Open Access Article
Open Access ArticleFolding of single-stranded circular DNA into rigid rectangular DNA accelerates its cellular uptake†
Shozo
Ohtsuki‡
 a,
Yukako
Shiba‡
a,
Tatsuoki
Maezawa
a,
Kumi
Hidaka
b,
Hiroshi
Sugiyama
a,
Yukako
Shiba‡
a,
Tatsuoki
Maezawa
a,
Kumi
Hidaka
b,
Hiroshi
Sugiyama
 bc,
Masayuki
Endo
bc,
Masayuki
Endo
 bc,
Yuki
Takahashi
a,
Yoshinobu
Takakura
a and
Makiya
Nishikawa
bc,
Yuki
Takahashi
a,
Yoshinobu
Takakura
a and
Makiya
Nishikawa
 *ad
*ad
aDepartment of Biopharmaceutics and Drug Metabolism, Graduate School of Pharmaceutical Sciences, Kyoto University, Sakyo-ku, Kyoto 606-8501, Japan. E-mail: makiya@rs.tus.ac.jp
bDepartment of Chemistry, Graduate School of Science, Kyoto University, Sakyo-ku, Kyoto 606-8502, Japan
cInstitute for Integrated Cell-Material Sciences, Kyoto University, Sakyo-ku, Kyoto 606-8501, Japan
dLaboratory of Biopharmaceutics, Faculty of Pharmaceutical Sciences, Tokyo University of Science, Noda, Chiba 278-8510, Japan
First published on 28th November 2019
Abstract
Despite the importance of the interaction between DNA and cells for its biological activity, little is known about exactly how DNA interacts with cells. To elucidate the relationship between the structural properties of DNA and its cellular uptake, a single-stranded circular DNA of 1801 bases was designed and folded into a series of rectangular DNA (RecDNA) nanostructures with different rigidities using DNA origami technology. Interactions between these structures and cells were evaluated using mouse macrophage-like RAW264.7 cells. RecDNA with 50 staple DNAs, including four that were Alexa Fluor 488-labeled, was designed. RecDNA with fewer staples, down to four staples (all Alexa Fluor 488-labeled), was also prepared. Electrophoresis and atomic force microscopy showed that all DNA nanostructures were successfully obtained with a sufficiently high yield. Flow cytometry analysis showed that folding of the single-stranded circular DNA into RecDNA significantly increased its cellular uptake. In addition, there was a positive correlation between uptake and the number of staples. These results indicate that highly folded DNA nanostructures interact more efficiently with RAW264.7 cells than loosely folded structures do. Based on these results, it was concluded that the interaction of DNA with cells can be controlled by folding using DNA origami technology.
Introduction
The self-assembling nature of DNA has been used to construct uniquely structured DNA nanostructures, including immobile Holliday junctions, cubic cages, and DNA nanodevices.1–3 DNA origami technology, which comprises a long single-stranded ‘scaffold’ DNA and many short ‘staple’ DNAs (staples) complementary to regions of the scaffold DNA, has allowed the creation of precisely designed, two- or three-dimensional DNA nanostructures with sophisticated functions.4–6 These DNA origami-based nanostructures have been used as sensors for single nucleotide polymorphisms and RNA sequences.7,8DNA is a natural, biodegradable, and biologically active molecule; therefore, the therapeutic applications of such DNA nanostructures have been explored. Our previous studies reported that DNA nanostructures consisting of three or more oligodeoxynucleotides (ODNs) were efficiently taken up by immune cells, including mouse macrophage-like RAW264.7 cells and mouse bone marrow-derived dendritic cells. When these DNA nanostructures contained immunostimulatory unmethylated cytosine-phosphate-guanine (CpG) DNA, they efficiently induced the release of proinflammatory cytokines from immune cells expressing Toll-like receptor 9, the intracellular receptor that recognizes CpG DNA.9–12 This enhanced cytokine release was found to be effective at inhibiting tumor growth in tumor-bearing mouse models.13,14
Increasing the activity of CpG DNA by constructing DNA nanostructures also resulted in an increase in cellular uptake. We demonstrated that the uptake of single stranded, double stranded, and branched, polypod-like structured DNA was dependent on structural complexity.9 Another study using tetrapod-like structured DNA (tetrapodna), tetrahedral DNA (tetrahedron), and tetragonal DNA (tetragon), all of which were prepared using four ODNs of the same length, also showed that the uptake of these DNA nanostructures by RAW264.7 cells was quite different.12 These differences in cellular uptake could be attributed to their structural properties. Although there is little information about the relationship between the structural properties and cellular uptake of DNA nanostructures, it is possible that their rigidity or flexibility plays a role, because structural flexibility was found to affect the penetration of DNA nanoribbons through plasma membranes.15 To examine the importance of the rigidity or flexibility of DNA nanostructures on their cellular uptake, it is indispensable to use DNA nanostructures whose structural properties can be strictly controlled. In our previous studies,9,12 several parameters, such as size and shape, were different among the samples. To test this hypothesis, DNA origami technology was used to construct a series of DNA nanostructures with different structural properties. Rectangular DNA (RecDNA) was designed with a scaffold DNA and 50 staples. Additional structures with fewer staples were also prepared, which had the same shape but were more flexible. The DNA nanostructures were added to RAW264.7 cells and the degree of cellular uptake was evaluated.
