Open Access Article
Open Access ArticleStructural determinants of mechanical resistance against breakage of a virus-based protein nanoparticle at a resolution of single amino acids†
María
Medrano
,
Alejandro
Valbuena
,
Alicia
Rodríguez-Huete
and
Mauricio G.
Mateu
*
Centro de Biología Molecular “Severo Ochoa” (CSIC-UAM), Universidad Autónoma de Madrid, 28049 Madrid, Spain. E-mail: mgarcia@cbm.csic.es
First published on 23rd April 2019
Abstract
Virus particles and other protein-based supramolecular complexes have a vast nanotechnological potential. However, protein nanostructures are “soft” materials prone to disruption by force. Whereas some non-biological nanoparticles (NPs) may be stronger, for certain applications protein- and virus-based NPs have potential advantages related to their structure, self-assembly, production, engineering, and/or inbuilt functions. Thus, it may be desirable to acquire the knowledge needed to engineer protein-based nanomaterials with a higher strength against mechanical breakage. Here we have used the capsid of the minute virus of mice to experimentally identify individual chemical groups that determine breakage-related properties of a virus particle. Individual amino acid side chains that establish interactions between building blocks in the viral particle were truncated using protein engineering. Indentation experiments using atomic force microscopy were carried out to investigate the role of each targeted side chain in determining capsid strength and brittleness, by comparing the maximum force and deformation each modified capsid withstood before breaking apart. Side chains with major roles in determining capsid strength against breakage included polar groups located in solvent-exposed positions, and did not generally correspond with those previously identified as determinants of mechanical stiffness. In contrast, apolar side chains buried along the intersubunit interfaces that generally determined capsid stiffness had, at most, a minor influence on strength against disruption. Whereas no correlated variations between strength and either stiffness or brittleness were found, brittleness and stiffness were quantitatively correlated. Implications for developing robust protein-based NPs and for acquiring a deeper physics-based perspective of viruses are discussed.
Introduction
Protein-based nanostructures are raising high expectations for widespread application in biomedicine and nanotechnology.1,2 In particular, engineered viruses and their protein capsids3–5 are eliciting a rapidly growing, trans-disciplinary interest as nanoparticles (NPs) for targeted drug delivery; contrast agents for medical imaging; nanobiosensors; nanocages for confined enzymatic reactions; light-harvesting devices; ferrofluids; templates to produce metallic NPs for novel catalysts, batteries, electronic circuits or memory devices; etc.6–12 Viruses and viral capsids are also guiding the design of artificial protein-based NPs and other nanomaterials.1,2,13–17Protein- and virus-based NPs possess a number of potentially advantageous features compared to other NPs for many biomedical or industrial applications:12 (i) ability to self-assemble; (ii) monodispersity and a precisely defined, reproducible structure and composition; (iii) inbuilt, useful chemical and biological functionalities (e.g., inducible conformational transitions, specific molecular recognition or cell targeting); (iv) suitability for readily engineering collectively precise structural or functional modifications by chemical or genetic means; (v) economical large-scale production; (vi) biocompatibility; (vii) lack of toxicity; (viii) biodegradability.
Confronting these practical strengths stands the fact that protein- and virus-based NPs are collectively considered “soft” materials,18 characterised by weak noncovalent interactions between and within polymeric components and prone to permanent structural alteration. The physical instability of many virus particles provides a biological advantage during the infection process, and may also be convenient for certain biomedical applications such as gene therapy or drug delivery.12 However, easy breakage of some protein- and virus-based NPs by mechanical forces acting during compression, tension and/or shear stress is a major concern when considering other medical or technological uses for which virus-based NPs may otherwise be most suitable.19,20 For example, when considering implanted biomaterials for cell tissue regeneration, that will be repeatedly bent and stretched.19 Or in cases where the NPs may be under stress due to high centrifugal forces (during purification),21 internal pressure (by packaged molecules),22 hydrostatic pressure,23 osmotic shock24 or dessication.25
Whereas some non-biological NPs may be mechanically stronger than those based on protein, they may not necessarily provide a better choice for many applications because they may not possess some (or most) of the advantageous features of protein- and virus-based NPs mentioned above.12 Thus, it may be desirable to acquire the fundamental knowledge needed to guide the engineering of protein- and virus-based nanostructures with increased resistance against mechanical breakage.12,17 In addition, growing experimental evidence indicates that certain mechanical properties of virus particles (such as stiffness) are important for their biological function.26–29 Thus, a detailed understanding of the relationship between atomic structure and breakage-related properties of virus particles may also provide novel physics-based insights into virus biology.
In recent years, investigation of the mechanical properties of virus particles has been enabled by the development of both theoretical approaches including coarse-grained molecular dynamics (MD) simulations,30–40 and experimental single-molecule techniques such as atomic force microscopy (AFM) under near-physiological conditions.26–29,41–43 In particular, our group is using AFM to provide insights into the relationships between the fine structure of model virus particles, their mechanical properties, and their function.44–50 By exploring the effects on a certain mechanical property (such as stiffness) of small structural alterations in the virus particle by single mutations introduced by protein engineering, knowledge about the specific chemical groups and interactions that determine that property can be acquired.45–50
A particularly suited model for these studies is the icosahedral capsid of the minute virus of mice (MVM).51,52 The MVM capsid (Fig. 1A and B), one of the smallest (25 nm in diameter) and structurally simplest viral NPs known, is made of a minimum number (60) of structurally equivalent protein monomers that have the same sequence and fold. There are no conformational switches related to quasi-equivalence that occur in icosahedral capsids made of more than 60 subunits.53,54
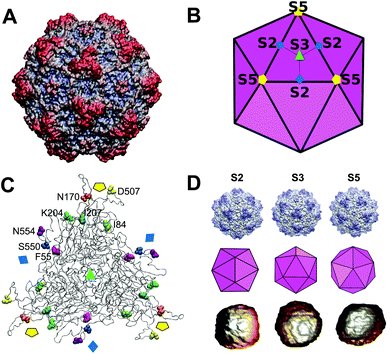 | ||
| Fig. 1 Atomic structure of the MVM capsid and residues chosen for mechanical analysis. (A) Atomic structure of the MVM capsid54 (PDB id 1Z14), represented as a surface model. (B) Scheme of the icosahedral architecture of the MVM capsid. Orientation is approximately the same as in panel A. Some symmetry axes of each type (S2, S3, S5) are labelled. Trimeric subunits (the stable CBBs the capsid is made of, and dissociated into) are delineated using thick lines. Three subunits in one trimer are delineated using thin lines. (C) Structure of a trimer in the MVM capsid, represented as a ribbon model. The positions of capsid symmetry axes are indicated by the same symbols used in panel (B). The 8 residues per capsid monomer located at intertrimer interfaces and chosen for analysis in this study are represented as spacefilling models, each of them using a specific color. Residues in one monomer are labelled. (D) Atomic models, icosahedral schemes and actual AFM images of individual MVM capsid particles oriented with a S2 (left), S3 (center) or S5 (right) symmetry axis close to the top. | ||
Using the MVM capsid, we have recently uncovered at high resolution some structural determinants of mechanical stiffness in a model virus particle.48 Specific groups of amino acid side chains located at interfaces between capsid building blocks (CBBs) were individually removed through site-directed mutagenesis; and changes in the elastic constant of the viral particle upon mutation were determined by indentation of different capsid regions with the AFM tip under the elastic regime, without breaking the particle. It was found that, in general, hydrophobic side chains in secondary structure elements buried along the interfaces between CBBs contribute to maintain most regions of the viral capsid in a state of comparatively low stiffness; their individual removal led to a large, frequently anisotropic stiffening of different capsid regions.48 This effect was specific, as other side chains (generally located in solvent-exposed protein loops) had a minor or insignificant role in determining capsid stiffness. Individual substitutions of other amino acid residues surrounding pores or cavities in the MVM capsid46,47,49 or in other viral particles50,55–57 had also stiffening effects.
These and other studies provided evidence that introduction of even very small structural changes in viral capsids by individual mutation can lead to largely stiffened NPs. However, comparison between different virus species or different biological states of a same viral particle indicated that increased stiffness does not necessarily involve increased resistance against mechanically induced breakage.27,58
Whereas a number of studies on virus particles have focused on analyzing structural, physical and/or biological aspects related with stiffness,26–50 much less attention has been directed towards uncovering the relationships between atomic structure and other important mechanical properties such as resistance against disruption by force (here termed mechanical strength) or brittleness.
