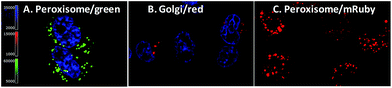Open Access Article
Open Access ArticleDevelopment of ergosterol peroxide probes for cellular localisation studies†
Taotao
Ling‡
a,
Walter H.
Lang‡
a,
Michelle M.
Martinez-Montemayor
 *b and
Fatima
Rivas
*b and
Fatima
Rivas
 *a
*a
aDepartment of Chemical Biology and Therapeutics, St Jude Children's Research Hospital, Memphis, Tennessee 38105-3678, USA. E-mail: Fatima.Rivas@stjude.org
bDepartment of Biochemistry, Universidad Central del Caribe-School of Medicine., P.O. Box 60327, Bayamón, Puerto Rico, 00960-6032, USA. E-mail: michelle.martinez@uccaribe.edu
First published on 26th April 2019
Abstract
Ergosterol peroxide selectively exhibits biological activity against a wide range of diseases; however, its mode of action remains unknown. Here, we present an efficient synthesis of ergosterol peroxide chemical probes for in vitro anticancer evaluation, live cell studies and proteomic profiling. Ergosterol peroxide analogues show promising anti-proliferation activity against triple negative breast cancer cellular models, revealing information on the structure–activity relationship of this natural product in order to develop superior analogues. The combined cellular studies demonstrate that ergosterol peroxide is distributed across the cytosol with significant accumulation in the endoplasmic reticulum (ER). These chemical probes support our efforts towards uncovering the potential target(s) of ergosterol peroxide against triple negative breast cancer cell lines.
Introduction
The ergosterol peroxide natural product (2, Fig. 1) is a compound isolated from plants,1 algae,2 lichens,3 anemonae,4 corals,5 and mushrooms6 such as Ganoderma lucidum among others.7 Ergosterol peroxide has been reported as an anti-tumour agent,8 and shows proapoptotic9 and anti-inflammatory/immunosuppressive effects,10 and anti-mycobacterial and anti-proliferative activities against cancer cell lines.6,11–13 Ergosterol peroxide triggers apoptosis and modulates the cell cycle of cancer cell models in a dose-dependent manner.8,13 We have shown that ergosterol peroxide shows anti-proliferative effects through G1 phase cell cycle arrest, apoptosis induction via caspase 3/7 activation, and PARP cleavage. It decreases migratory and invasive effects of triple negative cancer cells while inhibiting the expression of total AKT1, AKT2, BCL-XL, Cyclin D1 and c-Myc in the tested breast cancer cells.11We and others have studied the biological activity of the extract of Ganoderma lucidum and shown that this extract induces tumour reduction in in vivo breast cancer models.11a Ergosterol peroxide belongs to the steroidal family of natural products, sharing a cholesterol core and an unexpected endoperoxide bridge as the reactive center of the molecule (Fig. 1). Derivatives lacking a peroxidic bond have no significant activity (EC50 > 50 μM). Although the apoptosis induction presumably arises from this motif and several studies have reported the affected potential pathways, the underlying molecular mechanism of ergosterol peroxide in cancer (in vitro and in vivo murine cancer models) is not fully understood.14,15
We are particularly interested in studying the properties of ergosterol peroxide against triple-negative breast cancer cellular models. Triple-negative breast cancer is diagnosed when breast cancer cells lack the three common types of protein receptors: oestrogen (ER), progesterone (PR), and human epidermal growth factor receptor 2 (HER-2). Triple-negative breast cancer is associated with the advanced disease stage, higher-grade tumours at diagnosis, an increased recurrence risk, and a poor five-year survival rate (relative to other breast cancer subtypes).16
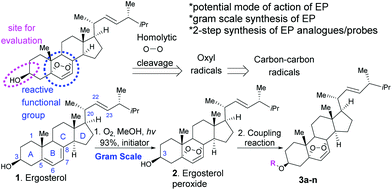
Most cancer-related deaths are a result of metastasis, and metastatic tumour cells are genetically unstable, with no single dominant pathway likely to control metastasis in most cancers. Thus, there is growing interest in developing new pharmacological treatments from natural products, which might induce their effects via primary and secondary molecules that cross-talk in cancer-dependent signaling pathways rather than affecting a single biological target. There is a unanimous agreement that ergosterol peroxide induces cell death in cancer cell models, but the specific biological target(s) remain elusive. Several reports indicate that ergosterol peroxide might influence various pathways across cancer subtypes.17 Wu and our group have shown that ergosterol peroxide induces reactive oxygen species (ROS), which are radicals with a sole unpaired electron in the outermost shell of electrons.11,17 Accordingly, the expression of ROS-detoxifying antioxidant proteins is altered in cancer cells. However, ROS show anticancer properties by decreasing cell proliferation, damaging DNA, and inducing apoptosis, among other mechanisms.18 Endoperoxides are known for generating radicals. It is hypothesized that oxyl radicals are generated by the homolytic cleavage of the peroxide, which ultimately lead to a stable carbon–carbon radical and presumably bind to specific proteins (Fig. 1). Although such a reactivity is expected to be promoted by iron(II),19 we did not detect changes in cytotoxicity by combined treatments of iron chelators and ergosterol peroxide (data not shown).