Materials and methods
Chemicals
RPMI1640 medium was obtained from Nissui Pharmaceutical Co., Ltd (Tokyo, Japan). Fetal bovine serum (FBS) was obtained from Equitech-Bio, Inc. (Kerrville, TX, USA). Opti-modified Eagle's medium (Opti-MEM) was purchased from Invitrogen (Carlsbad, CA, USA). All other chemicals were of the highest grade available and used without further purification.Cell culture
Murine macrophage-like RAW264.7 cells were obtained from ATCC (Manassas, VA, USA) and cultured in RPMI1640 medium supplemented with 10% heat-inactivated FBS, 0.2% NaHCO3, 100 units per mL penicillin, 100 μg mL−1 streptomycin and 2 mM L-glutamine at 37 °C in humidified air containing 5% CO2. Cells were seeded onto 96-well culture plates at a density of 5 × 104 cells per well and cultured for 24 h prior to use.Scaffold
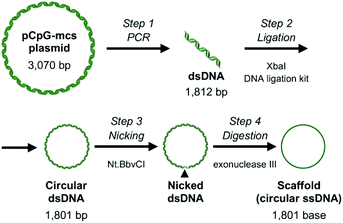
A 1800 bp sequence from a pCpG-mcs plasmid (InvivoGen, San Diego, CA, USA) was amplified by polymerase chain reaction using the following primers: forward: 5′-ACTTCTAGATACAAATGTGGTATGGAATTCAGTC-3′, reverse: 5′-CATTCTAGACCTCAGCGAAACAGAGAGCTGAACAAAGAG-3′. Xba I recognition site is underlined; bold letters indicate Nt·BbvcI recognition site. Both ends of the PCR product were digested with Xba I (Takara Bio, Otsu, Japan), and ligated using a DNA ligation kit ver.2.1 (Takara Bio). Escherichia coli strain GT115 was transformed with the double-strand circular DNA, miniprep was performed for amplification, then the double-strand circular DNA was purified using GenElute Plasmid Miniprep Kit (Sigma-Aldrich, St Louis, MO, USA). All products were sequenced three times using a BigDye Terminator v3.1 Cycle Sequencing Kit (Thermo Fisher Scientific Inc., Waltham, MA, USA) by ABI3000xl (Thermo Fisher Scientific Inc.) using the following primers: primer 1: 5′-GGAATTCAGTCAATATGTTCAC-3′ primer 2: 5′-AGAGAATTGTAATATGCAGATTAT-3′ primer 3: 5′-GCAACTGTGTGCACTTTGTG-3′. Maxiprep was performed to obtain a sufficient amount of double-stranded DNA, which was then purified using a PureLink HiPure Plasmid Maxiprep Kit (Thermo Fisher Scientific Inc.). One strand of the double-strand DNA was digested with