6S [M + Na]+: calc.: 530.2301, found: 530.2298 |
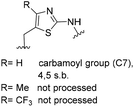
|
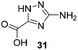
|
C26H41N5NaO7 [M + Na]+: calc.: 558.2904, found: 558.2911 |
|
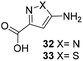
|
For X = S C25H39N3NaO6S [M + H]+: calc.: 532.2457, found: 532.2451 |
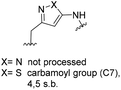
|
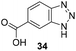
|
C28H43N4O7 [M + H]+: calc.: 547.6730, found:547.6725 |
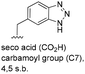
|
Finally, the benzotriazole derivative 34, which bears a meta orientation between the carboxylate and in this case a hetarene NH group, was also processed to the amidinated seco acid. We did not encounter macrolactamisation, likely due to the presence of a secondary sp2-hybridised amine instead of an aniline.
At this point, one can conclude that the PKS machinery involved in the biosynthesis of geldanamycin shows a rather broad substrate flexibility with respect to structural alterations in the aromatic starter group. Additional substituents at the 4, 5 and 6 positions as well as disubstitution of selected 3-aminobenzoic acids are tolerated as long as these substituents are not too large. In several cases, we encountered seco acid formation which included formation of carboxamides, a process that stops macrolactamisation. The open chain metabolites indicate that the amide synthase can act as a bottle neck for further processing, while substrate tolerance of the PKS up to that point must have been rather broad. Advanced intermediates, whether they are acyclic or cyclic, all serve as substrates for the tailoring amidination. Other post PKS transformations, such as oxidations of the aromatic core as well as 4,5-desaturation, are much more sensitive to structural variation in progeldanamycin precursors and in most cases, they do not occur.
Mutasynthetic experiments (preparative scale)
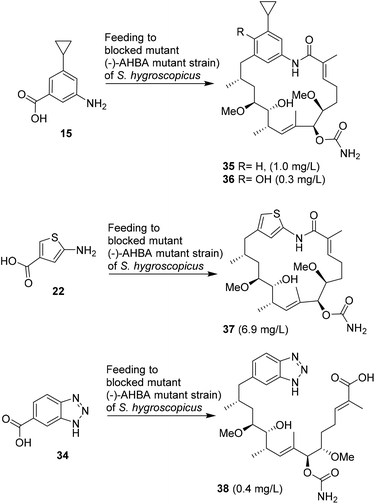
Principally, all new seco acids and ansa macrolactams that are listed in Tables 1–3 should be accessible as pure compounds, provided that the fermentation is appropriately upscaled. However, substantial HPLC purification is required to separate all metabolites and mg quantities are needed for full structural characterisation as well as biological evaluation. These requirements may therefore necessitate fermentation at a 10–50 L scale.After close inspection of all chromatograms of the crude fermentation extracts, we decided that mutasynthons 15, 22 and 34 provided sufficient amounts of new reblastatin derivatives for complete structure elucidation and biological evaluation. For this purpose, the fermentations were repeated in up to 5 L scale.
After harvesting, the fermentation broths were extracted with ethyl acetate. Depending on the complexity of the metabolic spectrum, several purification steps using silica gel chromatography, Sephadex size exclusion chromatography and RP-HPLC were performed (see ESI†). In all cases, the fermentation yields of new mutaproducts were significantly lower than that obtained when the natural starter unit, AHBA 1, was fed to yield geldanamycin 3. Supplementation of cyclopropyl-substituted aminobenzoic acid 15 to a growing culture of the (−)AHBA mutant of Streptomyces hygroscopicus provided reblastatin derivatives 35 and 36 in sufficient amounts for isolation and characterisation (Scheme 2). Besides amidination of the alcohol at position 7, oxidation at C17 also occurred as a tailoring modification. Likewise, thiophene 22 provided reblastatin derivative 37 in excellent yield in comparison to those commonly observed for such mutasynthetic experiments.18 Finally, we carried out the supplementation of AHBA(−) S. hygroscopicus with benzotriazole 34 and were able to collect the chemically labile seco acid derivative 38. This is the first example of a bicyclic mutasynthon undergoing successful loading onto the starter module and being fully processed by the PKS; although this proceeded without macrolactamisation by the amide synthase.
Biological studies
Antiproliferative activities and Hsp binding affinities
For the evaluation of its antiproliferating potential the two new derivative 35 and 37 that were available in sufficient amounts in pure form were administered to cultured human tumour cell lines (Table 4). The data were compared to geldanamycin 3 and the 19-fluoro derivative 8 one of the most potent nonhydroquinone derivatives known so far. The branched cyclopropyl substituent at position 17 led to reduced activity. Remarkably, the thiophene analogue 37 of reblastatin shows good antiproliferative activity against all human cancer cell lines tested. Still it is not as active as geldanamycin 3. This can be ascribed to the fact that thiophene and benzene are bioisosteres and the sulfur atom is located at position 18 of geldanamycin 3 and reblastatin 4, respectively.| IC50 [nM] | ||||
|---|---|---|---|---|
| KB-3-1 | PC-3 | SK-OV-3 | A-431 | |
| a KB-3-1 = Cervix carcinoma; PC-3 = Prostate carcinoma; SK-OV-3 = Ovarian carcinoma; A-431 = Epidermoid carcinoma. | ||||
| 35 | 960 | 1500 | 1300 | 980 |
| 37 | 128 | 472 | 159 | 85 |
| 3 | 53 | 18 | 125 | 18 |
| 8 | 73 | 42 | 54 | 62 |
Recently, we reported a new displacement assay for the ATP domain of heat shock proteins such as for Hsp90α.20 The inhibition is monitored using fluorescence-labelled Cy3- or Cy5-ATP in the presence of a potential inhibitor that binds to the ATP binding domain. This approach allows to distinguish between the bound and unbound inhibitor. The assay is performed by incubating different Hsp-loaded microarrays for 16 h at 4 °C with a solution containing a mixture consisting of 100 nM of the fluorescent-labelled ATP derivatives Cy3-ATP or Cy5-ATP, the binding buffer and the potential Hsp90 inhibitor (500 nM), including 17-aminoallyl geldanamycin (17-AAG) 39 (line 2), reblastatin derivatives 40–43 (lines 3–6) and thiophene reblastatin 37 (line 7). To compare different heat shock proteins from different sources, we chose human Hsp90α (Fig. 2a), Hsp90β (b), Xanthomonas campestris XcHtpG (c) and Helicobacter pylori HpHtpG (d). The results are presented as a heat map (Fig. 2). The simultaneous screening of different purified Hsps reveals that most of all synthesised reblastatin derivatives, except for cyclopropyl derivative 35, are capable of inhibiting human and HpHtpG heat shock proteins. Remarkably, the clinical candidate 17-AAG 39 only shows a minor activity for HpHtpG. The EC50 of thiophene 37 against HpHtpG and HsHtpG were 59 nM and 128 nM, respectively. This comparison reveals that the thiophene group shows varying affinities to the ATP binding sites of different Hsps, which can serve as a starting point for developing Hsp specific inhibitors.
