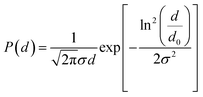Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Maghemite nanoparticles stabilize the protein corona formed with transferrin presenting different iron-saturation levels†
Ulrike
Martens
ab,
Dominique
Böttcher
a,
Delphine
Talbot
c,
Uwe
Bornscheuer
a,
Ali
Abou-Hassan
 c and
Mihaela
Delcea
c and
Mihaela
Delcea
 *ab
*ab
aInstitute of Biochemistry, University of Greifswald, Felix-Hausdorff-Str. 4, 17489 Greifswald, Germany. E-mail: delceam@uni-greifswald.de; Fax: +4938344204377; Tel: +4938344204423
bZIK HIKE – Center for Innovation Competence “Humoral Immune Reactions in Cardiovascular Diseases”, University of Greifswald, Fleischmannstr. 42-44, 17489 Greifswald, Germany
cSorbonne Université, CNRS, Physico-chimie des Electrolytes et Nanosystèmes InterfaciauX, F-75005 Paris, France
First published on 29th July 2019
Abstract
Magnetic nanoparticles are ideal candidates for biomedical applications given their potential use in magnetic resonance imaging, magnetic hyperthermia and targeted drug delivery. Understanding protein–nanoparticle interactions in the blood stream is of major importance due to their potential risks, especially immunogenicity (i.e. the ability to induce an immune response). Here, we report on the interaction of superparamagnetic maghemite (γ-Fe2O3) nanoparticles with human blood plasma protein transferrin presenting different iron-saturation levels: partially iron-saturated (i.e. transferrin) and iron-free transferrin (i.e. apotransferrin). The nanoparticle–protein interaction and the protein corona formation were studied using biophysical and chemical approaches based on dynamic light scattering, gel electrophoresis, circular dichroism spectroscopy and differential scanning fluorimetry. We found that iron content governs the protein corona formation and induces a strong effect on the thermal stability of the bound protein. Our results demonstrate a stabilizing effect of the nanoparticles with a change of the unfolding position of approximately 10 °C towards higher temperatures for transferrin. Our study may be relevant for the further development of magnetic nanoparticles as diagnostic and therapeutic tools.
Introduction
Magnetic nanoparticles (NPs) are highly in focus in biomedical research due to their unique properties which allow magnetic separation and target-oriented positioning. These features result in a large scale of applications which are summarized with three generic terms: separation, diagnosis and therapy. For example, magnetic NPs are well established as drug carriers and magnetic resonance imaging contrast agents as well as for hyperthermia therapy.1–8Several studies deal with the question of the processes that evolve after the injection of nanoparticles (e.g. magnetic NPs) into the blood stream.9–17 Upon entering the human body, NPs may interact with plasma proteins forming a protein shell called a “protein corona”. The NP–protein interaction may result in an alteration of the protein conformation which could lead to loss of protein function, aggregation, bio-incompatibility and thus, potential immunogenicity (i.e. the ability to induce an immune response).18–23 Therefore, to characterize and understand the NP–protein interactions in blood, knowledge needs to be gained about the identity and retention time of the protein corona. In addition, proteins are in a continuous dynamical cycle between adsorption on the surface of the NPs and desorption due to competing exchange processes.10,24,25 Understanding of such processes is a requirement for the design of functional and safe nanomaterials e.g. for drug delivery as changes in the protein structure and related reactions (e.g. immunogenicity of nanoparticles) can cause diseases.
Transferrin (Mw = 78 kDa) is a blood protein containing two distinct homologous domains (C- and N-lobe) each presenting an iron-binding site coupled by a short peptide linker. Its main function is to deliver iron to all biological tissues.26 The binding of iron includes bicarbonate binding acting as a synergistic anion.27 As a result, a red complex is formed which can be observed by the absorbance at 470 nm.27–30 Iron-uptake and release is a complex process which is also mediated by domain–domain communication.31 In healthy humans, the transferrin serum concentration is about 25–50 μM with a saturation rate of approximately 30%, composed of approximately 39% iron-free, 23% monoferric N-lobe, 11% monoferric C-lobe and 27% diferric.32
Furthermore, the combination of NPs and transferrin was studied in terms of tumor targeting in cancer research.33–35 Li et al.34 demonstrated a six-fold increase in cancerous cell-uptake due to the modification of gold NPs with transferrin compared to a four times increase in uptake in non-cancerous cells. It is well known that cancer cells have a higher demand for iron and thus, overexpress the transferrin receptor.36–38 Protein corona formation through the interaction of superparamagnetic iron oxide NPs and serum proteins was studied e.g. by Sakulkhu et al.13 who found that transferrin is present in the protein hard corona of positively, negatively and neutral charged polyvinyl alcohol (PVA)- and dextran-coated NPs. Besides these studies, little is known about the influence of the NP corona formation on the protein itself. Here we direct our focus to possible structural changes of the protein, especially the thermal stability of the protein in the bioconjugates with NPs.