In this study we have used the MVM capsid as a model to unveil single chemical groups and their interactions in particular structural contexts that determine breakage-related mechanical properties of a virus particle, including its strength and brittleness. In addition, by comparing the effects of specific structural changes on breakage-related properties with the previously determined effects of the same structural changes on stiffness,48 we have investigated possible quantitative linkages between different mechanical properties of a viral, protein-based NP. Implications of the results for the engineering of protein-based NPs with increased mechanical strength, and for a wider physical perspective on viruses, are discussed.
Results
Selection of amino acid side chains in the MVM capsid for analysis of chemical groups that determine capsid resistance against breakage under load
The natural MVM capsid is formed by two versions of the protein monomer (each viral particle contains 10 copies of VP1 and 50 copies of VP2). These versions differ in the length of their structurally disordered N-terminal segments (Nt), which do not contribute to capsid assembly or architecture. VP2-only MVM capsids, structurally indistinguishable from natural MVM capsids, can be assembled both ex vivo59 and in vitro,60 and have been used in this study.Trimers of protein monomers constitute stable capsid building blocks (CBBs) from which the MVM capsid is assembled60–63 (Fig. 1). Previous evidence showed that when this capsid is deeply indented with a AFM tip, fractures occur at the energetically weakest interfaces, those between trimers, leading to capsid fragmentation.64 Thus, to study molecular determinants of capsid resistance against breakage, we focused on different amino acid side chains in different structural contexts that establish noncovalent interactions between trimers in the viral particle.
Eight representative side chains per capsid subunit at the intertrimer interfaces (Table 1 and Fig. 1C) were chosen as targets based on several considerations: (i) their individual truncation by mutation to alanine does not prevent capsid assembly; (ii) nearly all of them participate mainly or exclusively in interactions between neighboring trimers, being virtually free from interactions within the same trimer; (iii) they are distributed along the intertrimer interfaces, from their centers (at capsid 2-fold symmetry (S2) axes) to their edges (at 5-fold (S5) axes), and they are all far from the centres of the trimers (at 3-fold (S3) axes) (Fig. 1C);65 (iv) they either form a part of exposed protein loops or are buried in the β barrel within each protein subunit; (v) they sample the different types of side chains and interactions involved in intertrimer association (Table 1).
| Residue | Intertrimer interactionsa | Intratrimer interactionsa | Relative infectivity of mutant virusd | ||||
|---|---|---|---|---|---|---|---|
| H-Bondsb | Ionicb | vdWc | H-Bondsb | Ionicb | vdWc | ||
| a Intertrimer or intratrimer noncovalent interactions on the crystal structure of the MVM capsid54 (PDB id 1Z14) were identified as described in ESI, using the following cutoff distances: hydrogen bonds: 3.6 Å; ionic interactions: 6 Å; van der Waals contacts: 0.8 Å + sum of van der Waals radii of the two atoms considered. b The interacting residue is indicated. (2×) indicates two hydrogen bonds are formed with residue Q548. c The number of total van der Waals contacts is indicated, and the number of carbon–carbon contacts is given in parenthesis. d Infectious titer of a mutant virus in which the indicated capsid residue was replaced by alanine, relative to the infectious titer of the non-mutated virus. Values were calculated by dividing the average absolute titer of each virus mutant by the average absolute titer of the nonmutated virus obtained in the same experiment, and averaging the results of two independent experiments. ND, not determined (mutation could not be introduced in the infectious recombinant clone). Infectivity data taken from ref. 46 and 65. | |||||||
| F55 | 6(5) | <8 × 10−6 | |||||
| I84 | 1 | 8(7) | 0.08 | ||||
| N170 | D171, T173 | 9(1) | <5 × 10−6 | ||||
| K204 | N80 | 3 | 1.05 | ||||
| I207 | 2(2) | 0.11 | |||||
| D507 | K166, Y168 | K166 | 6(2) | <2 × 10−5 | |||
| S550 | Q548(2×) | 7 | ND | ||||
| N554 | 5(2) | ND | |||||
The chosen amino acid residues were individually replaced by alanine using site-directed mutagenesis. In this way, in any mutant capsid the chemical group(s) in the targeted side chain (beyond the Cβ), and its interactions with other groups, was(were) removed from each of the 60 capsid subunits, without introducing any other group in the capsid.
Comparison of breakage-related events in wt and mutant capsids
The mutant capsids and the non-mutated, wild-type (wt) control capsid were produced, purified and subjected to mechanical analysis in nanoindentation experiments using AFM. Previous imaging by AFM of topographic features of each viral particle to be indented revealed its orientation (Fig. 1D), and allowed the application of mechanical force on a particular capsid region:44 namely, the spike at the center of each trimer (S3 region), the dimple at the center of each intertrimer interface (S2 region) or the cylinder that surrounds a capsid pore located where 5 intertrimer interfaces converge (S5 region) (Fig. 1).Single, deep indentations (Fig. 2A) were carried out in a physiological buffer (phosphate-buffered saline, PBS, pH = 7.5) at ambient temperature (25 °C) on individual MVM capsids whose integrity and orientation had previously been determined. Every indentation of wt or mutant capsids was carried out at a speed of 60 nm s−1 under the same conditions in parallel experiments.
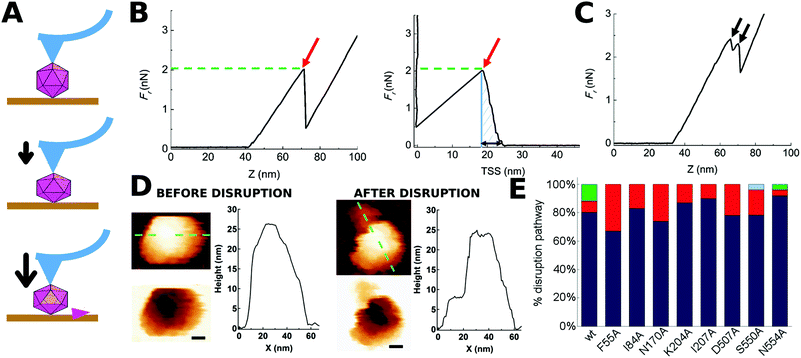 | ||
| Fig. 2 Indentation of individual MVM particles leading to their disruption. (A) Three stages during indentation of a single MVM capsid particle. Top: Tip-particle approach; middle: elastic deformation of the particle; bottom: particle disruption (in this example, a single trimeric CBB has been removed). (B) Force versus z-displacement (Fz) curve (left), and force versus indentation (Fd) curve (right) corresponding to a representative indentation of a MVM particle that was disrupted in a single event. The horizontal part of the curves corresponds to the tip-particle approach; the sloping part of the curve corresponds to the linear regime where the particle is being elastically deformed; a red arrow indicates the point where a drop in the force applied had occurred. The maximum force the particle withstood before breaking (Fr) is indicated by the green dotted line. The total amount of work carried out during the complete indentation process until the particle was disrupted (Wt) was estimated from the shadowed area in the Fd plot. The maximum deformation the particle withstood before breaking (dr) is indicated by a black double arrow in the Fd plot. (C) Fz curve corresponding to a different MVM particle that was disrupted during two separate events (black arrows). In B and C no smoothing was applied beyond the default implemented in the WSxM program. (D) AFM images of the same particle analyzed in panel B, before (left) and after (right) indentation. A scale bar (10 nm) is included. Height profiles following the green dotted line on the particle are shown. See Fig. S1† for other examples of single MVM particles disrupted by indentation. (E) Total percentages of wt or mutant capsids disrupted during a single nonlinear event (blue), two (red), three (green) or five (light blue) events. See Fig. S2† for equivalent results obtained when particles indented on different regions were separately considered. | ||
For most (81%) individual wt capsids tested (N = 26), the disruption process was associated with a single, large nonlinear event in the corresponding Fz curve, whereas for a minor fraction of capsids it involved several nonlinear events (Fig. 2E). When the same analysis was separately carried out for wt capsids that had been indented on different regions (S2, S3 or S5), qualitatively similar results were obtained (Fig. S2†).
AFM imaging of broken wt capsids (irrespective of the number of nonlinear events observed in the Fz curve) revealed that the final (disrupted) state consisted of either an incomplete particle in which one or a few trimers had been released (type I, 42% of cases) or, more frequently, several fragments that resulted from a more complete disintegration event (type II, 58% of cases) (Fig. 2D and S1†). Separate analysis of wt capsids indented on different regions revealed that indentation on a S2 or S3 region favors a more complete fragmentation (type II, 71% or 62% of cases, respectively) whereas indentation on a S5 region favors the loss of a few subunits only (type I, 60% of cases). Similar proportions were obtained when only those particles disrupted through a single detectable nonlinear event were considered (data not shown).