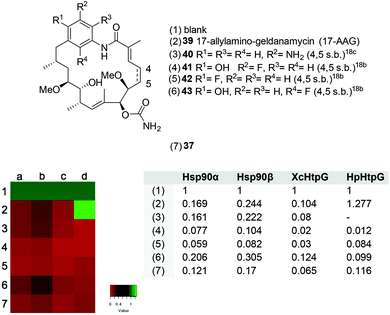 | ||
| Fig. 2 Top: Structures of geldanamycin and reblastatin derivatives screened. Bottom left: Heat map presentations of competitive Hsp activities tested for compounds 37 and 39–43 dedicated to lines 2–7 using a microarray-based assay with purified heat shock proteins in the presence of ATP. The normalised signals are ranked from 0 to 1 (line 1, blank) displaying different colours. Low signal values (red) indicate stronger competition against fluorescence dye labelled ATP and hence higher affinity for Hsps. No competition shown in position 1 provides a green colour. The concentrations of potential binders are 500 nM against (a) human Hsp90a, (b) Hsp90b, (c) Xanthomonas campestris XcHtpG and (d) Helicobacter pylori HpHtpG. These were spotted onto a microarray as described before.20a Bottom left: Corresponding normalised values derived from the optical measurements. | ||
Additionally, the measurements were conducted at different concentrations (50 μM, 5 μM, 0.5 μM, 50 nM, 5 nM, 0.5 nM, 50 pM) to estimate the IC50 value. Unbound fluorescently-labelled ATP was removed by several washing steps and then the amount of bound Cy3-labelled ATP was determined. The signal intensity was analysed and normalised for the unbound value and hence visualised. These studies18,20 reveal that the inhibitory properties found for new reblastatin derivatives 35 and 37 match those of the antiproliferative activity (Tables 4 and 5).
Except for the cyclopropyl reblastatin 35, all derivatives show pronounced displacement properties for the heat shock proteins tested. Reblastatin derivative 39 bearing the simplest possible substitution pattern at the aromatic ring demonstrates that non-quinone and even non phenolic geldanamycin derivatives are very effective in replacing ATP in human Hsp90α (39, IC50Hsp90α = 30 nM). However, in no case have we encountered a relevant selectivity difference between human Hsp90α and Hsp90β (Fig. 2). Interestingly, thiophene reblastatin 37 exhibits a stronger activity for the bacterial Hsp homologue HpHtpG than for human Hsp90α. Therefore, it can serve as a starting point for developing antibiotics that target bacterial heat shock proteins, especially because it was recently shown that HtpG is required for adaptation through environmental stress.24
Conclusions
In conclusion, we reported on the mutasynthetic generation and biological evaluation as heat shock protein inhibitors of several new reblastatin derivatives. The cyclopropane derivatives 35 and 36 are remarkable in that this is the first example of introducing such structural elements into complex natural products by a mutasynthetic approach. More striking is the high yielding uploading and PKS-processing of 5-aminothiophene-3-carboxylic acid 22 to the corresponding thiophene reblastatine derivative 37, the first mutaproduct of this kind reported so far.25Importantly, this broad study also demonstrates the scope and limitations of mutasynthesis as a synthetic tool in natural product chemistry. The structural diversity of aromatic building blocks including hetarenes that are accepted by the starting module of Gdm-PKS and fully processed up to that stage of the macrolactam is astounding. A promising new candidate is the thiophene derivative of reblastatin. It shows inhibitory activity against HtpG from Helicobacter pylori and this derivative clearly can serve as a starting point to develop new selective inhibitors against bacterial heat shock proteins. Finally, the limitation of producing sufficient quantities of new derivatives by mutasynthesis has to be kept in mind26–29 and is also in issue in the present study. This becomes particularly evident when calculating the isolated yields of new mutaproducts based on the amount of mutasynthons fed: 35 (0.15% from 15), 36 (0.04% from 15), 37 (1.1% from 22) and 38 (0.09% from 34).
Experimental procedures
Mutasynthetic experiments
Feeding experiments for small scale transformations
Benzoic acid derivatives 10–20 and hetarenes 21–34 (37.5 μmol, in 2 mL sterile solution in DMSO/H2O = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) were added in three 670 μL portions at 48, 72 and 96 h after inoculation to 30 mL of the main culture of S. hygroscopicus mutant K390-61-1. After having fed precursors according to the fermentation conditions described above, UPLC-ESI-HRMS analysis was employed for detecting new products as [M + H]+ or [M + Na]+ peaks.
1) were added in three 670 μL portions at 48, 72 and 96 h after inoculation to 30 mL of the main culture of S. hygroscopicus mutant K390-61-1. After having fed precursors according to the fermentation conditions described above, UPLC-ESI-HRMS analysis was employed for detecting new products as [M + H]+ or [M + Na]+ peaks.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) was fed in three equal portions at 48, 72 and 96 h after inoculation to sixteen 75 mL main cultures of S. hygroscopicus mutant K390-61-1. Extraction with EtOAc afforded 17-demethoxy-18-cyclopropyl-reblastatin 35 (1.3 mg, 2.3 μmol) and 17-demethyl-18-cyclopropyl-reblastatin 36 (0.3 mg, 0.5 μmol) after silica gel filtration and three HPLC purification steps.