In this study, we investigated the interactions of superparamagnetic maghemite (γ-Fe2O3) NPs with human transferrin presenting different iron-saturation levels: the partially iron-saturated (i.e. transferrin) and the iron-free transferrin (i.e. apotransferrin). Superparamagnetic iron oxide nanoparticles (SPIONs) obtained by the co-precipitation method were characterized by transmission electron microscopy (TEM) and dynamic light scattering (DLS). Furthermore, the NP–protein interaction and the protein corona formation upon magnetic separation by an external magnetic field were analysed by various biophysical methods including DLS, gel electrophoresis, circular dichroism spectroscopy (CD) and differential scanning fluorimetry (DSF). Here, also the influence of applied static magnetic fields (SMF) on the protein corona formation was evaluated. In addition, the effect of NPs on the structure and thermal stability of the protein in relation to the applied temperature was studied. Research reported in this work gives new insight into the influence of iron-loading of transferrin on the interaction with NPs. It is shown that the iron content not only plays a decisive role in protein corona formation, furthermore bioconjugation also has a strong effect on the thermal stability of the bound protein which should be considered when using magnetic NPs for e.g. drug delivery applications, especially in combination with hyperthermia.
Experimental
Two different types of transferrin, which differ in iron-loading level, were used in this study. Partially iron-saturated as well as iron-free transferrin (apotransferrin) were compared in the experiments. Partially iron-saturated human transferrin (purity ≥98%) was purchased from Sigma (Merck, Darmstadt, Germany). Human apotransferrin was ordered from neoLab (neoFroxx, Einhausen, Germany).The different iron-loading levels of the proteins used for the interaction studies were verified by urea gel analysis which is a common method to determine the iron distribution in transferrin.26,32,39 Denaturing urea gel (6% TBE-Urea gel, Invitrogen by Thermo Fisher) was used to monitor the iron-binding status of the purchased proteins. Proteins were loaded in different concentrations into the gel. The gel was electrophoresed for 135 min at 125 V.
Protein concentrations were determined based on the Beer–Lambert equation using a spectrophotometer (NanoDrop 2000, Thermo Fischer Scientific, Darmstadt, Germany) to monitor the absorbance at 280 nm.
Citrate-stabilized maghemite nanoparticles
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ξ − 1/ξ), where ξ = μ0msVH/kT is the Langevin parameter with the vacuum magnetic permeability μ0, the saturation magnetization of the magnetic material ms, the magnetic field H, the Boltzmann constant k, the temperature T, the particle magnetic volume V, and the volume fraction of particles in the suspension φ. For a polydisperse sample, M(H) was well adjusted by weighting the Langevin expression by a log-normal distribution of the particle diameter d.
ξ − 1/ξ), where ξ = μ0msVH/kT is the Langevin parameter with the vacuum magnetic permeability μ0, the saturation magnetization of the magnetic material ms, the magnetic field H, the Boltzmann constant k, the temperature T, the particle magnetic volume V, and the volume fraction of particles in the suspension φ. For a polydisperse sample, M(H) was well adjusted by weighting the Langevin expression by a log-normal distribution of the particle diameter d.The fit of the theoretical expression to the experimental magnetization curve allowed us to determine the characteristic magnetic diameter d0,mag and the polydispersity index σmag.
Protein corona formation
The synthesized γ-Fe2O3 nanoparticles were diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 in phosphate buffered saline (PBS, pH 7.4) and a volume of 100 μL was added to 500 μL of 2 mg mL−1 protein stock solution of transferrin (Tf) or apotransferrin (ApoTf) and filled with PBS to a total volume of 1 mL. Samples were incubated overnight at 4 °C without shaking. Of note, the used high protein concentration, as well as the long incubation time were carefully selected to reach a high saturation level of the nanoparticles, which does not mimic physiological conditions. The incubation temperature was chosen to protect the protein from unfolding, based on the storage temperature emphasized in the data sheet of the distributing company. DLS measurements via multi-angle detection (Zetasizer Ultra, Malvern Panalytical, Kassel, Germany) revealed a content of ∼1.5 × 1011 maghemite NPs of the samples with an iron content of 1.17 mM. The bare NPs and their conjugates with proteins before and after purification via magnetic separation were characterized by dynamic light scattering (DLS). In addition, the bioconjugates consisting of nanoparticles with attached proteins were analysed after magnetic purification using gel electrophoresis (SDS PAGE), western blot, bicinchoninic acid assay (BCA), circular dichroism spectroscopy (CD) and differential scanning fluorimetry (nanoDSF).
100 in phosphate buffered saline (PBS, pH 7.4) and a volume of 100 μL was added to 500 μL of 2 mg mL−1 protein stock solution of transferrin (Tf) or apotransferrin (ApoTf) and filled with PBS to a total volume of 1 mL. Samples were incubated overnight at 4 °C without shaking. Of note, the used high protein concentration, as well as the long incubation time were carefully selected to reach a high saturation level of the nanoparticles, which does not mimic physiological conditions. The incubation temperature was chosen to protect the protein from unfolding, based on the storage temperature emphasized in the data sheet of the distributing company. DLS measurements via multi-angle detection (Zetasizer Ultra, Malvern Panalytical, Kassel, Germany) revealed a content of ∼1.5 × 1011 maghemite NPs of the samples with an iron content of 1.17 mM. The bare NPs and their conjugates with proteins before and after purification via magnetic separation were characterized by dynamic light scattering (DLS). In addition, the bioconjugates consisting of nanoparticles with attached proteins were analysed after magnetic purification using gel electrophoresis (SDS PAGE), western blot, bicinchoninic acid assay (BCA), circular dichroism spectroscopy (CD) and differential scanning fluorimetry (nanoDSF).