Most amino acid substitutions had no significant effect on the ratio between type I and type II states observed after capsid disruption. The exceptions were I207A, which may favor extensive fragmentation (type II), and S550A, which may favor dissociation of a few subunits only (type I) (data not shown).
To sum up, directional application of enough force on many individual wt or mutant MVM capsids led, in a large majority of cases, to a single nonlinear event in the Fz curve, that was physically associated to capsid breakage through either loss of a few subunits or (more frequently) extensive fragmentation. In view of these results, only viral particles that were broken through a single detectable event were considered in the subsequent analysis of the effect of mutations on capsid resistance against breakage under load.
Influence on capsid strength of specific individual side chains at intertrimer interfaces
The force Fr required to break the capsid was compared for the wt and the 8 chosen mutant capsids under the same experimental conditions as described in Methods. Higher Fr values were taken as an indication of a higher mechanical strength.| Capsid | Indentations | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Global | S2 | S3 | S5 | |||||||||
| F r (nN) | N | p | F r (nN) | N | p | F r (nN) | N | p | F r (nN) | N | p | |
| a Breaking force (average ± standard deviation). b Number of particles used for analysis. c p value obtained in a Student's t-test with a significance level of 0.05. | ||||||||||||
| wt | 3.12 ± 0.42 | 21 | — | 3.31 ± 0.26 | 6 | — | 3.27 ± 0.33 | 10 | — | 2.60 ± 0.33 | 5 | — |
| F55A | 2.70 ± 0.43 | 18 | 0.004 | 2.81 ± 0.53 | 6 | 0.077 | 2.64 ± 0.41 | 7 | 0.002 | 2.66 ± 0.39 | 5 | 0.772 |
| I84A | 2.98 ± 0.41 | 20 | 0.286 | 3.01 ± 0.41 | 8 | 0.134 | 2.87 ± 0.42 | 7 | 0.019 | 3.08 ± 0.46 | 5 | 0.096 |
| N170A | 2.54 ± 0.64 | 20 | 0.002 | 2.46 ± 0.52 | 8 | 0.002 | 3.07 ± 0.38 | 7 | 0.089 | 1.92 ± 0.53 | 5 | 0.046 |
| K204A | 2.62 ± 0.55 | 20 | 0.003 | 2.73 ± 0.35 | 7 | 0.006 | 2.88 ± 0.54 | 8 | 0.064 | 2.06 ± 0.46 | 5 | 0.068 |
| I207A | 3.10 ± 0.69 | 19 | 0.894 | 3.15± 0.17 | 7 | 0.256 | 3.44± 0.65 | 7 | 0.851 | 2.53 ± 0.91 | 5 | 0.892 |
| D507A | 2.61 ± 0.61 | 21 | 0.003 | 2.68 ± 0.57 | 9 | 0.014 | 2.80 ± 0.30 | 7 | 0.002 | 2.21 ± 0.91 | 5 | 0.419 |
| S550A | 2.88 ± 0.54 | 22 | 0.104 | 3.05± 0.30 | 7 | 0.128 | 3.07± 0.50 | 10 | 0.287 | 2.26± 0.48 | 5 | 0.233 |
| N554A | 2.54 ± 0.82 | 23 | 0.005 | 2.46 ± 0.81 | 11 | 0.007 | 3.16 ± 0.66 | 7 | 0.408 | 1.87 ± 0.36 | 5 | 0.010 |
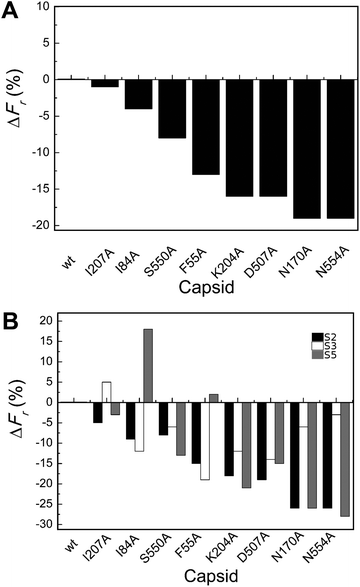 | ||
| Fig. 3 Percent variation in Fr (maximum force withstood without breaking apart) for wt or mutant MVM capsids. For each mutant capsid, bars indicate the % increase (positive values) or decrease (negative values) in Fr relative to the wt capsid. (A) Values obtained considering all capsids analyzed. (B) Values obtained when particles indented on different regions (S2, black bars; S3, white bars; S5, grey bars) were separately considered. Average values are represented in panels A and B; the corresponding standard deviations and statistical analysis are indicated in Table 2. | ||
Local Fr values determined for each mutant capsid by separately averaging data obtained with particles indented on different regions revealed that most mutations reduce capsid strength anisotropically (to different extents depending on the indented region) (Table 2 and Fig. 3B). The data allowed the statistical validation of many but not all of these differences (Table 2). For example, Fr was significantly reduced (by ∼20–25%) by substitutions N170A and N554A when the capsids were indented on a S2 or S5 region, but not when indented on a S3 region.
To summarize, most tested substitutions led to capsid breakage at significantly lower loads, especially when the force was applied at the interface between two trimers (and not at the center of a trimer). Thus, certain amino acid side chains at the intertrimer interfaces in the MVM capsid constitute major determinants of capsid strength against breakage under load.
Influence on capsid brittleness of specific individual side chains at intertrimer interfaces
The maximum deformation accepted by the capsid without breaking apart was then compared for the wt and the 8 chosen mutant capsids under the same experimental conditions, as described in Methods. The parameter dr/D was used, where dr is the maximum deformation accepted without detectable breakage, and D the external diameter of the roughly spherical particle. Lower dr/D values were taken as an indication of higher brittleness.| Capsid | Indentations | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Global | S2 | S3 | S5 | |||||||||
| d r/Da | N | p | d r/Da | N | p | d r/Da | N | p | d r/Da | N | p | |
| a Maximum relative deformation accepted (dr/D) (average ± standard deviation). b Number of particles used for analysis. c p value obtained in a Student's t-test with a significance level of 0.05. | ||||||||||||
| wt | 0.28 ± 0.08 | 21 | — | 0.23 ± 0.06 | 6 | — | 0.33 ± 0.05 | 10 | — | 0.22 ± 0.06 | 5 | — |
| F55A | 0.24 ± 0.05 | 18 | 0.060 | 0.22 ± 0.05 | 6 | 0.757 | 0.25 ± 0.07 | 7 | 0.035 | 0.24 ± 0.04 | 5 | 0.697 |
| I84A | 0.25 ± 0.08 | 20 | 0.259 | 0.24 ± 0.09 | 8 | 0.940 | 0.27 ± 0.08 | 7 | 0.141 | 0.24 ± 0.07 | 5 | 0.751 |
| N170A | 0.20 ± 0.08 | 20 | 0.004 | 0.19 ± 0.08 | 8 | 0.230 | 0.21 ± 0.05 | 7 | 0.001 | 0.22 ± 0.12 | 5 | 0.978 |
| K204A | 0.27 ± 0.11 | 20 | 0.878 | 0.28 ± 0.09 | 7 | 0.312 | 0.30 ± 0.14 | 8 | 0.575 | 0.22 ± 0.07 | 5 | 0.910 |
| I207A | 0.22 ± 0.09 | 19 | 0.045 | 0.29 ± 0.10 | 7 | 0.244 | 0.18 ± 0.05 | 7 | 2.2 × 10−4 | 0.19 ± 0.03 | 5 | 0.387 |
| D507A | 0.24 ± 0.09 | 21 | 0.193 | 0.24 ± 0.10 | 9 | 0.863 | 0.30 ± 0.08 | 7 | 0.382 | 0.17 ± 0.04 | 5 | 0.165 |
| S550A | 0.32 ± 0.11 | 22 | 0.118 | 0.33 ± 0.13 | 7 | 0.127 | 0.34 ± 0.10 | 10 | 0.768 | 0.28 ± 0.11 | 5 | 0.345 |
| N554A | 0.22 ± 0.08 | 23 | 0.029 | 0.22 ± 0.90 | 11 | 0.810 | 0.23 ± 0.09 | 7 | 0.029 | 0.22 ± 0.04 | 5 | 0.857 |
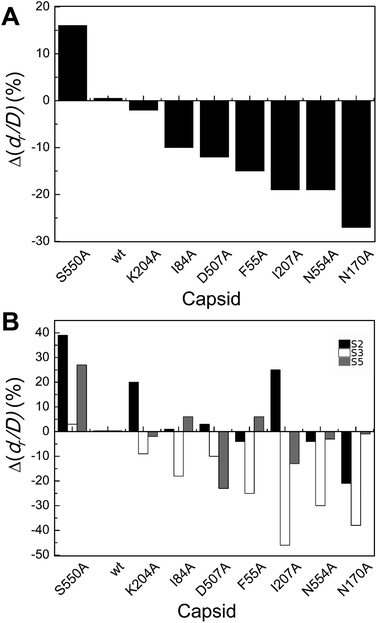 | ||
| Fig. 4 Percent variation in dr/D (maximum deformation withstood without breaking apart) for wt or mutant MVM capsids. For each mutant capsid, bars indicate the % increase (positive values) or decrease (negative values) in dr/D relative to the wt capsid. (A) Values obtained considering all capsids analyzed. (B) Values obtained when particles indented on different regions (S2, black bars; S3 white bars; S5, grey bars) were separately considered. Average values are represented in panels A and B; the corresponding standard deviations and statistical analysis are indicated in Table 3. | ||
Local dr/D values determined by separately averaging data obtained with particles indented on different regions revealed that effects of the substitutions on capsid brittleness are generally different depending on the region indented (Table 3 and Fig. 4B). However, the limited number of data thus obtained allowed the statistical validation of some of these differences only (Table 3). In general larger, significant increases in brittleness relative to the wt were observed for mutant capsids F55A, N170A, I207A and N554A indented on a S3 region (ΔdrS3/Dmut–wt = −25% to −46%). In most cases, indentation on other regions led to minor or no variations in dr/D.