The objectives of our study are: (a) to generate a focus library of ergosterol peroxide probes to evaluate their cytotoxic/apoptotic potential via CellTiter-Glo and propidium iodide viability assays against breast cancer cell models [epithelial cell model: MDA-MB-231 (ER–/PR–/HER2–), luminal A cell model: T-47D (ER+/PR+/HER2–) both of which are P53 mutant, and SUM149 (invasive ductal carcinoma, ER–/PR–/HER2-deactivated)], both of which are P53 mutant, SUM149 (invasive ductal carcinoma, ER−/PR−/HER2-deactivated), and the non-cancerous cell models: BJ (fibroblast) and HMEC (primary mammary epithelial), (b) to assess the subcellular accumulation of ergosterol peroxide probes, and (c) their potential biological target(s) in triple negative breast cancer cell models.
Results and discussion
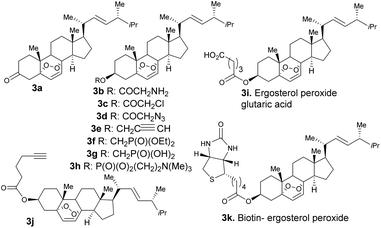
First, we generated ergosterol peroxide by the treatment of the natural product ergosterol with a catalytic radical initiator (phloxine B) in the presence of oxygen and visible light, to undergo a hetero Diels–Alder reaction in excellent yields (93% isolated yield, Fig. 1).The hydroxyl functional group at C3 represents a feasible site for functionalization strategies, while maintaining the rest of the molecule intact. Thus, we generated several ergosterol peroxide analogues (3a–3k, Fig. 2) to investigate the biological effects of a diverse group of linkers at the C3-hydroxyl group. We have previously shown that benzyl carbamates of ergosterol peroxide exhibited superior biological activity to ergosterol peroxide, presumably due to the improved aqueous solubility.11
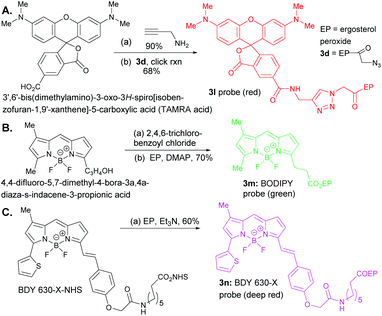
The oxidation of compound 2 with TPAP/NMO at RT provided the enone system 3a. The synthesis of 3b, 3c, 3h, 3i, 3j, and 3k involved esterification reactions of ergosterol peroxide with the corresponding carboxylic acid, acyl chloride, or anhydride (see the ESI† for detailed information). The synthesis of 3d–3g involved SN2-type reactions to provide the desired compounds in good yields. For the synthesis of the biotinylated compound, ergosterol peroxide was subjected to the mixed anhydride of biotin to afford 3k in good yield (Fig. 2). The synthesized compounds were characterized by NMR (1H and 13C) and mass spectrometry (see the ESI†). Next, the ergosterol peroxide fluorescent probes (3l–3n) were synthesized for the anticipated live-cell analysis, including organelle accumulation and proteomic profiling studies (Scheme 1). We envisioned that ergosterol peroxide conjugates of fluorescent dyes would enable live-cell imaging to explore their subcellular interactions. Furthermore, the fluorescent probes were rationally designed to be spectrally orthogonal to organelle-fluorescent trackers for co-localisation studies using fluorescence microscopy. The syntheses of ergosterol peroxide probes with the corresponding fluorescent reagents tetramethylrhodamine (TAMRA) and boron-dipyrromethene (Bodipy)20 were evaluated. Recently, Bu et al. have generated a coumarin fluorescent probe of ergosterol peroxide that accumulates in the mitochondria.10 However, our unbiased fluorescent ergosterol