1) was fed in three equal portions at 48, 72 and 96 h after inoculation to sixteen 75 mL main cultures of S. hygroscopicus mutant K390-61-1. Extraction with EtOAc afforded 17-demethoxy-18-cyclopropyl-reblastatin 35 (1.3 mg, 2.3 μmol) and 17-demethyl-18-cyclopropyl-reblastatin 36 (0.3 mg, 0.5 μmol) after silica gel filtration and three HPLC purification steps.
HPLC conditions employed for the first purification step were (a) column: Reprosil-Pur 120 C18 AQ, 250 × 25 mm, 5 μm, endc.; (b) guard: Reprosil-Pur 120 C18 AQ, 30 × 20 mm, 10 μm, endc.; (c) solvents: methanol/H2O and (d) program: H2O![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH = 60
MeOH = 60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40 {5 min}, 60
40 {5 min}, 60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40 → 50
40 → 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 {15 min}, H2O
50 {15 min}, H2O![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH = 50
MeOH = 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 → 30
50 → 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 70 {65 min}, H2O
70 {65 min}, H2O![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH = 30
MeOH = 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 70 → 0
70 → 0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 {15 min}, H2O
100 {15 min}, H2O![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH = 0
MeOH = 0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 {10 min}, (15.0 ml min−1) (tR = 65.0 min). For the second purification step the conditions were (a) column: Reprosil-Pur 120 C18-ISIS AQ, 250 × 8.0 mm, 5 μm, endc.; (b) guard: Reprosil-Pur 120 C18-ISIS AQ, 40 × 8 mm, 5 μm, endc.; (c) solvents: MeOH/H2O and (d) program: H2O
100 {10 min}, (15.0 ml min−1) (tR = 65.0 min). For the second purification step the conditions were (a) column: Reprosil-Pur 120 C18-ISIS AQ, 250 × 8.0 mm, 5 μm, endc.; (b) guard: Reprosil-Pur 120 C18-ISIS AQ, 40 × 8 mm, 5 μm, endc.; (c) solvents: MeOH/H2O and (d) program: H2O![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH = 80
MeOH = 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 → 65
20 → 65![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 35 {5 min}, H2O
35 {5 min}, H2O![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH = 65
MeOH = 65![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 35 → 40
35 → 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 60 {65 min}, H2O
60 {65 min}, H2O![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH = 40
MeOH = 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 60 → 0
60 → 0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 {10 min}, H2O
100 {10 min}, H2O![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH = 0
MeOH = 0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 {10 min}, (2.5 mL min−1) (tR = 63.0 min). For the third purification step the conditions were (a) column: Reprosil-Pur 120 C18-ISIS AQ, 250 × 8.0 mm, 5 μm, endc.; (b) guard: Reprosil-Pur 120 C18-ISIS AQ, 40 × 8 mm, 5 μm, endc.; (c) solvents: methanol/H2O and (d) program: H2O
100 {10 min}, (2.5 mL min−1) (tR = 63.0 min). For the third purification step the conditions were (a) column: Reprosil-Pur 120 C18-ISIS AQ, 250 × 8.0 mm, 5 μm, endc.; (b) guard: Reprosil-Pur 120 C18-ISIS AQ, 40 × 8 mm, 5 μm, endc.; (c) solvents: methanol/H2O and (d) program: H2O![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH = 80
MeOH = 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 → 65
20 → 65![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 35 {5 min}, H2O
35 {5 min}, H2O![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH = 65
MeOH = 65![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 35 → 40
35 → 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 60 {80 min}, H2O
60 {80 min}, H2O![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH = 40
MeOH = 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 60 → 0
60 → 0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 {5 min}, (3.0 ml min−1) (tR = 64.4 min).
100 {5 min}, (3.0 ml min−1) (tR = 64.4 min).
17-Demethoxy-18-cyclopropyl-reblastatin 35
1H NMR (500 MHz, T = 323 K, THF-d8, THF-d7 = 1.73 ppm): δ 8.43 (s, 1H, NH), 7.02 (s, 1H, Ar–H), 6.61 (s, 1H, Ar–H), 6.58 (s, 1H, Ar–H), 5.88–5.80 (m, 1H, 3-H), 5.69 (br. s, 2H, CONH2), 5.30 (d, J = 9.9 Hz, 1H, 9-H), 5.06 (d, J = 5.8 Hz, 1H, 7-H), 4.54 (br. s., 1H, 11-OH), 3.54–3.48 (m, 1H, 11-H), 3.35 (s, 3H, 6-OMe), 3.31 (s, 3H, 12-OMe), 3.30–3.24 (m, 1H, 6-H), 3.15–3.08 (m, 1H, 12-H), 2.65 (dd, J = 13.2, 5.0 Hz, 1H, 15-Ha), 2.47–2.40 (m, 1H, 10-H), 2.37–2.25 (m, 1H, 15-Hb), 2.37–2.25 (m, 1H, 4-Ha), 2.17–2.05 (m, 1H, 4-Hb), 1.91–1.82 (m, 1H, 14-H), 1.91–1.82 (m, 1H, CH-cyclopropyl) 1.79 (s, 3H, 2-Me), 1.60–1.52 (m, 1H, 13-Ha), 1.48 (s, 3H, 8-Me), 1.40–1.32 (m, 2H, 5-H), 1.27–1.20 (m, 1H, 13-Hb), 1.00 (d, J = 6.6 Hz, 3H, 10-Me), 0.90–0.86 (m, 2H, CH2-cyclopropyl), 0.82 (d, J = 6.8 Hz, 3H, 14-Me), 0.66–0.61 (m, 2H, CH2-cyclopropyl) ppm; 13C NMR (125 MHz, T = 323 K, THF-d8, THF-d8 = 25.5 ppm): δ 171.2 (s, C-1), 157.3 (s, CONH2), 145.4 (s, C-Ar), 141.8 (s, C-Ar), 141.3 (s, C-Ar), 134.7 (d, C-3), 133.0 (s, C-2), 133.0 (s, C-9), 131.7 (d, C-8), 124.3 (d, C-Ar), 123.5 (d, C-Ar), 116.3 (d, C-Ar), 82.7 (d, C-12), 81.6 (d, C-6), 81.3 (d, C-7), 75.1 (d, C-11), 58.9 (q, 6-OMe), 57.0 (q, 12-OMe), 43.9 (t, C-15), 35.5 (d, C-10), 34.6 (t, C-13), 32.8 (d, C-14), 31.0 (t, C-5), 25.5 (t, C-4), 19.8 (q, 14-Me), 17.3 (d, CH-cyclopropyl), 16.2 (q, 10-Me), 13.6 (q, 2-Me), 13.2 (q, 8-Me), 9.7 (t, CH2-cyclopropyl), 9.6 (t, CH2-cyclopropyl) ppm; HRMS (ESI) m/z for C31H46N2O6Na [M + Na]+: calculated: 565.3254 found: 565.3254.17-Demethyl-18-cyclopropyl-reblastatin 36