Dynamic light scattering measurements
The hydrodynamic diameter (calculated from the intensity weighted distribution) and the zeta potential of the samples were determined using a Zetasizer Nano-ZS (Malvern Instruments, Kassel, Germany). Samples were loaded into 10 mm pathlength cuvettes (BRAND, Wertheim, Germany) and left to equilibrate at 25 °C for 2 min before starting measurements. The instrument was used in automatic mode with three measurements for each sample and depending on quality criteria of the Zetasizer Nano, the number of runs was adapted. The data were collected with the detector positioned at an 173° angle to the sample cell. The recorded data were analyzed using the Zetasizer software ZS. Zeta potential measurements were carried out at an applied voltage of max. 10 V. Each measurement consisted of 20 runs with a run duration of 10 s. The presented data are averaged over six independent samples. Comparison between different sample groups was done by one-way-ANOVA in combination with the Tukey test.SDS-PAGE and western blot analysis
Protein binding to maghemite NPs was verified by sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE). For SDS-PAGE experiments, samples with a total volume of 1 mL were incubated in magnetic racks to isolate the protein loaded NPs. After pelleting and discarding the supernatant, the tube was refilled with PBS. The washing step was repeated three times and the final pellet was resuspended in a 20 μL mixture of PBS, reducing agent (NuPAGE, Invitrogen) and sample buffer (Tris-Glycin SDS, Novex). After 5 min heat-treatment at 95 °C, a sample volume of 14 μL was loaded into 8–16% gradient Tris-Glycin gel (Novex, Wedge Well, Invitrogen by Thermo Fisher Scientific, Darmstadt, Germany). The protein bands were visualized by SimplyBlue Coomassie stain (Thermo Fisher Scientific). All samples were run in triplicate.For western blot preparation, the bands were blotted to a nitrocellulose membrane (Amersham Protran 0.2 μm NC, GE Healthcare) for 40 min at 20 V. Blocking was done at room temperature with a 5% milk powder solution in Tris-buffered saline (TBS) with 0.05% Tween20 (TBST) for 1 h. Afterwards, the membrane was washed three times with TBST and incubated with the monoclonal anti-Tf antibody HTF-14 (ABIN94486, antibodies-online, Aachen, Germany) at 4 °C overnight in 3% bovine serum albumin and 10% NaN3 TBST. Next, three washing steps were applied followed by the incubation with the secondary goat anti-mouse antibody coupled with HRP in blocking buffer for 1 h at room temperature. After washing three times, the membrane was stained using a chemiluminescent substrate (SuperSignalR West Pico, Thermo Fischer Scientific) to visualize the specific antibody binding.
Circular dichroism spectroscopy
Circular dichroism (CD) spectroscopy was applied to monitor changes in the secondary structure of the protein which may occur after the interaction with the magnetic NPs. CD spectra were recorded using a Chirascan CD spectrometer (Applied Photophysics, Leatherhead, UK) together with a temperature controller (Quantum Northwest, Liberty Lake, USA). Measurements in the far-UV (198–270 nm) were carried out using 5 mm path length cuvettes (Hellma Analytics, Müllheim, Germany). CD spectra were recorded with a bandwidth of 1.0 nm, a scanning time of 1.5 s per point and five repetitions. For measurements in the absence of NPs, the protein concentration was adapted to 50 μg mL−1. All samples were diluted in PBS. In addition, all spectra were blank corrected. After incubation overnight at 4 °C, magnetic NPs with a protein corona were isolated from non-bound proteins using magnetic columns (MS MACS, Miltenyi Biotec, Teterow, Germany) and resuspended in 1 mL PBS. For CD measurements, samples were diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 (sample
2 (sample![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) PBS). The CD spectra as a function of temperature were tracked at the maximum wavelengths of 210 nm and 218 nm. Samples were heated at a rate of 0.3 °C min−1, while the temperature in the sample cuvette was continuously probed. The data were traced in 0.5 °C steps in three repeats while the applied temperature was set from 40 °C to 94 °C. In addition, the CD spectra were recorded before and after each heating cycle at 25 °C. All measurements were repeated at least three times. The protein concentration of the protein-loaded NPs as well as the flow through the fraction were determined using the bicinchoninic acid assay (BCA kit, Sigma Aldrich).
PBS). The CD spectra as a function of temperature were tracked at the maximum wavelengths of 210 nm and 218 nm. Samples were heated at a rate of 0.3 °C min−1, while the temperature in the sample cuvette was continuously probed. The data were traced in 0.5 °C steps in three repeats while the applied temperature was set from 40 °C to 94 °C. In addition, the CD spectra were recorded before and after each heating cycle at 25 °C. All measurements were repeated at least three times. The protein concentration of the protein-loaded NPs as well as the flow through the fraction were determined using the bicinchoninic acid assay (BCA kit, Sigma Aldrich).