peroxide probes were intended to uncover the specific subcellular accumulation sites of ergosterol peroxide with minimum influence from the fluorescent dye to provide insight into the ergosterol peroxide mode of action. The chemical nature of fluorescent dyes can affect the cellular permeability of the compound and the site of accumulation; thus we minimized the size of the linker to avoid excessive changes to the properties of ergosterol peroxide. First, the tetramethylrhodamine probe 3l (Scheme 1) was synthesized by the treatment of commercially available TAMRA acid, propargyl amine (2 equiv.) with PyBOP and Hünig's base to afford the corresponding fluorescein-propargyl amide (90% yield). Next, the synthesis of the corresponding coupling partner was achieved via (a) acylation of ergosterol peroxide with acyl chloride, followed by (b) azide displacement to provide the desired intermediate (see the ESI†). The copper-catalysed azide–alkyne cycloaddition (CuAAC)21 was mediated by CuSO4 and sodium ascorbate in a solvent mixture of t-BuOH and water (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) to provide 3I in 68% yield. Next, we generated the desired probe 3m (with a short linker between the fluorescent core and ergosterol peroxide) via acylation of Bodipy propionic acid with 2,4,6-trichlorobenzoyl chloride in the presence of Et3N, followed by ergosterol peroxide addition, to provide 3m in 70% yield. The synthesis of compound 3n consisted of a single-step condensation of ergosterol peroxide with Bodipy N-hydroxysuccinimide ester (BDY 630-X-NHS, Scheme 1) and Et3N to afford the expected compound 3n in 60% yield.
1) to provide 3I in 68% yield. Next, we generated the desired probe 3m (with a short linker between the fluorescent core and ergosterol peroxide) via acylation of Bodipy propionic acid with 2,4,6-trichlorobenzoyl chloride in the presence of Et3N, followed by ergosterol peroxide addition, to provide 3m in 70% yield. The synthesis of compound 3n consisted of a single-step condensation of ergosterol peroxide with Bodipy N-hydroxysuccinimide ester (BDY 630-X-NHS, Scheme 1) and Et3N to afford the expected compound 3n in 60% yield.
Previous biological studies of ergosterol peroxide11 suggested that the endoperoxide bridge is required for biological activity against cancer cell models. To obtain further information on the structure–activity relationship of ergosterol peroxide, modifications were performed at the C3 position and their cytotoxicity was assessed by an established cell viability assay described previously.22 A summary of the cytotoxicity findings is shown in Table 1, and cell death was validated by the propidium iodide apoptosis assay (ESI†). The triple-negative breast cancer cell lines MDA-MB-231 and SUM149 and the ER-positive cell line T-47D were evaluated along with the non-cancerous cell lines BJ and HMEC to determine the therapeutic index.22 Our study shows that the triple-negative breast cancer cell model, SUM149, was the most sensitive cell line towards these compounds, while the estrogen receptor positive cell line T-47D was the least responsive under the evaluated conditions.