HPLC conditions employed for the first purification step were (a) column: Reprosil-Pur 120 C18 AQ, 250 × 25 mm, 5 μm, endc.; (b) guard: Reprosil-Pur 120 C18 AQ, 30 × 20 mm, 10 μm, endc.; (c) solvents: methanol/H2O and (d) program: H2O![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH = 60
MeOH = 60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40 {5 min}, 60
40 {5 min}, 60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40 → 50
40 → 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 {15 min}, H2O
50 {15 min}, H2O![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH = 50
MeOH = 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 → 30
50 → 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 70 {65 min}, H2O
70 {65 min}, H2O![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH = 30
MeOH = 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 70 → 0
70 → 0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 {15 min}, H2O
100 {15 min}, H2O![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH = 0
MeOH = 0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 {10 min}, (15.0 ml min−1) (tR = 65.0 min). For the second purification step the conditions were (a) column: Reprosil-Pur 120 C18-ISIS AQ, 250 × 8.0 mm, 5 μm, endc.; (b) guard: Reprosil-Pur 120 C18-ISIS AQ, 40 × 8 mm, 5 μm, endc.; (c) solvents: MeOH/H2O and (d) program: H2O
100 {10 min}, (15.0 ml min−1) (tR = 65.0 min). For the second purification step the conditions were (a) column: Reprosil-Pur 120 C18-ISIS AQ, 250 × 8.0 mm, 5 μm, endc.; (b) guard: Reprosil-Pur 120 C18-ISIS AQ, 40 × 8 mm, 5 μm, endc.; (c) solvents: MeOH/H2O and (d) program: H2O![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH = 80
MeOH = 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 → 65
20 → 65![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 35 {5 min}, H2O
35 {5 min}, H2O![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH = 65
MeOH = 65![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 35 → 40
35 → 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 60 {65 min}, H2O
60 {65 min}, H2O![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH = 40
MeOH = 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 60 → 0
60 → 0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 {10 min}, H2O
100 {10 min}, H2O![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH = 0
MeOH = 0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 {10 min}, (2.5 mL min−1) (tR = 61.0 min). For the third purification step the conditions were (a) column: Reprosil-Pur 120 C18-ISIS AQ, 250 × 8.0 mm, 5 μm, endc.; (b) guard: Reprosil-Pur 120 C18-ISIS AQ, 40 × 8 mm, 5 μm, endc.; (c) solvents: MeOH/H2O and (d) program: H2O
100 {10 min}, (2.5 mL min−1) (tR = 61.0 min). For the third purification step the conditions were (a) column: Reprosil-Pur 120 C18-ISIS AQ, 250 × 8.0 mm, 5 μm, endc.; (b) guard: Reprosil-Pur 120 C18-ISIS AQ, 40 × 8 mm, 5 μm, endc.; (c) solvents: MeOH/H2O and (d) program: H2O![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH = 80
MeOH = 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 → 70
20 → 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30 {5 min}, H2O
30 {5 min}, H2O![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH = 70
MeOH = 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30 → 40
30 → 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 60 {80 min}, H2O
60 {80 min}, H2O![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH = 40
MeOH = 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 60 → 0
60 → 0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 {5 min}, (3.0 ml min−1) (tR = 60.2 min).
100 {5 min}, (3.0 ml min−1) (tR = 60.2 min).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) was fed in three equal portions at 48, 72 and 96 h after inoculation to sixteen 75 mL main cultures of S. hygroscopicus mutant K390-61-1. Extraction with EtOAc afforded thiageldanamycin-derivative 37 (8.3 mg, 16.3 μmol) after silica gel filtration and three HPLC purification steps. HPLC conditions employed for the first purification step were (a) column: Reprosil-Pur 120 C18 AQ, 250 × 25 mm, 5 μm, endc.; (b) guard: Reprosil-Pur 120 C18 AQ, 30 × 20 mm, 10 μm, endc.; (c) solvents: methanol/H2O and (d) program: H2O
1) was fed in three equal portions at 48, 72 and 96 h after inoculation to sixteen 75 mL main cultures of S. hygroscopicus mutant K390-61-1. Extraction with EtOAc afforded thiageldanamycin-derivative 37 (8.3 mg, 16.3 μmol) after silica gel filtration and three HPLC purification steps. HPLC conditions employed for the first purification step were (a) column: Reprosil-Pur 120 C18 AQ, 250 × 25 mm, 5 μm, endc.; (b) guard: Reprosil-Pur 120 C18 AQ, 30 × 20 mm, 10 μm, endc.; (c) solvents: methanol/H2O and (d) program: H2O![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH = 60
MeOH = 60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40 {5 min}, 60
40 {5 min}, 60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40 → 50
40 → 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 {15 min}, H2O
50 {15 min}, H2O![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH = 50
MeOH = 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 → 30
50 → 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 70 {65 min}, H2O
70 {65 min}, H2O![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH = 30
MeOH = 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 70 → 0
70 → 0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 {15 min}, H2O
100 {15 min}, H2O![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH = 0