Differential scanning fluorimetry
For differential scanning fluorimetry (DSF) measurements, magnetic NPs with a protein corona were isolated from non-binding proteins using magnetic columns (MACS, Miltenyi Biotec). DSF studies were carried out using a Prometheus NT.48 equipment (NanoTemper technologies GmbH, München, Germany) in combination with high sensitivity capillaries which allow the detection of fluorescence based on tryptophan and tyrosine at protein concentrations ranging from 5 μg mL−1. The instrument was set to record the fluorescence signal at 330 nm and 350 nm within the temperature range from 20 to 95 °C with a ramp of 1 °C min−1. The thermal unfolding process of a protein can be described by a sigmoidal function. Protein unfolding causes changes in the emission intensity as well as in the wavelength maximum. To track changes and thus, the unfolding positions, the first derivative dy/dx of the fluorescence ratio (350 nm/330 nm) data is evaluated. Data analysis was performed using NT Melting Control software (NanoTemper Technologies GmbH). In detail, the melting temperature TM was determined by analysing the first derivative of the tryptophan fluorescence emission ratio of 350 nm/330 nm. The maximum peak of the first derivative curve indicates the unfolding position of the protein. For plotting the data of the first derivative of the fluorescence ratio, the high density of data points needs to be reduced by removing equidistant x-data to apply the analysis of the first derivative.Results and discussion
Maghemite nanoparticles and bioconjugation with apotransferrin and transferrin
The synthesized γ-Fe2O3-NPs dispersed in water have a rock-like shape with an average physical diameter of ≈10.2 ± 0.11 nm as deduced from TEM imaging (Fig. S1A and B†). Magnetization curve analysis (Fig. S1C and D†) provided a magnetic diameter d0,mag = 9 nm and a polydispersity index σmag = 0.23. The zeta potential of the citrate-stabilized NPs in ddH2O (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000) was ∼−85 mV ± 9 mV which indicates the high electrostatic stability of the colloidal suspension.
1000) was ∼−85 mV ± 9 mV which indicates the high electrostatic stability of the colloidal suspension.
The iron-binding status of the two different forms of transferrin used for the bioconjugate formation was verified by urea gel analysis. The Coomassie-stained gel (Fig. S2A†) displays one band for the iron-free apotransferrin (ApoTf), while for the partly iron-saturated transferrin (Tf) three bands appear. One band is attributed to the iron-free form, while the additional bands display more stable forms of transferrin due to the iron-binding which results in a more compact structure. The SDS gel analysis (Fig. S2B†) of the proteins in different concentrations confirmed the purity of transferrin in both the iron-free and the partly iron-saturated forms with one strong band with a molecular weight of around 80 kDa.
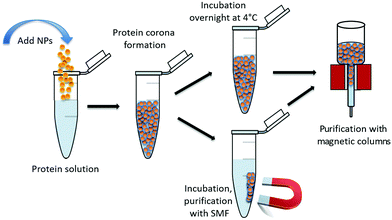
The experiments were carried out as schematically shown in Fig. 1 which indicates the protein corona formation, the incubation parameters and the used purification methods.
The hydrodynamic diameter and the zeta potential of the formed bioconjugates under physiological conditions (PBS, pH 7.4) after overnight incubation at 4 °C without further purification were determined.
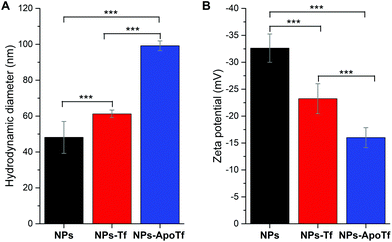
Fig. 2 displays the data for the hydrodynamic size (A) and the zeta potential (B) of the citrate-stabilized maghemite NPs and their bioconjugates with Tf or ApoTf in PBS (pH 7.4). The γ-Fe2O3-NPs reveal a hydrodynamic diameter of approximately 48 nm with no aggregation as supported by the autocorrelation function (data not shown). The difference between the nanoparticle's size obtained from TEM and DLS measurements is due to the fact that the hydrodynamic size (calculated from the intensity weighted distribution) measured by DLS represents the size of the nanoparticle plus the liquid layer around the particle while the size measured by TEM gives the physical size of the nanoparticle. Protein corona formation was confirmed as the size increases upon bioconjugation with partially saturated transferrin (to 61 nm) or with iron-free apotransferrin (to 99 nm). Furthermore, statistical analysis verified that the hydrodynamic diameter for NPs-Tf (p = 0.002) and for NPs-ApoTf (p < 10−14) in comparison with NPs significantly increased. The data for NPs-Tf and NPs-ApoTf (p < 10−14) also display a significant variation.
The electrophoretic mobility measurements (Fig. 2B) showed negatively charged citrate-stabilized magnetic particles and negatively charged bioconjugates of Tf or ApoTf. The zeta potential of the maghemite NPs in PBS decreased from −33 mV to −23 mV and −16 mV upon adsorption with Tf and ApoTf, respectively. Statistical tests showed that the zeta potential of the single groups (NPs to NPs-ApoTf, p < 10−14; NPs to NPs-Tf, p = 2.3 × 10−5 and NPs-Tf to NPs-ApoTf, p = 3.6 × 10−4) significantly differs. The change in the hydrodynamic diameter correlates directly with the zeta potential. The theoretical isoelectric point of transferrin is given as 6.8. Therefore, the charge of transferrin at pH 7.4 in PBS can be assumed as slightly negative or even neutral. As a result, the zeta potential of the bioconjugate decreases with increasing amount of attached protein. Furthermore, we can state that the nanoparticles are stabilized through steric instead of electrostatic interactions with the attached proteins.