| Number | MDA-MB-231 EC50 (μM) | SUM149 EC50 (μM) | T-47D EC50 (μM) | HMEC EC50 (μM) | TI (HMEC/SUM149) |
|---|---|---|---|---|---|
| Phosphoramide | >11 | >12 | ND | >10 | >1 |
| Doxorubicin | 1 | 1 | ND | >10 | >11 |
| Taxol | 2 | 0.01 | ND | >10 | >800 |
| Capecitabine | 5 | 5 | ND | 10 | >2 |
| 2 | 18 | 9 | 19 | >20 | >3 |
| 3a | 7 | 5 | 10 | >13 | >3 |
| 3b | 16 | 3 | >20 | >20 | >4 |
| 3c | >20 | 5 | >20 | >20 | >4 |
| 3d | >20 | 10 | >20 | >20 | >2 |
| 3e | 16 | 10 | >20 | >20 | >2 |
| 3f | 9 | 5 | >20 | >20 | >4 |
| 3g | 37 | 5 | >20 | >20 | >4 |
| 3h | 25 | 21 | 17 | >20 | >1 |
| 3i | 33 | 5 | 18 | >20 | >4 |
| 3j | 8 | 4 | 11 | >13 | >4 |
| 3k | 10 | 4 | >10 | >14 | >6 |
| 3l | 20 | 18 | >20 | >20 | >1 |
| 3m | 18 | 2 | 18 | >15 | >6 |
| 3n | 7 | 2 | 16 | >15 | >6 |
Ergosterol peroxide has been reported to have variable potency across cancer cell lines, which is attributed to modest aqueous solubility (<50 μM), limiting its bioavailability to the cell. In addition, at 5–10 mM concentrations in DMSO, precipitation occurs after 168 h at RT. Suitable formulation strategies are currently under development. Some of the generated compounds also exhibited poor aqueous solubility, indicating that liposomal delivery (formulation/prodrug approaches) will be necessary to overcome this potential liability. In terms of stability, ergosterol peroxide is surprisingly stable as we did not observe any degradation by MS/MS in PBS over 72 h at 37 °C; however, there is a report on the conversion of ergosterol peroxide to ergosterol,6b presumably via the retro-Diels–Alder reaction. Noteworthily, the oxidized compound 3a was homogeneously soluble at 100 μM in PBS; this compound exhibited improved activity, which was partially attributed to the improved aqueous solubility of this compound. We used the Caco-2 cell model permeability assay22 (derived from a human colon adenocarcinoma) for examining the permeability property of ergosterol peroxide, and found that it exhibits good cellular permeability (Avg Papp, A/B, nm/s = 342.21, and efflux ratio B2A/A2B = 0.93) as compared to the control drug carbamazepine (Avg Papp, A/B, nm/s = 375.73, and efflux ratio B2A/A2B = 0.74).
Next, we explored the bioactivity of the ergosterol peroxide analogues. Compound 3b, chloride 3c, and azide 3d showed promising activity against SUM149, with little or no activity observed at the tested concentrations against MDA-MB-231 and T-47D. The corresponding alkynyl ether 3e showed moderate bioactivity, and compound 3j which containing the longer side chain showed superior bioactivity against the tested cancer cell lines. The phosphonate ester 3f showed improved activity; however, the corresponding phosphonic acid (3g), phosphocholine (3h), and glutaric ester (3i) exhibited poor activity, presumably due to the carboxylic acid/charged moiety of these compounds that may limit passive diffusion across the membrane. The biotinylated compound 3k demonstrated cytotoxicity comparable to that of ergosterol peroxide. The cellular evaluation of compound 3l revealed that the compound did not accumulate in the cell and showed poor cytotoxicity. Further studies indicate that 3l exhibits poor solubility in both organic and inorganic solvents (as it readily precipitates in DMSO or PBS at 1 mM). Finally, the fluorescent probe 3m disclosed cytotoxicity comparable to that of ergosterol peroxide. However, 3n showed superior bioactivity across cancer cell lines. The combined cytotoxicity studies indicate that the introduction of lipophilic substituents at the C3 hydroxyl group can improve the potency of this natural product. The fluorescent tag significantly improves the potency of ergosterol peroxide, presumably due to its ability to accumulate in the cell.