MeOH = 0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 {10 min}, (15.0 ml min−1) (tR = 55.0 min). For the second purification step the conditions were (a) column: Reprosil-Pur 120 C18-ISIS AQ, 250 × 8.0 mm, 5 μm, endc.; (b) guard: Reprosil-Pur 120 C18-ISIS AQ, 40 × 8 mm, 5 μm, endc.; (c) solvents: MeOH/H2O and (d) program: H2O
100 {10 min}, (15.0 ml min−1) (tR = 55.0 min). For the second purification step the conditions were (a) column: Reprosil-Pur 120 C18-ISIS AQ, 250 × 8.0 mm, 5 μm, endc.; (b) guard: Reprosil-Pur 120 C18-ISIS AQ, 40 × 8 mm, 5 μm, endc.; (c) solvents: MeOH/H2O and (d) program: H2O![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH = 80
MeOH = 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 → 65
20 → 65![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 35 {5 min}, H2O
35 {5 min}, H2O![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH = 65
MeOH = 65![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 35 → 40
35 → 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 60 {65 min}, H2O
60 {65 min}, H2O![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH = 40
MeOH = 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 60 → 0
60 → 0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 {10 min}, H2O
100 {10 min}, H2O![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH = 0
MeOH = 0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 {10 min}, (2.5 mL min−1) (tR = 47.0 min). For the third purification step the conditions were (a) column: Reprosil-Pur 120 CN AQ, 250 × 8.0 mm, 5 μm, endc.; (b) guard: Reprosil-Pur 120 CN AQ, 40 × 8 mm, 5 μm, endc.; (c) solvents: MeCN/H2O and (d) program: H2O
100 {10 min}, (2.5 mL min−1) (tR = 47.0 min). For the third purification step the conditions were (a) column: Reprosil-Pur 120 CN AQ, 250 × 8.0 mm, 5 μm, endc.; (b) guard: Reprosil-Pur 120 CN AQ, 40 × 8 mm, 5 μm, endc.; (c) solvents: MeCN/H2O and (d) program: H2O![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeCN = 90
MeCN = 90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 → 85
10 → 85![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15 {20 min}, H2O
15 {20 min}, H2O![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeCN = 85
MeCN = 85![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15 {40 min}, H2O
15 {40 min}, H2O![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeCN = 85
MeCN = 85![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15 → 75
15 → 75![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 25 {20 min}, H2O
25 {20 min}, H2O![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeCN = 75
MeCN = 75![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 25 → 55
25 → 55![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 45 {20 min}, H2O
45 {20 min}, H2O![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeCN = 55
MeCN = 55![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 45 → 0
45 → 0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 {15 min}, (4.0 ml min−1) (tR = 40.0 min).
100 {15 min}, (4.0 ml min−1) (tR = 40.0 min).
Thiophene-reblastatin 37
1H-NMR (500 MHz, T = 323 K, THF-d8, THF-d7 = 1.73 ppm): δ 8.61 (s, 1H, NH), 6.57 (s, 1H, Ar), 6.38 (s, 1H, Ar), 5.72 (br. s., 1H, 3-H), 5.57 (br. s, 2H, CONH2), 5.22 (d, J = 10.0 Hz, 1H, 9-H), 4.99 (d, J = 7.0 Hz, 1H, 7-H), 3.56–3.51 (m, 1H, 11-H), 3.37 (s, 3H, 6-OMe), 3.31 (s, 3H, 12-OMe), 3.28–3.19 (m, 1H, 6-H), 3.11–3.05 (m, 1H, 12-H), 2.65 (dd, J = 13.6, 5.4 Hz, 1H, 15-Ha), 2.45–2.35 (m, 1H, 10-H), 2.45–2.35 (m, 1H, 15-Hb), 2.22–2.05 (m, 2H, 4-H), 2.05–1.91 (m, 1H, 14-H), 1.75 (s, 3H, 2-Me), 1.66–1.58 (m, 1H, 13-Ha), 1.55 (s, 3H, 8-Me), 1.24–1.14 (m, 2H, 5-H), 1.20–1.11 (m, 1H, 13-Hb), 1.00 (d, J = 6.6 Hz, 3H, 10-Me), 0.75 (d, J = 6.8 Hz, 3H, 14-Me) ppm; 13C-NMR (125 MHz, T = 323 K, THF-d8, THF-d8 = 25.5 ppm): δ 172.3 (s, C-1), 156.9 (s, CONH2), 143.2 (s, C-Ar), 139.6 (s, C-Ar), 135.3 (d, C-3), 134.0 (d, C-9), 131.8 (s, C-2), 131.7 (s, C-8), 124.4 (d, C-Ar), 117.3 (d, C-Ar), 82.1 (d, C-12), 81.8 (d, C-7), 81.4 (d, C-6), 74.2 (d, C-11), 59.1 (q, 6-OMe), 57.0 (q, 12-OMe), 39.0 (t, C-15), 35.7 (d, C-10), 33.3 (t, C-13), 31.4 (t, C-5), 30.5 (d, C-14), 24.5 (t, C-4), 18.7 (q, 14-Me), 17.9 (q, 10-Me), 14.1 (q, 2-Me), 12.7 (q, 8-Me) ppm; HRMS (ESI) m/z für C26H41N2O6S [M + H]+: calculated: 509.2685 found: 509.2666.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 was fed in three equal portions at 48, 72 and 96 h after inoculation to sixteen 75 mL main cultures of S. hygroscopicus mutant K390-61-1. Extraction with EtOAc afforded seco-progeldanamycin acid derivative 38 (0.5 mg, 0.9 μmol) after silica gel filtration and three HPLC purification steps. HPLC conditions employed for the first purification step were (a) column: Reprosil-Pur 120 C18-ISIS AQ, 250 × 25 mm, 5 μm, endc.; (b) guard: Reprosil-Pur 120 C18-ISIS AQ, 30 × 20 mm, 10 μm, endc.; (c) solvents: MeOH/H2O and (d) program: H2O
1 was fed in three equal portions at 48, 72 and 96 h after inoculation to sixteen 75 mL main cultures of S. hygroscopicus mutant K390-61-1. Extraction with EtOAc afforded seco-progeldanamycin acid derivative 38 (0.5 mg, 0.9 μmol) after silica gel filtration and three HPLC purification steps. HPLC conditions employed for the first purification step were (a) column: Reprosil-Pur 120 C18-ISIS AQ, 250 × 25 mm, 5 μm, endc.; (b) guard: Reprosil-Pur 120 C18-ISIS AQ, 30 × 20 mm, 10 μm, endc.; (c) solvents: MeOH/H2O and (d) program: H2O![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH = 60
MeOH = 60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40 {5 min}, 60
40 {5 min}, 60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40 → 50
40 → 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 {15 min}, H2O