nicking endonuclease Nt·BbvCI (New England BioLabs, Ipswich, MA, USA), which was then digested with Exonuclease III (New England BioLabs). Scaffold formation was analyzed by agarose gel electrophoresis, the band was extracted from the gel, and purified by NucleoSpin Gel and PCR Clean-up (Takara Bio).The sequence of the scaffold is as follows:
CTAGATACAAATGTGGTATGGAATTCAGTCAATATGTTCACCCCAAAAAAGCTGTTTGTTAACTTGCCAACCTCATTCTAAAATGTATATAGAAGCCCAAAAGACAATAACAAAAATATTCTTGTAGAACAAAATGGGAAAGAATGTTCCACTAAATATCAAGATTTAGAGCAAAGCATGAGATGTGTGGGGATAGACAGTGAGGCTGATAAAATAGAGTAGAGCTCAGAAACAGACCCATTGATATATGTAAGTGACCTATGAAAAAAATATGGCATTTTACAATGGGAAAATGATGATCTTTTTCTTTTTTAGAAAAACAGGGAAATATATTTATATGTAAAAAATAAAAGGGAACCCATATGTCATACCATACACACAAAAAAATTCCAGTGAATTATAAGTCTAAATGGAGAAGGCAAAACTTTAAATCTTTTAGAAAATAATATAGAAGCATGCCATCAAGACTTCAGTGTAGAGAAAAATTTCTTATGACTCAAAGTCCTAACCACAAAGAAAAGATTGTTAATTAGATTGCATGAATATTAAGACTTATTTTTAAAATTAAAAAACCATTAAGAAAAGTCAGGCCATAGAATGACAGAAAATATTTGCAACACCCCAGTAAAGAGAATTGTAATATGCAGATTATAAAAAGAAGTCTTACAAATCAGTAAAAAATAAAACTAGACAAAAATTTGAACAGATGAAAGAGAAACTCTAAATAATCATTACACATGAGAAACTCAATCTCAGAAATCAGAGAACTATCATTGCATATACACTAAATTAGAGAAATATTAAAAGGCTAAGTAACATCTGTGGCTTAATTAAAACAGGTAGTTGACAATTAAACATTGGCATAGTATATCTGCATAGTATAATACAACTCACTATAGGAGGGCCATCATGGCCAAGTTGACCAGTGCTGTCCCAGTGCTCACAGCCAGGGATGTGGCTGGAGCTGTTGAGTTCTGGACTGACAGGTTGGGGTTCTCCAGAGATTTTGTGGAGGATGACTTTGCAGGTGTGGTCAGAGATGATGTCACCCTGTTCATCTCAGCAGTCCAGGACCAGGTGGTGCCTGACAACACCCTGGCTTGGGTGTGGGTGAGAGGACTGGATGAGCTGTATGCTGAGTGGAGTGAGGTGGTCTCCACCAACTTCAGGGATGCCAGTGGCCCTGCCATGACAGAGATTGGAGAGCAGCCCTGGGGGAGAGAGTTTGCCCTGAGAGACCCAGCAGGCAACTGTGTGCACTTTGTGGCAGAGGAGCAGGACTGAGGATAACCTAGGAAACCTTAAAACCTTTAAAAGCCTTATATATTCTTTTTTTTCTTATAAAACTTAAAACCTTAGAGGCTATTTAAGTTGCTGATTTATATTAATTTTATTGTTCAAACATGAGAGCTTAGTACATGAAACATGAGAGCTTAGTACATTAGCCATGAGAGCTTAGTACATTAGCCATGAGGGTTTAGTTCATTAAACATGAGAGCTTAGTACATTAAACATGAGAGCTTAGTACATACTATCAACAGGTTGAACTGCTGATCTTAATTAAAATTATCTCTAAGGCATGTGAACTGGCTGTCTTGGTTTTCATCTGTACTTCATCTGCTACCTCTGTGACCTGAAACATATTTATAATTCCATTAAGCTGTGCATATGATAGATTTATCATATGTATTTTCCTTAAAGGATTTTTGTAAGAACTAATTGAATTGATACCTGTAAAGTCTTTATCACACTACCCAATAAATAATAAATCTCTTTGTTCAGCTCTCTGTTTCGCTGAGGT.