To validate the anti-proliferative effects of these compounds (3k, 3m and 3n), their activity was validated via the propidium iodide assay (ESI, Fig. S1–3†). Significantly, ergosterol peroxide and its analogues, particularly compounds 3k, 3m and 3n, displayed a moderate therapeutic index comparable to capecitabine, a chemotherapy approved drug.
Next, we evaluated the co-localisation of ergosterol peroxide with orthogonal probes for cellular organelles. When possible, we used commercially available chemical probes for live cell staining organelles (endoplasmic reticulum, mitochondria, and lysosomes). However, to study the Golgi apparatus or peroxisomes, no orthogonal dye was available for our chemical probes 3m and 3n. Therefore, transiently transfected with Red Fluorescent Protein (RFP) or Green Fluorescent Protein (GFP) organelle-labelled cellular models were generated as shown in Fig. 3 (Fig. 3A and B for MDA-MB-231, and see ESI† for SUM149).23a Also, stably transformed cancer cell lines transfected with mRuby-peroxisomes-2 plasmid23b were generated (Fig. 3C).
We hypothesized that ergosterol peroxide would accumulate in the peroxisome due to its endoperoxide chemical nature. Probe 3l was the first evaluated; as previously mentioned, no cell staining was observed. Either the compound did not penetrate the cell or it was pumped out of the cell too rapidly for measurement. As shown in washout experiments with probe 3m and the corresponding BodipyFL methyl ester (Fig. S1–4.1†), 3m distributes across the cytosol and appears to accumulate in cellular substructures. Then, compound 3m was evaluated in co-localisation studies utilizing an array of organelle markers. A pattern opposite to our hypothesis was captured, no co-localisation was detected with the RFP-labelled peroxisome cellular model (Fig. 4A). Similarly, no co-localisation was observed with the Golgi apparatus (Fig. 4B), lysosomes, (Fig. 4C) or mitochondria (MitoTracker Deep Red, Fig. 4D). However, substantial co-localisation was detected with the endoplasmic reticulum using the blue/white endoplasmic reticulum tracker along with 3m (Fig. 4E). Consistent observations were recorded for both MDA-MB-231 and SUM149 cancer cell lines (see the ESI†). The antiproliferative activity of 3m was comparable to that of ergosterol peroxide; thus the combined results suggest that ergosterol peroxide is likely to accumulate in the cytosol and partially co-localise to the endoplasmic reticulum.
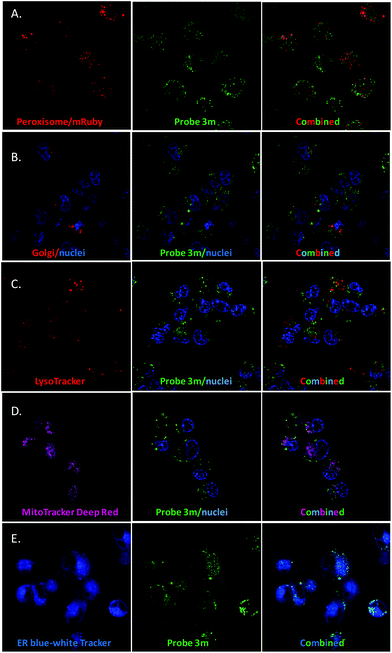 | ||
| Fig. 4 Co-localisation studies of 3m in MDA-MB-231 (see the ESI† for SUM149 cells). A. Peroxisome. B. Golgi. C. Lysosome. D. Mitochondria. E. Endoplasmic reticulum. | ||
After generating the probe 3n (MW = 1030 g mol−1), which carries a longer linker between the core of ergosterol peroxide and the fluorescent tag than 3m (MW = 702 g mol−1), we tested the hypothesis that such a chemical change would affect the accumulation of the compound. Washout experiments with probe 3n and the corresponding BDY 630-X ester (Fig. S1–4.2†) were conducted; 3n permeated the cell and accumulated in cellular substructures. Interestingly, a different subcellular accumulation pattern was observed for 3n in the GFP-peroxisome cell lines (Fig. 5A) as previously recorded for probe 3m. Then to validate the findings, the orthogonal MitoTracker Green was utilized to investigate the compound distribution in the cell. To our surprise, 3n demonstrated selective accumulation in the mitochondria. This finding explained our cytotoxicity data, showing 3n as the most potent compound of the evaluated ergosterol peroxide analogues across cancer cell lines, as accumulation in the mitochondria presumably directly influences the apoptotic program.