50 {15 min}, H2O![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH = 50
MeOH = 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 → 30
50 → 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 70 {65 min}, H2O
70 {65 min}, H2O![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH = 30
MeOH = 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 70 → 0
70 → 0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 {15 min}, H2O
100 {15 min}, H2O![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH = 0
MeOH = 0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 {10 min}, (15.0 ml min−1) (tR = 55.0 min). For the second purification step the conditions were (a) column: Reprosil-Pur 120 C18-ISIS AQ, 250 × 8.0 mm, 5 μm, endc.; (b) guard: Reprosil-Pur 120 C18-ISIS AQ, 40 × 8 mm, 5 μm, endc.; (c) solvents: methanol/H2O and (d) program: H2O
100 {10 min}, (15.0 ml min−1) (tR = 55.0 min). For the second purification step the conditions were (a) column: Reprosil-Pur 120 C18-ISIS AQ, 250 × 8.0 mm, 5 μm, endc.; (b) guard: Reprosil-Pur 120 C18-ISIS AQ, 40 × 8 mm, 5 μm, endc.; (c) solvents: methanol/H2O and (d) program: H2O![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH = 80
MeOH = 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 → 65
20 → 65![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 35 {5 min}, H2O
35 {5 min}, H2O![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH = 65
MeOH = 65![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 35 → 40
35 → 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 60 {65 min}, H2O
60 {65 min}, H2O![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH = 40
MeOH = 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 60 → 0
60 → 0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 {10 min}, H2O
100 {10 min}, H2O![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH = 0
MeOH = 0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 {10 min}, (2.5 mL min−1) (tR = 52.0 min). For the third purification step the conditions were (a) column: Reprosil-Pur 120 CN AQ, 250 × 8.0 mm, 5 μm, endc.; (b) guard: Reprosil-Pur 120 CN AQ, 40 × 8 mm, 5 μm, endc.; (c) solvents: MeCN/H2O and (d) program: H2O
100 {10 min}, (2.5 mL min−1) (tR = 52.0 min). For the third purification step the conditions were (a) column: Reprosil-Pur 120 CN AQ, 250 × 8.0 mm, 5 μm, endc.; (b) guard: Reprosil-Pur 120 CN AQ, 40 × 8 mm, 5 μm, endc.; (c) solvents: MeCN/H2O and (d) program: H2O![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeCN = 90
MeCN = 90![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 → 85
10 → 85![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15 {20 min}, H2O
15 {20 min}, H2O![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeCN = 85
MeCN = 85![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15 {40 min}, H2O
15 {40 min}, H2O![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeCN = 85
MeCN = 85![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15 → 75
15 → 75![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 25 {20 min}, H2O
25 {20 min}, H2O![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeCN = 75
MeCN = 75![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 25 → 55
25 → 55![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 45 {20 min}, H2O
45 {20 min}, H2O![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeCN = 55
MeCN = 55![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 45 → 0
45 → 0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 {15 min}, (4.0 ml min−1) (tR = 53.5 min).
100 {15 min}, (4.0 ml min−1) (tR = 53.5 min).
1H-NMR (500 MHz, T = 323 K, THF-d8, THF-d7 = 1.73 ppm): δ 7.72 (d, J = 7.0 Hz, 1H, Ph), 7.56 (s, 1H, Ph), 7.20 (d, J = 8.0 Hz, 1H, Ph), 6.77–6.69 (m, 1H, 3-H), 5.53 (br. s, 2H, CONH2), 5.28 (d, J = 9.4 Hz, 1H, 9-H), 4.90 (dd, J = 8.6, 2.9 Hz, 1H, 7-H), 3.86 (d, J = 6.3 Hz, 1H, 11-H), 3.42 (s, 3H, 6-OMe), 3.30 (s, 3H, 12-OMe), 3.35–3.25 (m, 1H, 6-H), 3.25–3.12 (m, 1H, 12-H), 2.84 (dd, J = 13.5, 5.7 Hz, 1H, 15-Ha), 2.79–2.69 (m, 1H, 10-H), 2.61–2.50 (m, 1H, 15-Hb), 2.28–2.17 (m, 2H, 4-H), 2.14–1.98 (m, 1H, 14-H), 1.80 (s, 3H, 2-Me), 1.75–1.02 (m, 1H, 5-Ha), 1.70–1.62 (m, 1H, 13-Ha), 1.60 (s, 3H, 8-Me), 1.50–1.36 (m, 1H, 13-Hb), 1.17–1.07 (m, 1H, 5-Hb), 0.97 (d, J = 6.6 Hz, 3H, 10-Me), 0.83 (d, J = 6.6 Hz, 3H, 14-Me) ppm; HRMS (ESI) m/z for C28H43N4O7 [M + H]+: calculated: 547.3132 found: 547.3137.
Preparation of Hsps and competitive microarray assay
Recombinant Hsps were produced as previously reported.18,20 Hsp microarrays were produced and the binding with fluorescently labelled ATP and displacement with inhibitors was performed as described before.18,20Experimental details on assay (Fig. 2)
The assay was performed by incubating the Hsp-loaded microarrays for 16 h at 4 °C with a solution containing a mixture consisting of 100 nM Cy3-ATP or Cy5-ATP, the binding buffer and the potential Hsp90 inhibitor at 500 nM for comparative screen on different Hsps or at different concentrations (50 μM, 5 μM, 0.5 μM, 50 nM, 5 nM, 0.5 nM, 50 pM) for estimation of the IC50 value. Unbound fluorescently-labelled ATP was removed by several washing steps and then the amount of bound labelled ATP was determined. The signal intensity was analysed and normalised for the unbound value.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the Deutsche Forschungsgemeinschaft. For funding (Ki397/13-1). We thank Wera Collisi (HZI) for performing the cell proliferation assays. We are indepted to M. Norris for helpful discussions.References
- (a) K. L. Rinehart Jr., Pure Appl. Chem., 1977, 49, 1361–1384 Search PubMed; (b) K. L. Rinehart Jr., Jpn. J. Antibiot., 1979, 32(Suppl), S32–S46 Search PubMed.