To summarize, most but not all tested amino acid substitutions reduced the maximum deformation allowed by the capsid without breaking apart, especially when it was indented at the center of a trimer (and not at an interface between trimers). Thus, certain side chains at the intertrimer interfaces in the MVM capsid contribute to limit particle brittleness. If any of these groups is removed, the capsid becomes more brittle (i.e., it will break apart at a lower deformation).
Total mechanical work carried out during nanoindentation leading to deformation and breakage of wt and mutant capsids
The total work (Wt) carried out during indentation events that first led to the deformation of the MVM capsid and ended with its breakage was estimated by integrating the area under the Fd curve up to the point that marks the disruption event (Fig. 2B).| Capsid | Indentations | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Global | S2 | S3 | S5 | |||||||||
| W t (kBT × 10−3)a | N | p | W t (kBT × 10−3)a | N | p | W t (kBT × 10−3)a | N | p | W t (kBT × 10−3)a | N | p | |
| a Total work carried out (average ± standard deviation). b Number of particles used for analysis. c p value obtained in a Student's t-test with a significance level of 0.05. | ||||||||||||
| wt | 2.96 ± 1.04 | 21 | 2.70 ± 0.81 | 6 | 3.65 ± 0.77 | 10 | 1.88 ± 0.73 | 5 | ||||
| F55A | 2.28 ± 0.84 | 18 | 0.031 | 2.31 ± 0.77 | 6 | 0.408 | 2.42 ± 1.00 | 7 | 0.013 | 2.04 ± 0.81 | 5 | 0.744 |
| I84A | 2.58 ± 0.11 | 20 | 0.276 | 2.50 ± 1.26 | 8 | 0.725 | 2.60 ± 0.94 | 7 | 0.020 | 2.68 ± 1.38 | 5 | 0.295 |
| N170A | 1.84 ± 0.90 | 20 | 7.2 × 10−4 | 1.54 ± 1.01 | 8 | 0.034 | 2.21 ± 0.66 | 7 | 0.001 | 1.81 ± 0.97 | 5 | 0.295 |
| K204A | 2.42 ± 1.43 | 20 | 0.181 | 2.57 ± 1.17 | 7 | 0.809 | 2.84 ± 1.74 | 8 | 0.199 | 1.55 ± 0.99 | 5 | 0.567 |
| I207A | 2.39 ± 1.26 | 19 | 0.132 | 3.20 ± 1.41 | 7 | 0.444 | 2.11 ± 0.85 | 7 | 0.002 | 1.65 ± 1.04 | 5 | 0.699 |
| D507A | 2.43 ± 1.18 | 21 | 0.130 | 2.57 ± 1.36 | 9 | 0.817 | 2.93 ± 0.72 | 7 | 0.043 | 1.46 ± 0.87 | 5 | 0.436 |
| S550A | 3.40 ± 1.62 | 22 | 0.288 | 3.63 ± 1.28 | 7 | 0.145 | 3.99 ± 1.75 | 10 | 0.580 | 1.90 ± 0.73 | 5 | 0.961 |
| N554A | 2.11 ± 1.25 | 23 | 0.019 | 2.16 ± 1.53 | 11 | 0.352 | 2.53 ± 0.98 | 7 | 0.029 | 1.41 ± 0.58 | 5 | 0.302 |
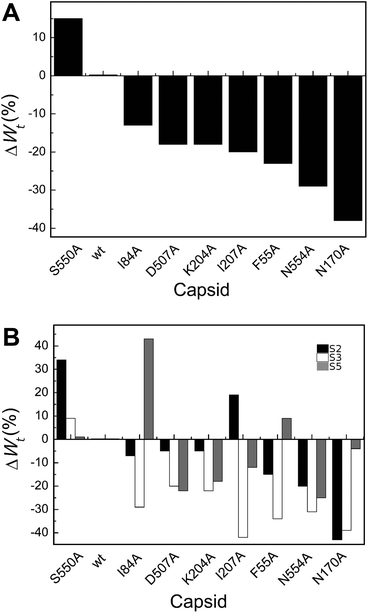 | ||
| Fig. 5 Percent variation in Wt (total work carried out during indentation until disruption) for wt or mutant MVM capsids. For each mutant capsid, bars indicate the % increase (positive values) or decrease (negative values) in Wt relative to the wt capsid. (A) Values obtained considering all capsids analyzed. (B) Values obtained when particles indented on different regions (S2, black bars; S3, white bars; S5, grey bars) were separately considered. Average values are represented in panels A and B; the corresponding standard deviations and statistical analysis are indicated in Table 4. | ||
Local Wt values determined by separately averaging data obtained with particles indented on different regions revealed that most amino acid substitutions led to variations in Wt that are dependent on the indented region (Table 4 and Fig. 5B). The largest differences were generally (though not universally) observed for particles indented on a S3 region, with most mutants showing significant reductions in Wt relative to the wt (ΔWmut–wttS3 = −20% to −42%). Variations in Wt between mutants and wt were lower or insignificant when other capsid regions were indented.
To summarize, for most MVM mutant capsids the average amount of total mechanical work associated with the indentation process leading to capsid deformation and ending with capsid breakage was lower than for the wt, especially when the capsid was indented on the center of a trimer.
Quantitative correlations between mechanical properties of a virus capsid
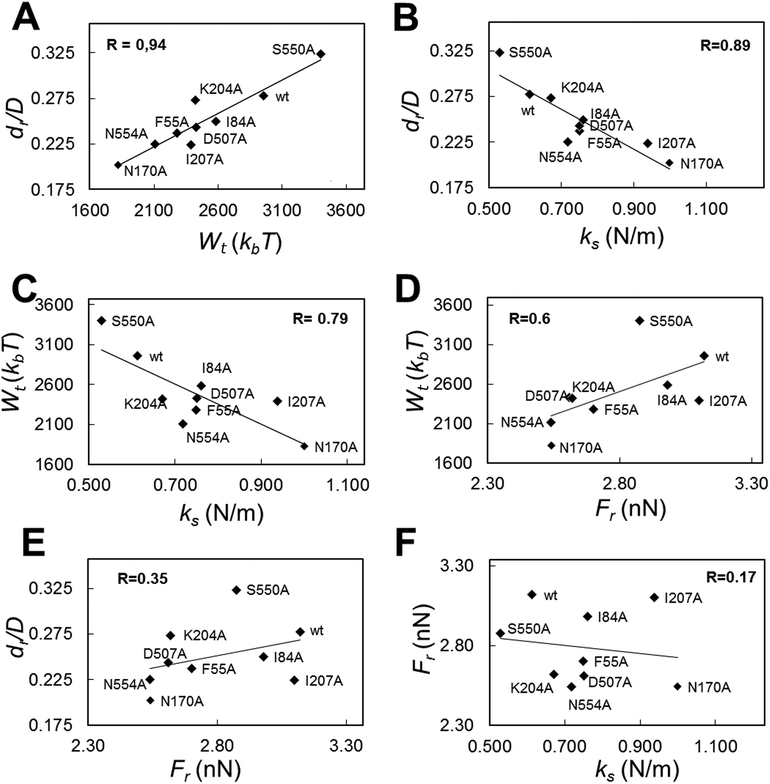
By taking advantage of the results described above, we were able to look for correlations between different mechanical properties in a same virus particle. In addition to values of breakage-related parameters (Fr, dr/D), or of a parameter related to the sum of deformation plus breakage (Wt), values for a stiffness-related parameter (the elastic constant ks) were obtained for each tested viral particle. ks was determined by analyzing the linear part of each Fz curve before the disruption event occurred. The global and local ks values thus obtained were statistically indistinguishable from the corresponding ks values previously obtained for the wt and those same mutants as a part of a different study focused on MVM stiffness.48 The average Fr, dr/D, Wt and ks values obtained for wt and mutant capsids were then plotted against each other (Fig. 6 and S3†).The highest correlation coefficients (R) between global values were found for dr/D vs. Wt (R = 0.94) (Fig. 6A); dr/D vs. ks (R = 0.89) (Fig. 6B); and Wtvs. ks (R = 0.79) (Fig. 6C); the R value obtained for Wtvs. Fr was lower (R = 0.60) (Fig. 6D), and quite low R values were obtained for dr/D vs. Fr (R = 0.35) (Fig. 6E) and Frvs. ks (R = 0.17) (Fig. 6F).