The effect of magnetic field on protein corona formation
Previous studies42–44 demonstrated that an applied static magnetic field (SMF) can affect the proteins and the protein corona. In particular, it was shown that SMFs influence the composition and the amount of proteins attached to the nanoparticles. For this reason, we evaluated the influence of an applied magnetic field during the incubation in our self-made magnetic racks as well as the separation method with magnetic columns to prove possible effects of different separation methods on the protein corona.The size of the maghemite NPs and their bioconjugates (incubated overnight at 4 °C) before and after magnetic separation with MACS columns is displayed in the ESI (Fig. S3A†). No significant difference in size was found between samples before and after purification with magnetic columns. The size was also monitored after incubation with an applied static magnetic field (SMF) to evaluate the influence on the protein corona. Because no variations in the hydrodynamic diameter of the samples with and without an applied static magnetic field were measured, it can be concluded that the SMF did not affect the protein corona. This was also supported by zeta potential measurements (Fig. S3B†) which indicate as well no significant differences between samples with and without purification with magnetic columns (around −33 mV for the NPs in PBS, −23 mV for the conjugates with Tf and around −17 mV for ApoTf).
In addition to size measurements which confirmed the protein corona formation, SDS-PAGE analyses (Fig. S4A†) and BCA assays were carried out to validate protein corona formation. Gel electrophoresis exhibits a typical band at ∼80 kDa for transferrin in the bioconjugation samples. Furthermore, several additional bands with a lower molecular weight appeared which was already observed by Pitek et al.45 Western blot analysis (Fig. S4B†) confirmed for all bioconjugate samples only one band corresponding to a molecular weight of 80 kDa. The protein concentration of the isolated NPs–protein corona complexes as analysed using a BCA kit revealed an ApoTf concentration of ∼120 μg mL−1 and a Tf concentration of ∼100 μg mL−1. These data show good agreement with the size measurements. The conjugates with ApoTf show a larger increase in size as well as a higher determined protein concentration than the conjugates with partially iron-saturated Tf and thus, a higher affinity of the iron-free protein to the maghemite NPs.
Protein corona thermal stability analysis
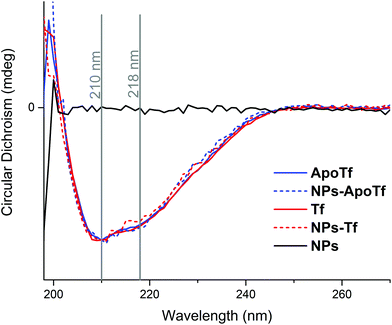
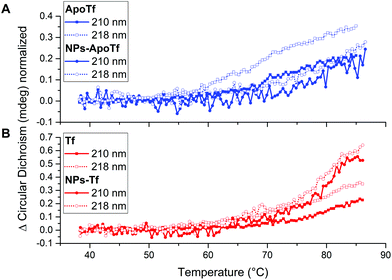
The thermal stability of the protein and NP–protein complexes was monitored by CD spectroscopy and differential scanning fluorimetry. Fig. 3 depicts the normalized CD spectra in the range of 198 nm to 270 nm of ApoTf and Tf as well as their bioconjugates formed after overnight incubation with NPs at 4 °C and purification with magnetic columns. The CD data detected at 25 °C show no signal differences, which indicates that the iron loading does not significantly alter the secondary structure of transferrin. In general, the composition of Tf slightly differs with iron-saturation. Iron-free Tf (ApoTf) is known to consist of 17% α-helix and 68% β-sheet, while fully saturated Tf displays a content of 23% α-helix and 62% β-sheet.46,47 The CD data in Fig. 3 display the spectra for iron-free and partly saturated transferrin.The saturation-level can be suggested to be insufficient to display differences to the iron-free form. Furthermore, the attachment of the protein to the NPs and their interactions did not lead to changes in the detectable structure. In comparison, a CD study of the run-through fraction of magnetic column separation by Mahmoudi et al.48 showed that irreversible changes of the iron-saturated transferrin conformation appeared after the interaction with SPIONs. They interpreted the structural change as a result of iron loss. Here, we analyzed the isolated fraction of NPs with a protein corona where the initial proteins present different iron-loading levels. The data display two maximum positions, at 210 nm and 218 nm. At these wavelengths, the CD signal was measured in dependence to a heating cycle up to an applied temperature of 94 °C. Here it has to be mentioned that the probed sample temperature reached a maximum of around 86 °C. The corresponding data are shown in Fig. 4.
The normalized signal change for ApoTf and the bioconjugates is displayed in Fig. 4A. The data for 210 nm are illustrated by a solid line, while 218 nm data are depicted by a dotted line. All samples display a change in signal and a gradually changing protein structure starting at 60 °C. ApoTf undertook more drastic changes in the CD signal in relation to the applied temperature than the ApoTf attached to maghemite NPs.
The corresponding recorded data are shown in Fig. 4B. Here, also differences concerning the CD signal change are observable. The conjugated Tf displays more rapid changes than the protein alone. It has to be noted, that the structural changes appeared steadily without defined turning points. Those changes in the CD signal can be interpreted in different ways e.g. concentration differences (protein and NP) and the influence of the NP itself. However, CD results do not enable one to make statements concerning the stability of the protein or the conjugated protein. Moreover, the data do not show a plateau displaying the denaturized state of the protein and therefore the determination of the melting temperature TM of the protein via a sigmoidal fit is not possible.