Staples
Staples, short DNA of 24–40 bases in length, were purchased from Eurofin Genomics Japan K.K. (Tokyo, Japan). Sequences of staples used in RecDNA are listed in ESI Table S1.† Each staple was identified with a code, for example 0[68]-1[52], according to the following rule: helix (0) base ([68]) numbers of position of 5′-end and helix (1) and base ([52]) numbers of the position of its 3′-end. Four ODNs labeled with Alexa Fluor488 at the 5′-end were purchased from Japan BioService Co., Ltd (Saitama, Japan).Preparation of DNA nanostructure
Each structure was prepared by mixing equimolar amounts of scaffold and staples. Briefly, scaffold and staples dissolved in an annealing buffer (20 mM Tris-HCl, 1 mM ethylenediaminetetraacetic acid (EDTA), and 10 mM magnesium chloride, pH 7.3) were mixed at a final concentration of 60 nM for each DNA. Mixtures were then heated to 85 °C, cooled to 35 °C at −0.2 °C min−1, cooled to 20 °C at −1 °C min−1, then gradually cooled to 4 °C using a thermal cycler (PC-818S Program Temp Control System; ASTEC Co., Ltd, Fukuoka, Japan). The staples excluded from the full set listed in ESI Table S1† to construct each DNA nanostructure are described in ESI Table S2.† Products were analyzed at 4 °C by agarose gel electrophoresis in 0.5× TBE buffer (45 mM Tris borate, 1 mM EDTA, supplemented with 11 mM MgCl2), stained with SYBR Gold and observed using a LAS3000 (Fujifilm, Tokyo, Japan). To determine the amount of fluorescent-labeled staples incorporated into each structure, Alexa Fluor488-labeled staples were imaged after agarose gel electrophoresis, using a LAS3000 with excitation wavelength 460 nm. The size of each DNA nanostructure was measured by dynamic light scattering (DLS) using a Zetasizer Nano ZS (Malvern, Worcestershire, UK).AFM imaging of DNA nanostructures
Atomic force microscopy (AFM) images of each structure were obtained using an AFM system (Nano Live Vision, RIBM, Tsukuba, Japan) using a silicon nitride cantilever (BL-AC10EGS; Olympus, Tokyo, Japan). Briefly, a DNA sample was adsorbed on a freshly cleaved mica plate pretreated with 0.1% aqueous 3-aminopropyltriethoxysilane for 5 min at room temperature then washed three times with a buffer containing 20 mM Tris and 10 mM MgCl2. Scanning was performed in the same buffer.Confocal fluorescence microscopic observation
Alexa Fluor488-labeled single stranded DNA (ssDNA), each rectangular DNA (RecDNA), or Scaffold4 at 9 nM in 200 μL of Opti-MEM was added to RAW264.7 cells in an 8-well chamber slide at a density of 5 × 105 cells per mL and incubated for 4 h at 37 °C. RAW264.7 cells were washed twice with phosphate buffer saline (PBS), fixed with 4% paraformaldehyde for 20 min, and washed twice with PBS. To stain nuclei, cells were incubated with 4′,6-diamino-2-phenylindole (DAPI) for 5 min, and washed with PBS. Cells were coverslipped with SlowFade Gold (Life Technologies, Tokyo, Japan), and analyzed using a confocal fluorescence microscope (Nikon A1R MP, Nikon, Tokyo, Japan).Flow cytometry
RecDNA, or Scaffold4 at 5.5 nM in 50 μL Opti-MEM was added to RAW264.7 cells on a 96 well plate at a density of 5 × 105 cells per mL and incubated for 4 h at 37 °C. RAW264.7 cells were washed three times with PBS and harvested. Fluorescence intensity was determined by flow cytometry (Gallios Flow Cytometer, BD Biosciences, NJ, USA) using Kaluza software (version 1.0, BD Biosciences), and mean fluorescence intensity (MFI) was calculated.Statistical analysis
Differences were statistically evaluated using one-way analysis of variance (ANOVA) followed by the Tukey–Kramer test for multiple comparisons. A P value of <0.05 was considered statistically significant.Graphical abstracts
Computational simulation images of DNA nanostructures were obtained by CanDo software (https://cando-dna-origami.org/).Results and discussion
Construction of DNA nanostructures
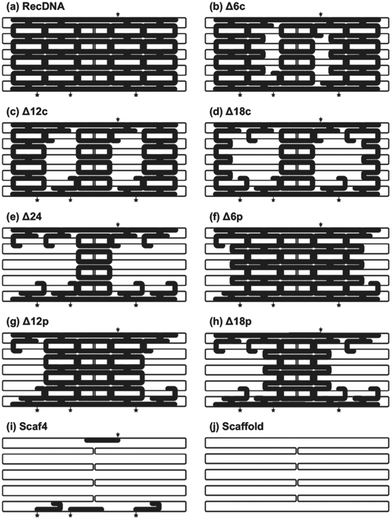