By separately plotting average values obtained with particles indented on different regions, it was found that correlations detected between certain pairs of mechanical parameters using global average values could be mostly traced to some capsid regions but not others: (i) for dr/D vs. Wt, to S2 (R = 0.93) and S3 (R = 0.92) regions; (ii) for dr/D vs. ks and Wtvs. ks, to S3 (R = 0.95 or 0.85, respectively); (iii) for Wtvs. Fr, to S2 (R = 0.71) and S5 (R = 0.82) regions (Fig. S3†).
To sum up, a clear relationship was found for a virus capsid between increased brittleness, and reduced total work during indentations leading to capsid deformation and breakage; and also between increased stiffness and increased brittleness. In contrast, no correlation was found between variations in brittleness vs. strength, or strength vs. stiffness.
Capsid structure and molecular determinants of breakage-related mechanical properties
We wondered whether the different effects of tested mutations on breakage-related mechanical properties of the MVM capsid could be connected to structural differences. No clear relationship was found between either reduced strength or increased brittleness on mutation, and type or number of intertrimer interactions lost (Table 1), or (estimated) variations in affinity between trimers (according to calculated association free energy values (ΔGa) for each specific residue in the VIPER data base67). However, mapping the targeted amino acid side chains in the atomic structure of the MVM capsid revealed that (i) side chains with major roles in determining capsid strength against breakage are polar and located in solvent-exposed positions (either in large loops protruding from each trimer and intertwined with loops from neighboring trimers (N554, D507, K204), or at the base of the capsid pores (N170)); (ii) in contrast, most side chains with no or a minor influence on strength (S550 excepted) are apolar and located in structural elements buried along each interface core (I207, I84, F55) (Fig. 7A).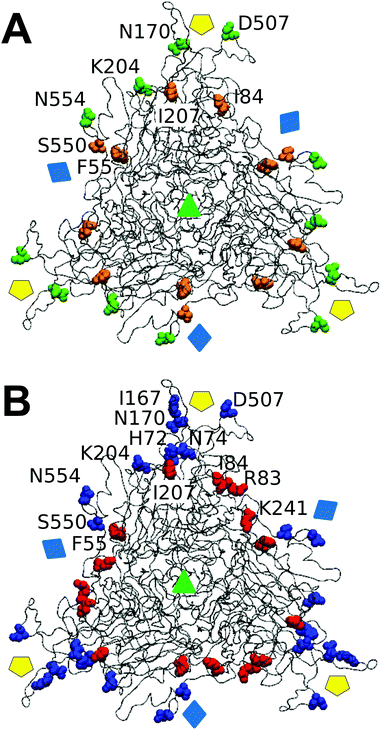 | ||
| Fig. 7 Spatial distribution in the MVM capsid of residues at intertrimer interfaces according to their role in determining different mechanical properties. Only a trimeric subunit in the MVM capsid is represented, using a ribbon model (compare Fig. 1). The location of capsid symmetry axes is indicated by colored symbols. (A) 8 interfacial residues tested in this study are represented in spacefilling models and colored according to their involvement in mechanical properties related to capsid resistance against breakage. Green, residues whose side chains constitute major determinants of capsid strength against breakage; orange, residues with no or minor roles on capsid strength. (B) 12 interfacial residues (including the same 8 depicted in panel A) represented in spacefilling models and colored according to their involvement in maintaining a limited stiffness, particularly at the central region (S2) in each intertrimer interface.48 Red: residues whose side chains constitute major determinants for preserving limited stiffness; blue, residues with no or minor roles in preserving limited stiffness. Residues in one monomer are labelled. | ||
Comparing differences in capsid mechanical properties and virus infectivity
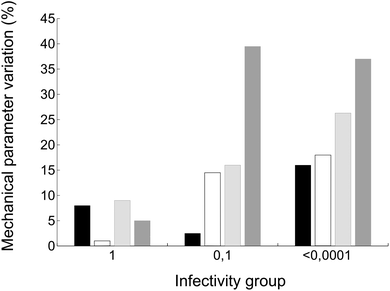
Finally, we wondered whether the same mutations that altered breakage-related mechanical properties of the MVM capsid could have also some effect on the infectivity of the virus (as indicated by comparing relative viral progeny titers obtained on parallel infections of host cells by wt and mutant viruses). Viruses carrying amino acid substitutions tested in this study could be grouped according to infectivity as follows: (i) close to normal infectivity: wt, K204A; (ii) moderately reduced infectivity (by one order of magnitude): I84A, I207A; (iii) drastically reduced infectivity (by >4 orders of magnitude): F55A, N170A, D507A (the infectivity of virus mutants S550A and N554A could not be tested). Global values of capsid mechanical parameters Fr, dr/D, Wt and ks were averaged for viruses of each infectivity group and compared with infectivity data (Fig. 8). Despite the limitations of this necessarily coarse analysis (see Discussion) the results reveal that, on average, the capsids of the less infectious mutant viruses are not only stiffer, but also qualitatively less resistant against breakage under load and more brittle.Discussion
Linked vs. independent mechanical properties of a protein-made virus NP
By comparing under the same experimental conditions mutation-related changes in mechanical strength and brittleness of a virus capsid (as determined in this study) and its stiffness (previously determined48), we were able to assess whether any linkage may exist between variations in different mechanical properties that may result from minor structural alterations49 in a protein-based particle. Good direct correlations were found between brittleness and reduced total work during indentations leading to capsid deformation and breakage, and between stiffness and brittleness; no correlations were found between brittleness and strength, or between strength and stiffness.The total estimated work Wt during indentation (deformation plus breakage) of MVM capsids is much higher that the calculated association energy for icosahedral virus capsids.66 Even though the MVM capsid may require a particularly high energy for dissociation relative to other icosahedral capsids (as reflected in the very high temperatures required for its disassembly59,65), the very large Wt values obtained clearly indicate that a large part of the total work during indentation is related to capsid deformation before breaking apart, and not to capsid disruption. Thus, ΔWt values should not be taken at all as an estimate of the contribution to binding energy of the chemical groups removed. A quantitative understanding of the different contributions to Wt, and of the physical basis for the inverse correlation found between Wt and dr/D, may require the development of a complex model that considers variations in Fr, ks, Wt, Wd, Wr and the capsid binding energy ΔGa.
The good correlation found between mutation-related variations in dr/D (brittleness) and ks (stiffness) (and between Wt and ks as a consequence of the dr/D–Wt correlation) may be quite relevant, both from fundamental and applied perspectives. With few exceptions, stiffer mutant capsids broke apart at lower deformations. At the other end of the spectrum S550A, the only tested mutant capsid that is more flexible (less stiff) than the wt capsid (ks reduced by 13%), was also proportionally less brittle (dr/D increased by 14%). This latter observation held, even quantitatively, when results were compared for particles indented on different regions: for the S550A capsid indented at S2, S5 or S3, ks was reduced by 17%, 23% or 5%, and dr/D increased by 43%, 27% or 3%, respectively.
Linked changes in stiffness and brittleness as observed here for a virus capsid have been found also with certain polymeric but structurally very different materials, including elastomers. However, the underlying mechanisms must be quite different. For example, in a hollow rubber ball stiffening and increased brittleness during aging are caused by oxidation and other chemical reactions, leading to molecular changes that affect extension and intertwining of random coil polymer chains. In the hollow, spherical virus capsid analyzed here, the same effects were unexpectedly achieved by removing certain chemical groups and their associated non-covalent interactions between well-folded polypeptides.