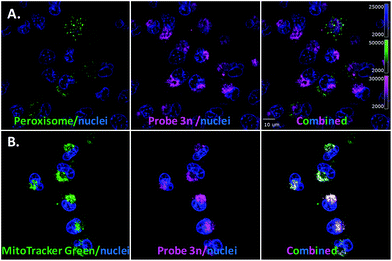 | ||
| Fig. 5 Evaluation of probe 3n. A. Cell light peroxisome-GFP/MDA-MB-231. B. MitoTracker Green and 3n. | ||
The target identification of natural products and small molecules remains one of the biggest challenges in chemical biology,24 and it is possible that ergosterol peroxide affects several biological pathways. Affinity-based proteomic target identification methods are still the most promising methods to elucidate biological targets in drug discovery. Thus, a pull-down experiment with the biotinylated probe 3k was designed. The compound exhibited biological activity analogous to ergosterol peroxide. Incubation of the biotinylated probe 3k bound to Dynabeads M-280 with cell lysates for 2 h at 4 °C overcomes some of the concerns related to compound solubility/bioavailability and potential secondary effects of the compound during cell death. Also, we generated a biotinylated cholesterol probe to identify nonspecific protein binders as a control. The bound protein species were identified by mass spectrometry using the spectral counting method and the complete data set is provided (ESI†).
The distribution of potential protein targets interacting with 3k in SUM149 and MDA-MB-231 cell models is shown in Fig. 6 respectively. They are represented in scatter plots displaying the ratios of Spectral Counts probe/Spectral Counts Control against the corresponding p-values. Excitingly, two potential biological targets, Rab interacting lysosomal protein like 1 (RIPL1) and E3 ubiquitin-protein ligase (UBR4), were identified in both SUM149 and MDA-MB-231 cell lines. These proteins are known to be distributed across the cytosol and the plasma membrane, which agrees with our co-localisation results. RIPL1 is involved in regulating the cell shape and polarity via cellular protein transport,25 while UBR4 is involved in ubiquitination and subsequent degradation of certain proteins, and interacts with the retinoblastoma-associated protein in the nucleus, calcium-bound calmodulin in the cytoplasm and regulates integrin-mediated signaling.26 Other relevant targets for these cell lines are shown in blue (PRKDC, FAS, EZRI, and GDIB for SUM149, and eIF4B and PP1A for MDA-MB-231) and they will be evaluated in future studies.
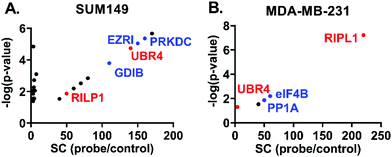 | ||
| Fig. 6 Scatter plots displaying potential protein targets of 3k, spectral counts (SC) against p-values. A. SUM149. B. MDA-MB-231. | ||
Conclusions
In summary, our study describes the synthesis of ergosterol peroxide probes from the readily accessible natural product ergosterol and evaluates their biological potential against breast cancer cell models. These chemical tools can be used for live cell imaging studies to delineate the mechanism of action of ergosterol peroxide in cancer cells. We disclose a short synthesis route to ergosterol peroxide probes and demonstrate that these probes accumulate in the cytosol with subcellular specificity. Our ergosterol peroxide probes show promising anti-proliferation activity against cancer cell models in alignment with reported studies,9,10–12,17 providing information on the role of substituent groups at the hydroxyl group and potential biological targets of this natural product. Furthermore, these chemical probes indicate that ergosterol peroxide is a promising molecular scaffold as an early lead compound for further therapeutic development