Most applications of DNA origami use M13mp18 single stranded DNA of 7249 bases as a scaffold, mainly because other sources of long single-stranded DNA are limited. A number of approaches have been explored to overcome this limitation.16,17 Considering that the upper size limit of endocytosis is around 100 nm,18 shorter single stranded DNAs would be suitable as a scaffold for evaluating the relationship between the structural properties and cellular uptake of DNA nanostructures. Based on this, in the current study, 1801 base-long single-stranded DNA was designed as a scaffold to construct RecDNA. Fig. 1 shows a schematic overview of the preparation process of the scaffold DNA, as previously reported.16 Briefly, a 1801 base circular single-strand DNA was produced by PCR amplification of a CpG-free pCpG-mcs plasmid (InvivoGen, San Diego, CA, USA) followed by enzyme treatments. Agarose gel electrophoresis analysis showed that the expected scaffold DNA was obtained (ESI Fig. S1†). The scaffold DNA was excised from the gel and purified.RecDNA was designed using caDNAno, a computer-aided design tool for DNA origami, as previously reported.19 Assuming the helical diameter of DNA, the interhelical gap, and the distance between bases are 2, 0.5, and 0.34 nm, respectively, RecDNA with the following parameters was designed using the scaffold DNA and 50 staples, including four Alexa Fluor 488-labeled ones: height 24.5 nm (10 helices), and width 54.1 nm (160 bases). Single-strand loops were left on both sides of the RecDNA to prevent self-sticking. Then, 6, 12, 18, or 24 staples located close to the central part of RecDNA were removed to obtain Δ6c, Δ12c, Δ18c, and Δ24, respectively (Δ(n)c). Staples located close to the periphery of the RecDNA were also removed to obtain Δ6p, Δ12p, and Δ18p (Δ(n)p). In addition, the scaffold DNA and four Alexa Fluor 488-staples only were annealed to obtain Scaf4 (Fig. 2). ESI Fig. S2† shows results of the agarose gel electrophoresis analysis for each structure. All preparations generated a major single band, indicating that each structure was formed correctly and with a high yield. Similar experiments were carried out using DNA nanostructures prepared with Alexa Fluor 488-labaled staples. The fluorescence intensity of the bands was comparable among the samples (ESI Fig. S3†), suggesting that Alexa Fluor 488-labeled staples bind to the scaffold irrespective of the structural properties of DNA nanostructures. Based on these results, the cellular uptake of DNA nanostructures was evaluated by measuring the fluorescence intensity of cells after addition of Alexa Fluor 488-labeled DNA nanostructures.
The size of the DNA nanostructures was measured using the dynamic light scattering method with a Malvern Zetasizer 3000HS (Malvern Instruments, Malvern, UK). The sizes of RecDNA, Δ(n)c, and Δ(n)p were about 30 nm, indicating that all DNA nanostructures were of similar size. The size of the Scaf4, which was hardly folded, was about 490 nm (Table 1).
| DNA | Size (nm) |
|---|---|
| RecDNA | 30.6 ± 6.7 |
| Δ6c | 33.0 ± 3.7 |
| Δ12c | 28.6 ± 3.7 |
| Δ18c | 27.1 ± 4.1 |
| Δ24 | 29.3 ± 7.5 |
| Δ6p | 32.7 ± 3.9 |
| Δ12p | 29.7 ± 3.8 |
| Δ18p | 31.1 ± 2.7 |
| Scaf4 | 488 ± 122 |
Cellular interaction of DNA nanostructures
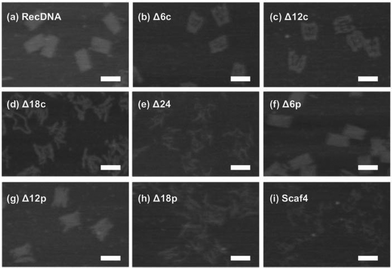
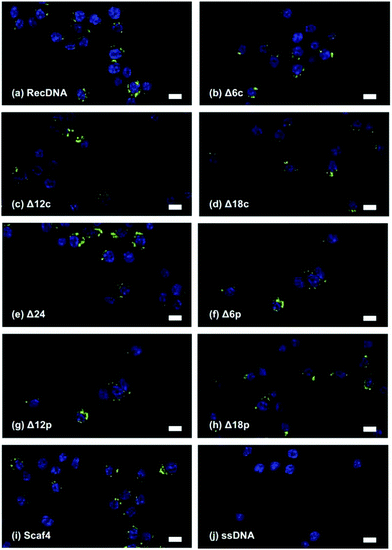
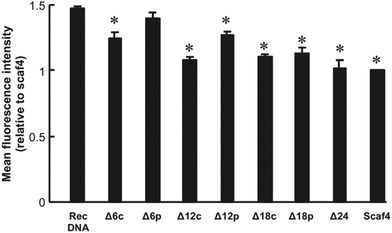