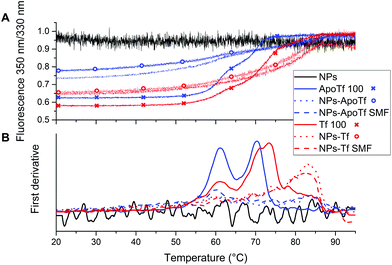
Because transferrin contains 26 tyrosine residues and 8 tryptophan residues, it allows the detection of its internal fluorescence. We used nanoDSF to determine the thermal unfolding characteristic of the protein by measuring the fluorescence at 350 nm and 330 nm. The first derivative of the integrated fluorescence ratio reveals the inflection points depicted in Fig. 5. The determined inflection points for all samples are listed in Table 1.
| Sample ID | Inflection point #1 (°C) | Inflection point #2 (°C) |
|---|---|---|
| NPs | — | — |
| ApoTf | 61.3 ± 0 | 69.8 ± 0 |
| NPs-ApoTf | 59.8 ± 0.4 | 83.1 ± 0.4 |
| NPs-ApoTf SMF | 60.3 ± 0.3 | 83.2 ± 0.3 |
| Tf | 60.6 ± 0.3 | 72.2 ± 0.1 |
| NPs-Tf | 61.3 ± 0.5 | 82.6 ± 0.3 |
| NPs-Tf SMF | 61.1 ± 0.6 | 82.4 ± 0.2 |
The data for the proteins alone with a concentration of 100 μg mL−1 are illustrated by solid lines. All samples reveal two unfolding positions attributed to the two domains. ApoTf unfolding took place at ∼60 °C and ∼70 °C. For Tf, the second inflection point is slightly shifted towards higher temperatures (72 °C). This slight change in the position is attributed to the more compact form of the partly iron-saturated Tf resulting in increased stability. The NP–protein conjugates exhibit changed characteristics. First, the fluorescence signal is subdued even though a similar protein concentration is present for the conjugates as shown by the BCA assay. In addition, the second inflection point illustrates a significant shift towards higher temperatures. Both, Tf and ApoTf conjugated with maghemite NPs display the second inflection point at the position of ∼83 °C which is a clear indicator of the stabilizing effect of the NPs, while bare NPs (black solid line) showed only a noise signal.
The highly sensitive nanoDSF reveals the unfolding positions at approximately 60 °C and the resolution of a second inflection point. While the CD results do not provide interpretable data concerning the effect of the NPs on the protein stability, nanoDSF displays a stabilizing effect for both proteins due to the shift of the second inflection position to higher temperatures.
NanoDSF reveals a high sensitivity to track unfolding positions of the protein and the protein–nanoparticle-conjugates.
However, the data for the pure protein are in good agreement with the literature. Lin et al.49 demonstrated by differential scanning calorimetry (DSC) the changing transition positions in dependence to the iron-loading of the protein and thus, the increasing thermal stability with increasing iron-saturation. This was also verified by Hadden et al.50 in other DSC studies which showed two transition states (at 61 °C and 73 °C) for ApoTf and one transition point at 93 °C for the fully saturated Tf.
The reasons for the stabilizing effect of the nanoparticles in our experiments cannot be fully explained by our data. We can hypothesise an increase in the iron-saturation level of the protein due to bioconjugate formation. Iron-uptake would then lead to changes in the secondary structure of the protein which was not observed in our CD spectra (Fig. 3). Most probably, those changes are insignificant.
In addition, transferrin needs a synergistic anion, usually bicarbonate, for iron-binding which is not present in the used buffer.
Another possible explanation involves the adsorption of amino acids onto the maghemite nanoparticle surface without affecting the protein structure. The adsorption characteristics for aspartic acid and lysine onto iron oxide nanoparticles are for example demonstrated by Pušnik et al.51 Consequently, a higher energy and thus a higher temperature are required to break the bonds which may explain the shift of +10 °C in the unfolding temperature deduced from DSF experiments.
Conclusions
In this study we have investigated the interaction of superparamagnetic maghemite (γ-Fe2O3) NPs with two different forms of iron-binding proteins (transferrin and apotransferrin). We used the unique advantage of magnetic nanoparticles which is they can be purified by magnetic separation. The purification method enabled to isolate the formed bioconjugates from free unbound proteins in the solution. The thermal stability of proteins attached to NPs in comparison with pure proteins was compared and studied for the first time over a broad temperature range. We found that iron content governs the protein corona formation. Iron-free transferrin forms a larger protein corona than the partially iron-saturated transferrin. In addition, we have demonstrated that the protein secondary structure is not significantly altered by the iron-saturation of the protein as shown by CD spectroscopy data. NanoDSF results verified a stabilizing effect of NPs for both proteins. Our study may be relevant for nanomedicine and biomedical field communities where surface engineered magnetic nanoparticles including a transferrin protein corona are used for targeting and drug delivery coupled or not to thermal therapies where temperatures up to 50 °C are generated in human tissues. Here, we present the stabilisation from thermal unfolding. However, the stabilizing effect is not assumed to be solely restricted to magnetic nanoparticles. Our investigations present the first study in this research direction. Future experiments should also address the stabilizing effect of e.g. gold and silver nanoparticles. It should also be considered that the stabilizing effect is not limited to thermal processes and can also be present for other stress parameters. Furthermore, the ability to stabilize proteins while conserving their structure is of main interest in studying protein misfolding diseases, e.g. Alzheimer's, Wilson and Fabry disease. Future experiments should also address the stabilizing effect of other types of nanoparticles and other proteins. In addition, it should be verified whether the function of the protein is preserved.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank Ina Buchholz for help with CD data interpretation. The financial support by the Bundesministerium für Bildung und Forschung (BMBF) within the NanoImmun Project (FKZ 03Z22C51) is gratefully acknowledged.References
- J. S. Weinstein, C. G. Varallyay, E. Dosa, S. Gahramanov, B. Hamilton, W. D. Rooney, L. L. Muldoon and E. A. Neuwelt, J. Cereb. Blood Flow Metab., 2010, 15–35 CrossRef CAS PubMed.