against breast cancer. Live cell imaging reveals significant co-localisation of 3m with endoplasmic reticulum fluorescent markers, while the probe 3n with a longer linker between the borane core and ergosterol peroxide showed accumulation in the mitochondria. We demonstrate that chemical modifications can lead to superior potency by causing accumulation in specific organelles to induce greater cellular damage. The identified potential targets warrant future validation studies via molecular biology/chemical biology approaches.Author contributions
M. M. M-M., T. L., and F. R. designed the study; T. L. performed all the synthetic chemistry; and W. L. conducted cellular experiments with the support of F. R. All authors interpreted the data and reviewed and edited the manuscript. All authors have approved the final version of the manuscript.Conflicts of interest
The authors declare that they have no competing interest.Acknowledgements
This study was supported by ALSAC St Jude Children's Research Hospital (F. R.) and NIH/NIGMS SC3GM111171 Universidad Central del Caribe School of Medicine (M. M. M-M). We thank the following core facilities for technical assistance: Analytical Technologies, Flow Cytometry and Cell Sorting, Cell and Tissue Imaging Core, Center for Proteomics and Metabolomics, and Bioinformatics core (Dr David Finkelstein), which are all supported fully or in part by ALSAC and Cancer Center Support Grant P30CA021765 from the National Cancer Institute.Notes and references
- T. Nakanishi, H. Murata, Y. Inatomi, A. Inada, J. Murata, F. A. Lang, K. Yamasaki, M. Nakano, T. Kawahata, H. Mori and T. Otake, Natural Medicines, 1998, 52, 521–526 CAS.
- K. Yasukawa, T. Akihisa, H. Kanno, T. Kaminaga, M. Izumida, T. Tamura and M. Takido, Biol. Pharm. Bull., 1996, 19, 573–576 CrossRef CAS.
- Y. Takahashi, M. Uda, T. Ohashi, K. Nakano, K. Murakami and T. Tomimatso, Phytochemistry, 1991, 30, 4117–4120 CrossRef.
- C. Jiménez, E. Quiñoá, R. Rignera, R. Vilalta and J. M. Quintela, J. Nat. Prod., 1989, 52, 619–622 CrossRef.
- M. C. Deghrigue, L. Ghribi and M. V. D'auria, et al. , J. Pharm. Sci., 2014, 22, 64 Search PubMed.
- (a) W. Qing-Ping, X. Yi-Zhen and D. Zhaoqun, et al. , PLoS One, 2012, 7, 44579 CrossRef PubMed; (b) D. B. Graca Sgarbi, A. J. Ribeiro-Silva, I. Z. Carlos, C. L. Silva, J. Angluster and C. S. Alviano, Mycopathologia, 1997, 139, 9–14 CrossRef.
- (a) J. M. Gao, M. Wang, L. P. Liu, G. H. Wei, A. L. Zhang and C. Draghici, et al. , Phytomedicine, 2007, 14, 821–824 CrossRef CAS PubMed; (b) A. Ramos-Ligonio, A. López-Monteon and Á. Trigos, Phytother. Res., 2012, 26, 938–943 CrossRef CAS PubMed; (c) L. Ma, H. Chen, P. Dong and X. Lu, Food Chem., 2013, 139, 503–508 CrossRef CAS PubMed.
- M. Kobori, M. Yoshida, M. Ohnishi-Kameyama and H. Shinmoto, Br. J. Pharmacol., 2007, 150, 209–219 CrossRef CAS PubMed.
- N. Duarte, M. U. Ferreira, M. Martins, M. Viveiros and L. Amaral, Phytother. Res., 2007, 21, 601–604 CrossRef CAS PubMed.
- (a) M. Bu, T. Cao, H. Li, M. Guo, B. B. Yang, C. Zeng and L. Hu, ChemMedChem, 2017, 12, 466–474 CrossRef CAS PubMed; (b) M. Bu, T. Cao, H. Li, M. Guo, B. B. Yang, C. Zeng, Y. Zhou, N. Zhang and L. Hu, Bioorg. Med. Chem. Lett., 2017, 27, 3856–3861 CrossRef CAS PubMed; (c) G. Battogtokh, Y. S. Choi, D. S. Kang, S. J. Park, M. S. Shim, K. M. Huh, Y.-Y. Cho, J.-Y. Lee, H. S. Lee and H. C. Kang, Acta Pharm. Sin. B, 2018, 8, 862–880 CrossRef PubMed.