Fig. 3 shows AFM images of the RecDNA and other prepared DNA nanostructures. The image of RecDNA (Fig. 3a) shows a rectangular shape of expected size; measurements from the image showed that it was 29.7 ± 2.5 nm in height and 54.5 ± 3.4 nm in width. Δ6c (Fig. 3b) and Δ12c (Fig. 3c) resembled black holes within white rectangles, and the structures of Δ18c (Fig. 3d) and Δ24 (Fig. 3e) were not rectangular. The AFM images of Δ(n)p showed different patterns from those of Δ(n)c. Both sides of Δ6p (Fig. 3f) and Δ12p (Fig. 3g) were disentangled, but the overall structure was still rectangular. Again, the structure of Δ18p (Fig. 3h) was not rectangular, comparable to that of Δ18c (Fig. 3i). The structure of Scaf4 was not rectangular and exhibited a variety of shapes.The cellular uptake of the DNA nanostructures was examined using RAW264.7 cells. Confocal microscopic images show that all of the Alexa Fluor 488-labeled DNA samples were efficiently taken up by the RAW264.7 cells, compared to Alexa Fluor 488-labeled ssDNA (Fig. 4). To quantitatively evaluate cellular uptake, the fluorescence intensity of RAW264.7 cells was measured and mean fluorescence intensity (MFI) values were calculated among the Alexa Fluor 488-labeled DNA samples for comparison. The MFI value was highest for Alexa Fluor 488-labeled RecDNA, and lowest for Alexa Fluor 488-labeled Scaf4 (Fig. 5). The data showed that the fewer the number of staples used, the lower the uptake by RAW264.7 cells. Removal of peripheral staples had less pronounced effects on cellular uptake than removal of central staples, although the differences were not significant.
In the present study, the cellular uptake was the highest for Alexa Fluor 488-labeled RecDNA. Alexa Fluor 488-labeled DNA nanostructures with fewer staples, which could be more flexible than RecDNA, showed lower uptake. In addition, removing central staples had a great impact on cellular uptake than removing peripheral ones (Fig. 5, between Δ6c and Δ6p, and between Δ12c and Δ12p). This result suggests that, besides the strength of negative charge, the structural properties, including flexibility, also affects the uptake by scavenger receptor-positive cells such as macrophages, which is consistent with our working hypothesis.
There have been several reports indicating that cell surface receptors for DNA contribute to the cellular uptake of DNA.20–26 Differences in cellular uptake efficiency could therefore be due to differences in the affinity of DNA nanostructures for these receptors. Further studies are needed, but it is possible that the folding of scaffold DNA with staples could have a strong impact on their affinity for the receptors.
Uptake of Alexa Fluor 488-labeled RecDNA was about 1.5 times higher than that of Alexa Fluor 488-labeled Scaf4. Structure-dependent differences in uptake were not so great compared with those observed in studies using three to eight ODNs of 30–55 nucleotide length.9,10,12 One reason for this could be differences in the number of nucleotides per structure; the DNA nanostructures used in the current study consisted of a 1801-base long circular DNA and many staples, which is much larger than the ODN-based DNA nanostructures.12 Another possible reason is the difference between 2D and 3D structures. While this study focused on the cellular uptake of a series of 2D DNA nanostructures with different flexibilities, the previous one used 3D DNA nanostructures, such as tetrapodna, tetrahedron, and tetragon, each of which could have different structural flexibility. Thus, using 3D DNA nanostructures with more structural flexibility could show even more remarkable differences in cellular uptake.
Conclusions
In conclusion, the present study showed that the cellular uptake of Alexa Fluor 488-labeled RecDNA, RecDNA with 50 staples, was more efficient than Alexa Fluor 488-labeled DNA nanostructures with fewer staples: Δ(n)c, Δ(n)p, and Scaf4. In addition, removing central staples had a greater impact on cellular uptake than removing peripheral staples. These results will provide useful information for the development of DNA nanostructure-based delivery systems for bioactive compounds, such as nucleic acid drugs.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported in part by JST CREST Grant Number JPMJCR1521, Japan, and Grants-in-Aid for Scientific Research (B) (23390010 and 26293008) from the Japan Society for the Promotion of Science.References
- N. C. Seeman, DNA in a material world, Nature, 2003, 421, 427–431 CrossRef PubMed.