- W. Wu, Z. Wu, T. Yu, C. Jiang and W.-S. Kim, Sci. Technol. Adv. Mater., 2015, 16, 23501 CrossRef PubMed.
- S. M. Dadfar, K. Roemhild, N. I. Drude, S. von Stillfried, R. Knüchel, F. Kiessling and T. Lammers, Adv. Drug Delivery Rev., 2019, 138, 302–325 CrossRef CAS PubMed.
- T. Neuberger, B. Schöpf, H. Hofmann, M. Hofmann and B. von Rechenberg, J. Magn. Magn. Mater., 2005, 293, 483–496 CrossRef CAS.
- H. Xu, Z. P. Aguilar, L. Yang, M. Kuang, H. Duan, Y. Xiong, H. Wei and A. Wang, Biomaterials, 2011, 32, 9758–9765 CrossRef CAS PubMed.
- A. Espinosa, M. Bugnet, G. Radtke, S. Neveu, G. A. Botton, C. Wilhelm and A. Abou-Hassan, Nanoscale, 2015, 7, 18872–18877 RSC.
- A. Espinosa, J. Kolosnjaj-Tabi, A. Abou-Hassan, A. Plan Sangnier, A. Curcio, A. K. A. Silva, R. Di Corato, S. Neveu, T. Pellegrino, L. M. Liz-Marzán and C. Wilhelm, Adv. Funct. Mater., 2018, 28, 1803660 CrossRef.
- A. Curcio, A. K. A. Silva, S. Cabana, A. Espinosa, B. Baptiste, N. Menguy, C. Wilhelm and A. Abou-Hassan, Theranostics, 2019, 9, 1288–1302 CrossRef CAS PubMed.
- M. A. Dobrovolskaia and S. E. McNeil, Nat. Nanotechnol., 2007, 2, 469–478 CrossRef CAS PubMed.
- O. Vilanova, J. J. Mittag, P. M. Kelly, S. Milani, K. A. Dawson, J. O. Rädler and G. Franzese, ACS Nano, 2016, 10, 10842–10850 CrossRef CAS PubMed.
- V. Gorshkov, J. A. Bubis, E. M. Solovyeva, M. V. Gorshkov and F. Kjeldsen, Environ. Sci.: Nano, 2019, 6, 1089–1098 RSC.
- L. Lartigue, C. Wilhelm, J. Servais, C. Factor, A. Dencausse, J.-C. Bacri, N. Luciani and F. Gazeau, ACS Nano, 2012, 6, 2665–2678 CrossRef CAS PubMed.
- U. Sakulkhu, M. Mahmoudi, L. Maurizi, J. Salaklang and H. Hofmann, Sci. Rep., 2014, 4, 5020 CrossRef CAS PubMed.
- M. Jansch, P. Stumpf, C. Graf, E. Rühl and R. H. Müller, Int. J. Pharm., 2012, 428, 125–133 CrossRef CAS PubMed.
- N. Feliu, D. Docter, M. Heine, P. Del Pino, S. Ashraf, J. Kolosnjaj-Tabi, P. Macchiarini, P. Nielsen, D. Alloyeau, F. Gazeau, R. H. Stauber and W. J. Parak, Chem. Soc. Rev., 2016, 45, 2440–2457 RSC.
- J. Kolosnjaj-Tabi, Y. Javed, L. Lartigue, J. Volatron, D. Elgrabli, I. Marangon, G. Pugliese, B. Caron, A. Figuerola, N. Luciani, T. Pellegrino, D. Alloyeau and F. Gazeau, ACS Nano, 2015, 9, 7925–7939 CrossRef CAS PubMed.
- B. Pelaz, G. Charron, C. Pfeiffer, Y. Zhao, J. M. de La Fuente, X.-J. Liang, W. J. Parak and P. Del Pino, Small, 2013, 9, 1573–1584 CrossRef CAS PubMed.
- P. Satzer, F. Svec, G. Sekot and A. Jungbauer, Eng. Life Sci., 2016, 16, 238–246 CrossRef CAS PubMed.
- H. Pan, M. Qin, W. Meng, Y. Cao and W. Wang, Langmuir, 2012, 28, 12779–12787 CrossRef CAS PubMed.
- L. Fei and S. Perrett, Int. J. Mol. Sci., 2009, 10, 646–655 CrossRef CAS PubMed.
- L. Shang, Y. Wang, J. Jiang and S. Dong, Langmuir, 2007, 23, 2714–2721 CrossRef CAS.
- I. Lynch and K. A. Dawson, Nano Today, 2008, 3, 40–47 CrossRef CAS.