- (a) I. J. Suarez-Arroyo, R. Rosario-Acevedo, A. Aguilar-Perez, P. L. Clemente, L. A. Cubano, J. Serrano, R. J. Schneider and M. M. Martínez-Montemayor, PLoS One, 2013, 8(2), e57431 CrossRef CAS PubMed; (b) M. M. Martínez-Montemayor, T. Ling, I. J. Suárez-Arroyo, G. Ortiz Soto, C. S. Negrón, M. Y. Lacourt-Ventura, A. Valentín-Acevedo, W. H. Lang and F. Rivas, Front. Pharmacol., 2019, 10, 115 CrossRef PubMed.
- I. Fernandez and A. Robert, Org. Biomol. Chem., 2011, 9, 4098–4107 RSC.
- M. Axelrod, V. L. Gordon, M. Conaway, A. Tarcsafalvi, D. J. Neitzke, D. Gioeli and M. J. Weber, Oncotarget, 2013, 4, 622–635 CrossRef PubMed.
- S. M. Mense and L. Zhang, Cell Res., 2006, 16, 681–692 CrossRef CAS PubMed.
- D. Chiabrando, F. Vinchi, V. Fiorito, S. Mercurio and E. Tolosano, Front. Pharmacol., 2014, 5, 1–24 CAS.
- (a) R. Chiorean, C. Braicu and I. Berindan-Neagoe, Breast, 2013, 22, 1026–1033 CrossRef PubMed; (b) K. J. Chavez, V. Sireesha, S. V. Garimella and S. Lipkowitz, Breast Dis., 2010, 32, 35–48 Search PubMed; (c) W. D. Foulkes, I. E. Smith and J. S. Reis-Filho, N. Engl. J. Med., 2010, 363, 1938–1948 CrossRef CAS PubMed.
- H. Y. Wu, F. L. Yang, L. H. Li, Y. K. Rao, T. C. Ju, W. T. Wong, C. Y. Hsieh, M. V. Pivkin, K. F. Hua and S. H. Wu, Sci. Rep., 2018, 8, 17956 CrossRef PubMed.
- Z. Zhang, L. Zhang, L. Zhou, Y. Lei, Y. Zhang and C. Huang, Redox Biol., 2018 DOI:10.1016/j.redox.2018.11.005.
- C. Florean, S. Song, M. Dicato and M. Diederich, Free Radical Biol. Med., 2019, 134, 177–189 CrossRef CAS PubMed.
- (a) H. M. Schipper, S. Cissé and P. A. Walton, Exp. Cell Res., 1993, 207, 62–67 CrossRef CAS PubMed; (b) Y. Han, M. Li, F. Qiu, M. Zhang and Y. H. Zhang, Nat. Commun., 2017, 8, 1307 CrossRef PubMed.
- C. S. McKay and M. G. Finn, Chem. Biol., 2014, 21, 1075–1101 CrossRef CAS PubMed.
- T. Ling, W. Lang, X. Feng, S. Das, J. Maier, C. Jeffries, A. Shelat and F. Rivas, Eur. J. Med. Chem., 2018, 146, 501–510 CrossRef CAS PubMed.
- (a) R. S. Ames, T. A. Kost and J. P. Condreay, Expert Opin. Drug Discovery, 2007, 12, 1669–1681 CrossRef PubMed , (BACMan techn); (b) Addgene plasmid #54840 deposited by Dr M. Davidson.
- T. Böttcher, M. Pitscheider and S. A. Sieber, Angew. Chem., Int. Ed., 2010, 49, 2680–2698 CrossRef PubMed.
- T. Wang, K. K. Wong and W. Hong, Mol. Biol. Cell, 2004, 15, 815–826 CrossRef CAS PubMed.
- K. W. Huh, J. DeMasi, H. Ogawa, Y. Nakatani, P. M. Howley and K. Münger, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 11492–11497 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ob00145j |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2019 |