- N. C. Seeman, Nucleic acid junctions and lattices, J. Theor. Biol., 1982, 99, 237–247 CrossRef CAS.
- S. Surana, A. R. Shenoy and Y. Krishnan, Designing DNA nanodevices for compatibility with the immune system of higher organisms, Nat. Nanotechnol., 2015, 10, 741–747 CrossRef CAS PubMed.
- P. W. K. Rothemund, Folding DNA to create nanoscale shapes and patterns, Nature, 2006, 440, 297–302 CrossRef CAS.
- T. Liedl, B. Högberg, J. Tytell, D. E. Ingber and W. M. Shih, Self-assembly of three-dimensional prestressed tensegrity structures from DNA, Nat. Nanotechnol., 2010, 5, 520–524 CrossRef CAS.
- E. S. Andersen, M. Dong, M. M. Nielsen, K. Jahn, R. Subramani, W. Mamdouh, M. M. Golas, B. Sander, H. Stark, C. L. P. Oliveira, J. S. Pedersen, V. Birkedal, F. Besenbacher, K. V. Gothelf and J. Kjems, Self-assembly of a nanoscale DNA box with a controllable lid, Nature, 2009, 459, 73–76 CrossRef CAS PubMed.
- H. K. K. Subramanian, B. Chakraborty, R. Sha and N. C. Seeman, The label-free unambiguous detection and symbolic display of single nucleotide polymorphisms on DNA origami, Nano Lett., 2011, 11, 910–913 CrossRef CAS PubMed.
- Y. Ke, S. Lindsay, Y. Chang, Y. Liu and H. Yan, Self-assembled water-soluble nucleic acid probe tiles for label-free RNA hybridization assays, Science, 2008, 319, 180–183 CrossRef CAS PubMed.
- K. Mohri, M. Nishikawa, N. Takahashi, T. Shiomi, N. Matsuoka, K. Ogawa, M. Endo, K. Hidaka, H. Sugiyama, Y. Takahashi and Y. Takakura, Design and development of nanosized DNA assemblies in polypod-like structures as efficient vehicles for immunostimulatory cpg motifs to immune cells, ACS Nano, 2012, 6, 5931–5940 CrossRef CAS.
- S. Uno, M. Nishikawa, K. Mohri, Y. Umeki, N. Matsuzaki, Y. Takahashi, H. Fujita, N. Kadowaki and Y. Takakura, Efficient delivery of immunostimulatory DNA to mouse and human immune cells through the construction of polypod-like structured DNA, Nanomedicine, 2014, 10, 765–774 CrossRef CAS.
- K. Mohri, E. Kusuki, S. Ohtsuki, N. Takahashi, M. Endo, K. Hidaka, H. Sugiyama, Y. Takahashi, Y. Takakura and M. Nishikawa, Self-Assembling DNA Dendrimer for Effective Delivery of Immunostimulatory CpG DNA to Immune Cells, Biomacromolecules, 2015, 16, 1095–1101 CrossRef CAS.
- S. Ohtsuki, N. Matsuzaki, K. Mohri, M. Endo, T. Emura, K. Hidaka, H. Sugiyama, Y. Takahashi, K. Ishiyama, N. Kadowaki, Y. Takakura and M. Nishikawa, Optimal Arrangement of Four Short DNA Strands for Delivery of Immunostimulatory Nucleic Acids to Immune Cells, Nucleic Acid Ther., 2015, 25, 245–253 CrossRef CAS PubMed.
- Y. Umeki, K. Mohri, Y. Kawasaki, H. Watanabe, R. Takahashi, Y. Takahashi, Y. Takakura and M. Nishikawa, Induction of Potent Antitumor Immunity by Sustained Release of Cationic Antigen from a DNA-Based Hydrogel with Adjuvant Activity, Adv. Funct. Mater., 2015, 25, 5758–5767 CrossRef CAS.
- T. Yata, Y. Takahashi, M. Tan, H. Nakatsuji, S. Ohtsuki, T. Murakami, H. Imahori, Y. Umeki, T. Shiomi, Y. Takakura and M. Nishikawa, DNA nanotechnology-based composite-type gold nanoparticle-immunostimulatory DNA hydrogel for tumor photothermal immunotherapy, Biomaterials, 2017, 146, 136–145 CrossRef CAS PubMed.