The lack of correlation between mutation-related variations in Fr (force required for breaking the particle) and dr/D (brittleness) or ks (stiffness) is equally interesting. Indeed, these mechanical properties are not necessarily linked when structurally very different macroscopic materials are compared. For example, a hollow sphere is mechanically strong and stiff but not brittle if made of steel, whereas it is stiff and brittle but not as strong if made of glass. However, when comparing mechanical effects of modest changes in molecular structure of a given material, a clear association between increased stiffness and reduced strength was found, at least in some cases. For example, molecular changes due to aging of the elastomer in a hollow rubber ball lead not only to increased stiffness and brittleness, but also to reduced strength against breakage. Regarding biological materials, aging of collagen fibers or cortical bone led to a gradual increase in stiffness and brittleness and to a gradual decrease in strength against fracture under load.68,69 These results contrast with the lack of correlation we observed in a protein-made virus capsid between stiffness and strength. Understanding the structural basis of this lack of correlation may be helped by further experiments (see below) and by theoretical modeling.
Structural changes that underlie variations in strength against breakage and/or brittleness of a protein-made virus NP
Another noteworthy result of this study is that individual removal of different chemical groups and the interactions they establish at the interface between protein subunits in the MVM capsid, invariably led to some reduction in the force required for breaking the particle apart (global Fr). The same was true (except for the S550A mutation) regarding the maximum deformation the capsid allowed without being disrupted (global dr/D). Although the smaller variations in global Fr or dr/D caused by a few mutations could not be statistically validated, it may be nonetheless revealing that even insignificant increases in global Fr or dr/D were not observed in any but the S550A mutant.A simple structural interpretation for this observation is that removal of some noncovalent interactions between capsid subunits, by reducing association energy, will facilitate the opening of fractures at the interfaces leading to capsid fragmentation at lower loads and/or deformations. If true, it could follow that, to permanently improve the strength of a virus-based NP against breakage (e.g., for any application involving mechanical stress), mutations designed to introduce further interactions between protein subunits should be engineered.
In fact, absence of a correlation between reduction in Fr or dr/D and (estimated) reduction in association free energy between trimers by different tested mutations suggests that the situation may be more complex than the scenario considered above. Comparison of the atomic structure of the N170A mutant capsid with that of the wt capsid revealed that this mutation leads to small structural rearrangements and reduced atomic mobility at different capsid regions (not only at the site of the mutation), and to a subtle overall compaction of the capsid structure.49 Thus, the quantitative effect of any mutation on the resistance of the virus particle against breakage may depend not only on the interactions directly lost or gained by substituting one residue for another, but also on any other intersubunit interactions that may be lost or gained during any subtle structural rearrangement of the particle induced by the mutation.
Different patterns in chemical nature and distribution of atomic groups that determine either strength against breakage or stiffness of a protein-made virus NP
The lack of correlation between Fr and dr/D or ks values reflects the finding that most specific side chains at the intertrimer interfaces in the MVM capsid that are more influential for preserving its strength against breakage are actually different from those side chains with major roles in limiting its brittleness or stiffness (Fig. 7).As described in Results, most interfacial side chains that constitute major determinants of capsid strength (N554, D507, K204) are polar and located in solvent-exposed loops that extend from the surface of each trimer and are intertwined with loops from other trimers (Fig. 7A). In contrast, these and other interfacial side chains located at these loops have minor roles in determining capsid stiffness48 (Fig. 7B).
Conversely, most interfacial side chains with major roles in limiting capsid stiffness, in particular those at intertrimer interfaces (I207, I84, F55, I167) are apolar and located at buried structural elements along the core of each interface48 (Fig. 7B). In contrast these residues (I207, I84, F55) showed a minor or insignificant influence in determining capsid strength (Fig. 7A).
The above observations open up several questions related to the uncoupling in a virus particle between structural determinants of strength against breakage vs. stiffness, and the different locations where determinants of strength or brittleness are located. Inspired by the overall structural rearrangement of the MVM capsid caused by the strengthening, stiffening N170A mutation49 a simplified, testable scenario may be proposed: (i) the observed, generally delocalized and anisotropic changes in capsid mechanical properties on removal of certain chemical groups may be a consequence of small but rather global structural rearrangements; (ii) such rearrangements may alter the number and strength of different interactions within and/or between protein subunits; (iii) minor structural rearrangements on removal of groups mostly located in exposed, intertwined loops (Fig. 7A, green residues) may lead to significant changes in association energy between building blocks that will reduce particle strength against breakage; (iv) minor structural rearrangements on removal of groups mostly located in buried structural elements (Fig. 7B, red residues) may lead to significant changes in the overall number of both inter- and intra-trimeric interactions between residues in the particle, and these changes together would increase particle stiffness. Comparison of atomic structures of other mechanically altered capsid mutants and MD simulations,70 as well as comparative studies using other viruses and/or protein particles, may help to support this proposal and to provide deeper insights into the relationships between atomic structure and mechanical properties of virus- and protein-based NPs.
Capsid resistance against mechanical disruption and virus biology
In other studies we had provided for MVM substantial experimental evidence for a complex linkage between certain region-specific changes in stiffness of a virus particle, changes in conformational dynamics and altered virus stability or infectivity.45–47 The present study on molecular determinants of the resistance of a virus NP against mechanical breakage was not aimed at establishing a biological connection of this particular physical feature with virus biology. The number of tested mutants was too limited to address this particular point. Moreover, the mutations tested had varying effects on capsid stiffness in addition to strength and brittleness.48Despite these qualifications, two observations are worth noting: (i) every tested mutation reduced strength against breakage of the biologically evolved MVM capsid (Fig. 3), and nearly all of them increased its brittleness (Fig. 4); (ii) mutations that clearly reduce capsid strength and increase brittleness invariably lead to a substantial reduction in MVM infectivity (Fig. 8). These observations encourage further investigation of a putative linkage between virus resistance against disruption by physical forces and viral biology. This is becoming an important subject in different fields: for those biotechnological or biomedical applications for which preservation of virus infectivity may be required, one should know whether an engineered change in a mechanical property of the virus may cause an undesirable reduction in its infectivity; from the perspective of molecular virology, further studies on any linkage between virus mechanics and infection may expand our understanding of virus biology from a physics-based perspective.
Experimental
Engineering, production and purification of mutant MVM particles
Protocols previously described were followed.48 Briefly, site-directed mutagenesis by inverse PCR (QuikChange, Stratagene) was used to introduce specific mutations in the VP2 gene of MVM cloned on plasmid pFB1-VP2.65,71 The presence of the mutations was confirmed by DNA sequencing. Then, a baculovirus expression system (Invitrogen) and pFB1-VP2 as donor plasmids were used to obtain BM-VP2 bacmids; BM-VP2 bacmids containing either the wt VP2 gene or mutants were used for the production in H5 insect cells of empty capsids made of VP2 subunits.65,71 Capsid purification was carried out as previously described.59,65,71 Pure capsid preparations were dialyzed against phosphate-buffered saline and kept at 4 °C or frozen at −70 °C. Purity and integrity were assessed by electrophoresis, electron microscopy, and/or AFM.Imaging by AFM of MVM particles
An atomic force microscope (Nanotec Electrónica) was used. Imaging and nanoindentation procedures followed are based on those previously described.41,44 Briefly, 20 μl of the sample in physiological buffer (PBS) at the desired protein concentration were placed on glass coverslips treated with hexamethyldisilazane (Sigma-Aldrich). After capsid adsorption for 30 minutes at room temperature (r.t.), the coverslips were washed four times with 20 μl PBS, and 20 μl PBS were then added. Imaging was carried out in jumping mode72 using RC800PSA cantilevers (Olympus) with a nominal spring constant of 0.1 N m−1. During imaging, the applied force was kept under ∼100 pN. Images were processed using WSxM software.73Determination by AFM of mechanical properties of MVM particles
Prior to mechanical analysis, AFM imaging of individual MVM capsids adsorbed on silanized glass and submerged in PBS was used to identify their topographic features and height as indicators of structural integrity, as well as their approximate orientation. Intact particles oriented with an axis of symmetry (S2, S3 or S5) close to the top were subjected to indentation with the AFM tip, using RC800PSA cantilevers with a nominal elastic constant kc = 0.1 N m−1. The actual spring constant of each cantilever used was determined using Sader's method.74The stiffness of different regions in the MVM capsid was determined by indenting capsid particles of known orientation at a rate of 60 nm s−1. Before and after each indentation on a viral particle, three indentations were performed on the glass substrate to confirm a consistent sensitivity of the optical detection system. The Fz curve obtained when a viral particle was indented was considered valid if the particle was deformed linearly in response to the applied force. Moreover, only data obtained well within the range of the linear elastic response47 were used for calculation of the elastic constant ks of the indented particle. ks was determined from the experimental ke value obtained by assuming that the viral particle and the AFM cantilever of elastic constant kc behave as two ideal springs in series.41,44
To determine mechanical parameters related to capsid breakage, a single indentation of greater depth was performed on the center of each individual particle selected, again using an appropriate number of particles oriented in different ways to obtain local and global average values. The indentation was carried out at a rate of 60 nm s−1. Particle disruption was consistently reflected in a nonlinear event during the tracing of the Fz curve, that was due to relaxation of the cantilever, and a sudden drop in the force being applied. Actual breakage of the indented particle was confirmed by AFM imaging after the nonlinear event had occurred. Only data obtained with particles that clearly showed partial or complete disruption after indentation were considered for analysis. The Fz and corresponding Fd curves obtained with many indented particles that were eventually disrupted were analyzed to obtain values for different mechanical breakage-related parameters (Fig. 2A) The breaking force, Fr, was taken as the force value corresponding to the point where the curve slope changes abruptly. The maximum deformation allowed without breakage, dr, was obtained from the Fd curve as the distance from the point the cantilever-particle contact is established to the point where the slope changes abruptly (the breaking point). The energy dissipated (work) during the entire indentation process until the particle is broken, Wt, was estimated following a method described before for other biological assemblies:75 by integrating the area defined under the Fd curve, from the point the cantilever-particle contact is established to the breaking point. An appropriate number of particles in different orientations were used to calculate average local and global ks, Fr, dr and Wt values (Tables 2–4).