- J. Wolfram, Y. Yang, J. Shen, A. Moten, C. Chen, H. Shen, M. Ferrari and Y. Zhao, Colloids Surf., B, 2014, 124, 17–24 CrossRef CAS PubMed.
- T. Cedervall, I. Lynch, S. Lindman, T. Berggård, E. Thulin, H. Nilsson, K. A. Dawson and S. Linse, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 2050–2055 CrossRef CAS PubMed.
- P. Aggarwal, J. B. Hall, C. B. McLeland, M. A. Dobrovolskaia and S. E. McNeil, Adv. Drug Delivery Rev., 2009, 61, 428–437 CrossRef CAS PubMed.
- P. Aisen and I. Listowsky, Annu. Rev. Biochem., 1980, 357–393 CrossRef CAS PubMed.
- S. Welch, Transferrin: the iron carrier, CRC Press, Boca Raton, Ann Arbor, London, Tokyo, 1992 Search PubMed.
- D. M. Surgenor, B. A. Koechlin and L. E. Strong, J. Clin. Invest., 1949, 28, 73–78 CrossRef CAS PubMed.
- J. Weaver and S. Pollack, Acta Haematol., 1990, 84, 68–71 CrossRef CAS PubMed.
- G. W. Bates, C. Billups and P. Saltman, J. Biol. Chem., 1967, 242, 2810–2815 CAS.
- S. L. Byrne and A. B. Mason, J. Biol. Inorg. Chem., 2009, 14, 771–781 CrossRef CAS PubMed.
- J. Williams and K. Moreton, Biochem. J., 1980, 185, 483–488 CrossRef CAS PubMed.
- C. H. J. Choi, C. A. Alabi, P. Webster and M. E. Davis, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 1235–1240 CrossRef CAS PubMed.
- J.-L. Li, L. Wang, X.-Y. Liu, Z.-P. Zhang, H.-C. Guo, W.-M. Liu and S.-H. Tang, Cancer Lett., 2009, 274, 319–326 CrossRef CAS PubMed.
- J. Wang, S. Tian, R. A. Petros, M. E. Napier and J. M. Desimone, J. Am. Chem. Soc., 2010, 132, 11306–11313 CrossRef CAS PubMed.
- T. R. Daniels, T. Delgado, G. Helguera and M. L. Penichet, Clin. Immunol., 2006, 121, 159–176 CrossRef CAS PubMed.
- D. Hoegemann-Savellano, E. Bos, C. Blondet, F. Sato, T. Abe, L. Josephson, R. Weissleder, J. Gaudet, D. Sgroi, P. J. Peters and J. P. Basilion, Neoplasia, 2003, 5, 495–506 CrossRef CAS.
- T. R. Daniels, E. Bernabeu, J. A. Rodríguez, S. Patel, M. Kozman, D. A. Chiappetta, E. Holler, J. Y. Ljubimova, G. Helguera and M. L. Penichet, Biochim. Biophys. Acta, 2012, 1820, 291–317 CrossRef CAS PubMed.
- R. W. Evans and J. Williams, Biochem. J., 1980, 541–546 CrossRef CAS PubMed.
- R. Massart, IEEE Trans. Magn., 1981, 17, 1247–1248 CrossRef.
- S. Lefebure, E. Dubois, V. Cabuil, S. Neveu and R. Massart, J. Mater. Res., 1998, 13, 2975–2981 CrossRef CAS.
- A. R. Jones, Mol. Phys., 2016, 114, 1691–1702 CrossRef CAS.
- Z. Liu, X. Zhan, M. Yang, Q. Yang, X. Xu, F. Lan, Y. Wu and Z. Gu, Nanoscale, 2016, 8, 7544–7555 RSC.
- Z. Liu, X. Zhan, X. Xu, Y. Wu and Z. Gu, Part. Part. Syst. Charact., 2018, 35, 1700418 CrossRef.
- A. S. Pitek, D. O'Connell, E. Mahon, M. P. Monopoli, F. Baldelli Bombelli and K. A. Dawson, PLoS One, 2012, 7, e40685 CrossRef CAS PubMed.
- J. Mazurier, J.-P. Aubert, M.-H. Loucheux-Lefevre and G. Spik, FEBS Lett., 1976, 66, 238–242 CrossRef CAS PubMed.
- B. Nagy and S. S. Lehrer, Arch. Biochem. Biophys., 1972, 148, 27–36 CrossRef CAS PubMed.
- M. Mahmoudi, M. A. Shokrgozar, S. Sardari, M. K. Moghadam, H. Vali, S. Laurent and P. Stroeve, Nanoscale, 2011, 3, 1127–1138 RSC.
- L. N. Lin, A. B. Mason, R. C. Woodworth and J. F. Brandts, Biochemistry, 1994, 33, 1881–1888 CrossRef CAS.
- J. M. Hadden, M. Bloemendal, P. I. Haris, S. K. S. Srai and D. Chapman, Biochim. Biophys. Acta, 1994, 1205, 59–67 CrossRef CAS.
- K. Pušnik, M. Peterlin, I. K. Cigić, G. Marolt, K. Kogej, A. Mertelj, S. Gyergyek and D. Makovec, J. Phys. Chem. C, 2016, 120, 14372–14381 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9nr04967c |
| This journal is © The Royal Society of Chemistry 2019 |