- G. Chen, D. Liu, C. He, T. R. Gannett, W. Lin and Y. Weizmann, Enzymatic synthesis of periodic DNA nanoribbons for intracellular pH sensing and gene silencing, J. Am. Chem. Soc., 2015, 137, 3844–3851 CrossRef CAS.
- M. Erkelenz, D. M. Bauer, R. Meyer, C. Gatsogiannis, S. Raunser, B. Saccà and C. M. Niemeyer, A facile method for preparation of tailored scaffolds for DNA-origami, Small, 2014, 10, 73–77 CrossRef CAS.
- E. Pound, J. R. Ashton, H. A. Becerril and A. T. Woolley, Polymerase chain reaction based scaffold preparation for the production of thin, branched DNA origami nanostructures of arbitrary sizes, Nano Lett., 2009, 9, 4302–4305 CrossRef CAS PubMed.
- R. A. Petros and J. M. Desimone, Strategies in the design of nanoparticles for therapeutic applications, Nat. Rev. Drug Discovery, 2010, 9, 615–627 CrossRef CAS.
- S. M. Douglas, A. H. Marblestone, S. Teerapittayanon, A. Vazquez, G. M. Church and W. M. Shih, Rapid prototyping of 3D DNA-origami shapes with caDNAno, Nucleic Acids Res., 2009, 37, 5001–5006 CrossRef CAS.
- Y. Kimura, K. Sonehara, E. Kuramoto, T. Makino, S. Yamamoto, T. Yamamoto, T. Kataoka and T. Tokunaga, Binding of oligoguanylate to scavenger receptors is required for oligonucleotides to augment NK cell activity and induce IFN, J. Biochem., 1994, 116, 991–994 CrossRef CAS.
- L. Benimetskaya, J. D. Loike, Z. Khaled, G. Loike, S. C. Silverstein, L. Cao, J. El Khoury, T. Q. Cai and C. A. Stein, Mac-1 (CD11b/CD18) is an oligodeoxynucleotide-binding protein, Nat. Med., 1997, 3, 414–420 CrossRef CAS.
- C. M. Sirois, T. Jin, A. L. Miller, D. Bertheloot, H. Nakamura, G. L. Horvath, A. Mian, J. Jiang, J. Schrum, L. Bossaller, K. Pelka, N. Garbi, Y. Brewah, J. Tian, C. Chang, P. S. Chowdhury, G. P. Sims, R. Kolbeck and A. J. Coyle, et al. RAGE is a nucleic acid receptor that promotes inflammatory responses to DNA, J. Exp. Med., 2013, 210, 2447–2463 CrossRef CAS.
- D. C. Siess, C. T. Vedder, L. S. Merkens, T. Tanaka, A. C. Freed, S. L. McCoy, M. C. Heinrich, M. E. Deffebach, R. M. Bennett and S. H. Hefeneider, A human gene coding for a membrane-associated nucleic acid-binding protein, J. Biol. Chem., 2000, 275, 33655–33662 CrossRef CAS.
- A. P. Moseman, E. A. Moseman, S. Schworer, I. Smirnova, T. Volkova, U. von Andrian and A. Poltorak, Mannose Receptor 1 Mediates Cellular Uptake and Endosomal Delivery of CpG-Motif Containing Oligodeoxynucleotides, J. Immunol., 2013, 191, 5615–5624 CrossRef CAS.
- M. H. Lahoud, F. Ahmet, J.-G. Zhang, S. Meuter, A. N. Policheni, S. Kitsoulis, C.-N. Lee, M. O'Keeffe, L. C. Sullivan, A. G. Brooks, R. Berry, J. Rossjohn, J. D. Mintern, J. Vega-Ramos, J. A. Villadangos, N. A. Nicola, M. C. Nussenzweig, K. J. Stacey and K. Shortman, et al. DEC-205 is a cell surface receptor for CpG oligonucleotides, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 16270–16275 CrossRef CAS.
- S. Ohtsuki, Y. Takahashi, T. Inoue, Y. Takakura and M. Nishikawa, Reconstruction of Toll-like receptor 9-mediated responses in HEK-Blue hTLR9 cells by transfection of human macrophage scavenger receptor 1 gene, Sci. Rep., 2017, 7, 13661 CrossRef.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9nr08695a |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2019 |