Numerical data analysis
For numerical data analysis, including statistical analysis, and some graphical representations, the programs OriginPro 8 (Origin Lab Corporation) and Excel (Microsoft Corporation) were used. Specifically, OriginPro 8 was used for calculations of indentations, breaking force values, dissipated energy and elastic constants, for Gaussian and Lorentzian fits, statistical validation of data, etc. To validate statistically that two values of a mechanical parameter are different, we used the two-tailed Student's t-test, with an α significance level equal to 0.05. No equal variance was assumed, thus providing a more stringent analysis.Molecular graphics and structural analysis
The Protein Data Bank coordinates of the MVMp empty capsid71 (PDB id 1Z14) were used. Programs used included Pymol76 and VMD77 for virtual mutagenesis and molecular graphics representations, and WHATIF78 for structural analyses.Conclusion
This study revealed that removal of different amino acid side chains that establish interactions between building blocks in a virus NP both reduced the force required to break the viral particle and increased its brittleness. However, quantitative effects on these breakage-related mechanical properties were quite dependent on the side chain that was removed. Most side chains that were identified as major determinants of strength against breakage are polar and located in solvent-exposed, intertwined loops; in contrast, most of those that had no or minor roles in maintaining strength are apolar and located in buried positions along the interfaces. In general, residues that had major roles in determining resistance against breakage were not important for determining capsid stiffness, and vice versa. These results support the view that viral and other protein-based NPs may be engineered for increased strength against breakage without necessarily increasing its brittleness or changing its stiffness (two undesirable effects for certain applications). Also, engineering protein NPs with altered stiffness may not entail any trade-off regarding strength. Achieving a lower stiffness could bring the added benefit of reduced brittleness, whereas engineering a stiffened NP could involve increased brittleness as a trade-off.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
M. M. and A. V. were the respective recipients of a FPI fellowship from Universidad Autónoma de Madrid and a postdoctoral contract from the Spanish Ministerio de Economía y Competitividad (MINECO). M. G. M. is an associate member of the Institute for Biocomputation and Physics of Complex Systems, Zaragoza, Spain. This work was funded by a grant from MINECO/FEDER EU (BIO2015-69928-R) and by an institutional grant from Fundación Ramón Areces. We also acknowledge support of the publication fee by the CSIC Open Access Support Initiative through its Unit of Information Resources for Research.References
- Protein Nanotechnology, ed. J. A. Gerrard, Humana Press, New York, NY, USA, 2nd edn, 2013; Methods in Molecular Biology. vol. 996 Search PubMed.
- Protein-based Engineered Nanostructures, ed. A. L. Cortajarena and T. Z. Grove, Springer, Switzerland, 2016; Advances in Experimental Medicine and Biology. vol. 940 Search PubMed.
- Structural Virology, ed. M. Agbandje-McKenna and R. McKenna, Royal Society of Chemistry Publishing, Cambridge, UK, 2011 Search PubMed.
- Viral Molecular Machines, ed. M. G. Rossmann and V. B. Rao, Springer, Boston, MA, USA, 2012; Advances in Experimental Medicine and Biology. vol. 726 Search PubMed.
- Structure and Physics of Viruses, ed. M. G. Mateu, Springer, Dordrecht, The Netherlands, 2013; Subcellular Biochemistry. vol. 68 Search PubMed.
- T. Douglas and M. Young, Science, 2006, 312, 873–875 CrossRef CAS PubMed.
- P. Roy and R. Noad, Hum. Vaccines, 2008, 4, 5–12 CrossRef CAS.
- I. Yildiz, S. Shukia and N. F. Steinmetz, Curr. Opin. Biotechnol., 2011, 22, 901–908 CrossRef CAS PubMed.
- A. M. Bittner, J. M. Alonso, M. L. Górzny and C. Wege, in Structure and Physics of Viruses, ed. M. G. Mateu, Springer, Dordrecht, The Netherlands, 2013, pp. 667–702 Search PubMed.
- J. Glasgow and D. Tullman-Ercek, Appl. Microbiol. Biotechnol., 2014, 98, 5847–5858 CrossRef CAS PubMed.
- F. Li and Q. Wang, Small, 2014, 2, 230–245 CrossRef PubMed.
- M. G. Mateu, in Protein-based Engineered Nanostructures, ed. A. L. Cortajarena and T. Z. Grove, Springer, Switzerland, 2016, pp. 83–120 Search PubMed.
- N. C. Seeman and A. M. Belcher, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 6451–6455 CrossRef CAS PubMed.
- C. C. DuFort and B. Dragnea, Annu. Rev. Phys. Chem., 2010, 61, 323–344 CrossRef CAS PubMed.
- S.-Y. Lee, J.-S. Lim and M. T. Harris, Biotechnol. Bioeng., 2012, 109, 16–30 CrossRef CAS PubMed.
- U. G. Wegst, H. Bai, E. Saiz, A. P. Tomsia and R. O. Ritchie, Nat. Mater., 2015, 14, 23–36 CrossRef CAS PubMed.
- R. Putri, J. J. Cornelissen and M. S. Koay, ChemPhysChem, 2015, 16, 911–918 CrossRef CAS PubMed.
- R. A. L. Jones, Faraday Discuss., 2009, 143, 9–14 RSC.
- F. J. O'Brien, Mater. Today, 2011, 14, 88–95 CrossRef.
- D. Guo, G. Xie and J. Luo, J. Phys. D: Appl. Phys., 2014, 47, 013001 CrossRef.
- S. K. Powell, M. A. Kaloss, A. Pinkstaff, R. McKee, I. Burimski, M. Pensiero, E. Otto, W. P. C. Stemmer and N.-W. Soong, Nat. Biotechnol., 2000, 18, 1279–1282 CrossRef CAS PubMed.
- D. E. Smith, S. J. Tans, S. B. Smith, S. Grimes, D. L. Anderson and C. Bustamante, Nature, 2001, 413, 748–752 CrossRef CAS PubMed.
- J. L. Silva and G. Weber, J. Mol. Biol., 1988, 199, 149–159 CrossRef CAS PubMed.
- P. Zavala-Rivera, K. Channon, V. Nguyen, E. Sivaniah, D. Kabra, R. H. Friend, S. K. Nataraj, S. A. Al-Muhtaseb, A. Hexemer, M. E. Calvo and H. Miguez, Nat. Mater., 2011, 11, 53–57 CrossRef PubMed.
- C. Carrasco, M. Douas, R. Miranda, M. Castellanos, P. A. Serena, J. L. Carrascosa, M. G. Mateu, M. I. Marqués and P. J. de Pablo, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 5475–5480 CrossRef CAS PubMed.
- W. H. Roos, R. Bruinsma and G. J. Wuite, Nat. Phys., 2010, 6, 733–743 Search PubMed.
- M. G. Mateu, Virus Res., 2012, 168, 1–22 CrossRef CAS PubMed.