- The first time, this concept was mentioned: (a) A. J. Birch, Pure Appl. Chem., 1963, 7, 527–537 CAS in a later paper this concept was specified: (b) W. T. Shier, K. L. Rinehart Jr. and D. Gottlieb, Proc. Natl. Acad. Sci. U. S. A., 1969, 63, 198–204 CrossRef CAS PubMed.
- Reviews: (a) S. Weist and R. D. Süssmuth, Appl. Microbiol. Biotechnol., 2005, 68, 141–150 CrossRef CAS; (b) A. Kirschning, F. Taft and T. Knobloch, Org. Biomol. Chem., 2007, 3245–3295 RSC; (c) A. Kirschning and F. Hahn, Angew. Chem., 2012, 124, 4086–4096 ( Angew. Chem. Int. Ed. , 2012 , 51 , 4012–4022 ) CrossRef.
- (a) M. A. Gregory, H. Petkovic, R. E. Lill, S. J. Moss, B. Wilkinson, S. Gaisser, P. Leadlay and R. M. Sheridan, Angew. Chem., 2005, 117, 4835–4838 ( Angew. Chem.Int. Ed. , 2005 , 44 , 4757–4760 ) CrossRef; (b) M. Ziehl, J. He, H.-M. Dahse and C. Hertweck, Angew. Chem., 2005, 117, 1226–1230 ( Angew. Chem.Int. Ed. , 2005 , 44 , 1202–1205 ) CrossRef.
- (a) I. Bułyszko, G. Dräger, A. Klenge and A. Kirschning, Chem. – Eur. J., 2015, 21, 19231–19242 CrossRef PubMed; (b) F. Taft, M. Brünjes, H. G. Floss, N. Czempinski, S. Grond, F. Sasse and A. Kirschning, ChemBioChem, 2008, 7, 1057–1060 CrossRef PubMed; (c) T. Kubota, M. Brünjes, T. Frenzel, J. Xu, A. Kirschning and H. G. Floss, ChemBioChem, 2006, 5, 1221–1225 CrossRef PubMed; (d) A. Meyer, M. Brünjes, F. Taft, T. Frenzel, F. Sasse and A. Kirschning, Org. Lett., 2007, 9, 1489–1492 CrossRef CAS PubMed; (e) K. Harmrolfs, M. Brünjes, G. Dräger, H. G. Floss, F. Sasse, F. Taft and A. Kirschning, ChemBioChem, 2010, 11, 2517–2520 CrossRef CAS.
- (a) J. M. Cassady, K. K. Chan, H. G. Floss and E. Leistner, Chem. Pharm. Bull., 2004, 52, 1–26 CrossRef CAS; (b) H. G. Floss and T. W. Yu, Chem. Rev., 2005, 105, 621–632 CrossRef CAS; (c) A. Kirschning, K. Harmrolfs and T. Knobloch, C. R. Chim., 2008, 11, 1523–1543 CrossRef CAS; (d) J. Franke, S. Eichner, C. Zeilinger and A. Kirschning, Nat. Prod. Rep., 2013, 30, 1299–1323 RSC; R. R. A. Kitson and C. J. Moody, J. Org. Chem., 2013, 78, 5117–5141 Search PubMed.
- W. Guo, P. Reigan, D. Siegel, J. Zirrolli, D. Gustafson and D. Ross, Cancer Res., 2005, 65, 10006–10015 CrossRef CAS PubMed.
- (a) A. Rascher, Z. Hu, N. Viswanathan, A. Schirmer, R. Reid, W. C. Nierman, M. Lewis and C. R. Hutchinson, FEMS Microbiol. Lett., 2003, 218, 223–230 CrossRef CAS PubMed; (b) Y.-S. Hong, D. Lee, W. Kim, J. K. Jeong, C. G. Kim, J. K. Sohng, J. H. Lee, S. G. Paik and J. J. Lee, J. Am. Chem. Soc., 2004, 126, 11142–11143 CrossRef CAS PubMed; (c) D. Lee, K. Lee, X. F. Cai, N. T. Dat, S. K. Boovanahalli, M. Lee, J. C. Shin, W. Kim, J. K. Jeong, J. S. Lee, C. H. Lee, J. H. Lee, Y.-S. Hong and J. J. Lee, ChemBioChem, 2006, 7, 246–248 CrossRef CAS PubMed.
- S. Eichner, T. Eichner, H. G. Floss, J. Fohrer, E. Hofer, F. Sasse, C. Zeilinger and A. Kirschning, J. Am. Chem. Soc., 2012, 134, 1673–1679 CrossRef CAS PubMed.
- W. Guo, P. Reigan, D. Siegel, J. Zirrolli, D. Gustafson and D. Ross, Mol. Pharmacol., 2006, 70, 1194–1203 CrossRef CAS PubMed.
- L. R. Kelland, S. Y. Sharp, P. M. Rogers, T. G. Myers and P. Workman, J. Natl. Cancer Inst., 1999, 91, 1940–1949 CrossRef CAS.
- A. C. Maroney, J. J. Marugan, T. M. Mezzasalma, A. N. Barnakov, T. A. Garrabrant, L. E. Weaner, W. J. Jones, L. A. Barnakova, H. K. Koblish, M. J. Todd, J. A. Masucci, I. C. Deckman, R. A. Galemmo Jr. and D. L. Johnson, Biochemistry, 2006, 45, 5678–5685 CrossRef CAS.
- H. G. Menzella, T.-T. Tran, J. R. Carney, J. Lau-Wee, J. Galazzo, C. D. Reeves, C. Carreras, S. Mukadam, S. Eng, Z. Zhong, P. B. M. W. M. Timmermans, S. Murli and G. W. Ashley, J. Med. Chem., 2009, 52, 1518–1521 CrossRef CAS PubMed.
- A. Rascher, Z. Hu, G. O. Buchanan, R. Reid and C. R. Hutchinson, Appl. Environ. Microbiol., 2005, 71, 4862–4871 CrossRef CAS PubMed.
- M. Q. Zhang, S. Gaisser, M. Nur-E-Alam, L. S. Sheehan, W. A. Vousden, N. Gaitatzis, G. Peck, N. J. Coates, S. J. Moss, M. Radzom, T. A. Foster, R. M. Sheridan, M. A. Gregory, S. M. Roe, C. Prodromou, L. Pearl, S. M. Boyd, B. Wilkinson and C. J. Martin, J. Med. Chem., 2008, 51, 5494–4947 CrossRef CAS PubMed.
- W. Kim, D. Lee, S. S. Hong, Z. Na, J. C. Shin, S. H. Roh, C. Z. Wu, O. Choi, K. Lee, Y. M. Shen, S. G. Paik, J. J. Lee and Y. S. Hong, ChemBioChem, 2009, 10, 1243–1251 CrossRef CAS PubMed.
- W. Kim, J. S. Lee, D. Lee, X. F. Cai, J. C. Shin, K. Lee, C.-H. Lee, S. Ryu, S.-G. Paik, J. J. Lee and Y.-S. Hong, ChemBioChem, 2007, 8, 1491–1494 CrossRef CAS.
- (a) S. Eichner, H. G. Floss, F. Sasse and A. Kirschning, ChemBioChem, 2009, 10, 1801–1805 CrossRef CAS PubMed; (b) J. Hermane, I. Bułyszko, S. Eichner, F. Sasse, W. Collisi, A. Poso, E. Schax, J.-G. Walter, T. Scheper, K. Kock, C. Herrmann, P. Aliuos, G. Reuter, C. Zeilinger and A. Kirschning, ChemBioChem, 2015, 16, 302–311 CrossRef CAS PubMed; (c) S. Mohammadi-Ostad-Kalayeh, F. Stahl, T. Scheper, K. Kock, C. Herrmann, F. A. H. Batista, J. C. Borges, F. Sasse, S. Eichner, J. Hermane, C. Zeilinger and A. Kirschning, ChemBioChem, 2018, 19, 562–574 CrossRef CAS.
- If not otherwise specified, numbering of atoms refers to the numbering of the natural product geldanamycin throughout the text.
- (a) E. Schax, J. Neunaber, F. Stahl, J.-G. Walter, T. Scheper, S. Eichner, A. Kirschning and C. Zeilinger, Biodiscovery, 2014, 14, 1–6 Search PubMed; (b) E. Schax, J.-G. Walter, H. Märzhäuser, F. Stahl, T. Scheper, D. A. Agard, S. Eichner, A. Kirschning and C. Zeilinger, J. Biotechnol., 2014, 180, 1–9 CrossRef CAS; (c) S. Mohammadi-Ostad-Kalayeh, V. Hrupins, C. Ahlbrecht, F. Stahl, T. Scheper, M. Preller, F. Surup, M. Stadler, A. Kirschning and C. Zeilinger, Bioorg. Med. Chem., 2017, 25, 6345–6352 CrossRef CAS; (d) Q. Yue, F. Stahl, O. Plettenburg, A. Kirschning, A. Warnecke and C. Zeilinger, Biochemistry, 2018, 57, 2601–2605 CrossRef CAS PubMed.
- The preparation of new aminobenzoic acids is described in: (a) S. Eichner, Ph.D. thesis, Leibniz Universität Hannover, 2011 Search PubMed; (b) J. Hermane, Ph.D. thesis, Leibniz Universität Hannover, 2013 Search PubMed; (c) L. Mancuso, Ph.D. thesis, Leibniz Universität Hannover, 2014 Search PubMed; (d) B. Schröder, Ph.D. thesis, Leibniz Universität Hannover, 2017 Search PubMed.
- G. A. Patani and E. J. LaVoie, Chem. Rev., 1996, 96, 3147–3176 CrossRef CAS PubMed.
- The Z-factor reported by Zhang et al. is a dimensionless parameter with values between −∞ < Z < 1, that depends on positive and negative controls. The quality of an assay is ideal, if the Z-factor has the value of 1. When the value is between 0.5 and 1 the assay is very reliable, while a value between 0 and 0.5 represents a poor assay that is difficult to reproduce: J. H. Zhang, T. D. Y. Chung and K. R. Oldenburg, J. Biomol. Screening, 1999, 4, 67–73 CrossRef.
- F. A. Honoré, V. Méjean and O. Genest, Cell Rep., 2017, 19, 680–687 CrossRef.
- Recent examples on aromatic mutasynthons employed: (a) Y. Yan, J. Chen, L. Zhang, Q. Zheng, Y. Han, H. Zhang, D. Zhang, T. Awakawa, I. Abe and W. Liu, Angew. Chem., 2013, 125, 12534–12538 ( Angew. Chem. Int. Ed. , 2013 , 52 , 12308–12312 ) CrossRef.
- J. A. Kalaitzis, M. Izumikawa, L. Xiang, C. Hertweck and B. S. Moore, J. Am. Chem. Soc., 2003, 125, 9290–9291 CrossRef CAS PubMed.
- W. C. Widdison, S. D. Wilhelm, E. E. Cavanagh, K. R. Whiteman, B. A. Leece, Y. Kovtun, V. S. Goldmacher, H. Xie, R. M. Steeves, R. J. Lutz, R. Zhao, L. Wang, W. A. Blättler and R. V. J. Char, J. Med. Chem., 2006, 4392–4408 CrossRef CAS.
- T. Knobloch, H. G. Floss, K. Harmrolfs, F. Sasse, F. Taft, B. Thomaszewski and A. Kirschning, ChemBioChem, 2011, 12, 540–547 CrossRef CAS.
- S. Eichner, T. Knobloch, H. G. Floss, J. Fohrer, K. Harmrolfs, J. Hermane, A. Schulz, F. Sasse, P. Spiteller, F. Taft and A. Kirschning, Angew. Chem., 2012, 124, 776–781 ( Angew. Chem. Int. Ed. , 2012 , 51 , 752–757 ) CrossRef.
Footnotes |
| † Electronic supplementary information (ESI) available: Syntheses of new amino acid precursors and analytical as well as spectroscopic data including copies of NMR spectra of new reblastatin derivatives. See DOI: 10.1039/c9ob00892f |
| ‡ These authors contributed equally to the work. |
| This journal is © The Royal Society of Chemistry 2019 |