- P. J. de Pablo and M. G. Mateu, in Structure and Physics of Viruses, ed. M. G. Mateu, Springer, Dordrecht, The Netherlands, 2013, pp. 519–551 Search PubMed.
- M. Marchetti, G. J. Wuite and W. H. Roos, Curr. Opin. Virol., 2016, 18, 82–88 CrossRef CAS PubMed.
- R. Zandi and D. Reguera, Phys. Rev. E: Stat., Nonlinear, Soft Matter Phys., 2005, 72, 021917 CrossRef PubMed.
- T. Guérin and R. Bruinsma, Phys. Rev. E: Stat., Nonlinear, Soft Matter Phys., 2007, 76, 061911 CrossRef PubMed.
- M. Zink and H. Grubmüller, Biophys. J., 2009, 96, 1350–1363 CrossRef CAS PubMed.
- W. S. Klug, W. H. Roos and G. J. Wuite, Phys. Rev. Lett., 2012, 109, 168104 CrossRef CAS PubMed.
- O. Kononova, J. Snijder, M. Brasch, J. J. Cornelissen, R. I. Dima, K. A. Marx, G. J. Wuite, W. H. Roos and V. Barsegov, Biophys. J., 2013, 105, 1893–1903 CrossRef CAS PubMed.
- M. Cieplak and M. O. Robbins, PLoS One, 2013, 8, e63640 CrossRef CAS PubMed.
- A. Luque and D. Reguera, in Structure and Physics of Viruses, ed. M. G. Mateu, Springer, Dordrecht, The Netherlands, 2013, pp. 553–595 Search PubMed.
- E. R. May, Mol. Simul., 2014, 40, 878–888 CrossRef CAS PubMed.
- O. Kononova, J. Snijder, Y. Kholodov, K. A. Marx, G. J. Wuite, W. H. Roos and V. Barsegov, PLoS Comput. Biol., 2016, 12, e1004729 CrossRef PubMed.
- A. Aggarwal, Phys. Rev. E, 2018, 97, 032414 CrossRef CAS PubMed.
- O. Kononova, F. Maksudov, K. A. Marx and V. Barsegov, J. Phys.: Condens. Matter, 2018, 30, 044006 CrossRef PubMed.
- I. L. Ivanovska, P. J. de Pablo, B. Ibarra, G. Sgalari, F. C. MacKintosh, J. L. Carrascosa, C. F. Schmidt and G. J. Wuite, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 7600–7605 CrossRef CAS PubMed.
- M. G. van Rosmalen, W. H. Roos and G. J. Wuite, Methods Mol. Biol., 2015, 1252, 115–137 CrossRef CAS PubMed.
- P. J. de Pablo, Semin. Cell Dev. Biol., 2018, 73, 199–208 CrossRef CAS PubMed.
- C. Carrasco, A. Carreira, I. A. Schaap, P. A. Serena, J. Gómez-Herrero, M. G. Mateu and P. J. de Pablo, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 13706–13711 CrossRef CAS PubMed.
- C. Carrasco, M. Castellanos, P. J. de Pablo and M. G. Mateu, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 4150–4155 CrossRef CAS PubMed.
- M. Castellanos, R. Pérez, C. Carrasco, M. Hernando-Pérez, J. Gómez-Herrero, P. J. de Pablo and M. G. Mateu, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 12028–12033 CrossRef CAS PubMed.
- M. Castellanos, P. J. P. Carrillo and M. G. Mateu, Nanoscale, 2015, 7, 5654–5664 RSC.
- P. J. P. Carrillo, M. Medrano, A. Valbuena, A. Rodríguez-Huete, M. Castellanos, R. Pérez and M. G. Mateu, ACS Nano, 2017, 11, 2194–2208 CrossRef CAS PubMed.
- P. Guerra, A. Valbuena, J. Querol-Audí, C. Silva, M. Castellanos, A. Rodríguez-Huete, D. Garriga, M. G. Mateu and N. Verdaguer, Sci. Rep., 2017, 7, 4101 CrossRef PubMed.
- A. Valbuena, A. Rodríguez-Huete and M. G. Mateu, Nanoscale, 2018, 10, 1440–1452 RSC.
- S. F. Cotmore and P. Tattersall, Annu. Rev. Virol., 2014, 1, 517–537 CrossRef PubMed.
- C. Ros, N. Bayat, R. Wolfisberg and J. M. Almendral, Viruses, 2017, 9, E313 CrossRef PubMed.
- M. Agbandje-McKenna, A. L. Llamas-Saiz, F. Wang, P. Tattersall and M. G. Rossmann, Structure, 1998, 6, 1369–1381 CrossRef CAS PubMed.
- M. Kontou, L. Govindasamy, H. J. Nam, N. Bryant, A. L. Llamas-Saiz, C. Foces-Foces, E. Hernando, M. P. Rubio, R. McKenna, J. M. Almendral and M. Agbandje-McKenna, J. Virol., 2005, 79, 10931–10943 CrossRef CAS PubMed.
- J. P. Michel, I. L. Ivanovska, M. M. Gibbons, W. S. Klug, C. M. Knobler, G. J. Wuite and C. F. Schmidt, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 6184–6189 CrossRef CAS PubMed.
- R. Ramalho, S. Rankovic, J. Zhou, C. Aiken and I. Rousso, Retrovirology, 2016, 13, 17 CrossRef PubMed.
- M. G. M. van Rosmalen, G. R. Nemerow, G. J. L. Wuite and W. H. Roos, J. Biol. Phys., 2018, 44, 119–132 CrossRef CAS PubMed.
- W. H. Roos, I. Gertsman, E. R. May, C. L. Brooks 3rd, J. E. Johnson and G. J. Wuite, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 2342–2347 CrossRef CAS PubMed.
- E. Hernando, A. L. Llamas-Saiz, C. Foces-Foces, R. McKenna, I. Portman, M. Agbandje-McKenna and J. M. Almendral, Virology, 2000, 267, 299–309 CrossRef CAS PubMed.
- M. Medrano, M. A. Fuertes, A. Valbuena, P. J. Carrillo, A. Rodríguez-Huete and M. G. Mateu, J. Am. Chem. Soc., 2016, 138, 15385–15396 CrossRef CAS PubMed.
- E. Lombardo, J. C. Ramírez, M. Agbandje-McKenna and J. M. Almendral, J. Virol., 2000, 74, 3804–3814 CrossRef CAS PubMed.
- L. Riolobos, J. Reguera, M. G. Mateu and J. M. Almendral, J. Mol. Biol., 2006, 357, 1026–1038 CrossRef CAS PubMed.
- J. M. Almendral., in Structure and Physics of Viruses, ed. M. G. Mateu, Springer, Dordrecht, the Netherlands, 2013, pp. 307–328 Search PubMed.
- M. Castellanos, R. Pérez, P. J. P. Carrillo, P. J. de Pablo and M. G. Mateu, Biophys. J., 2012, 102, 2615–2624 CrossRef CAS PubMed.
- J. Reguera, A. Carreira, L. Riolobos, J. M. Almendral and M. G. Mateu, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 2724–2729 CrossRef CAS PubMed.
- P. Ceres and A. Zlotnick, Biochemistry, 2002, 41, 11525–11531 CrossRef CAS PubMed.
- M. Carrillo-Tripp, C. M. Shepherd, I. A. Borelli, S. Venkataraman, G. Lander, P. Natarajan, J. E. Johnson, C. L. Brooks 3rd and V. Reddy, Nucleic Acids Res., 2009, 37, D436–D442 CrossRef CAS PubMed.
- G. E. Kempson, Ann. Rheum. Dis., 1982, 41, 508–511 CrossRef CAS PubMed.
- P. Zioupos and J. D. Currey, Bone, 1998, 22, 57–66 CrossRef CAS PubMed.
- V. Krishnamani, C. Globisch, C. Peter and M. Deserno, Eur. Phys. J.: Spec. Top., 2016, 225, 1757–1774 CAS.
- A. Carreira, M. Menéndez, J. Reguera, J. M. Almendral and M. G. Mateu, J. Biol. Chem., 2003, 279, 6517–6525 CrossRef PubMed.
- F. Moreno-Herrero, J. Colchero, J. Gómez-Herrero and A. M. Baró, Phys. Rev. E: Stat., Nonlinear, Soft Matter Phys., 2004, 69, 031915 CrossRef CAS PubMed.
- I. Horcas, R. Fernández, J. Gomez-Rodriguez, J. Colchero, J. Gómez-Herrero and A. Baró, Rev. Sci. Instrum., 2007, 78, 013705 CrossRef CAS PubMed.
- J. E. Sader, J. W. Chon and P. Mulvaney, Rev. Sci. Instrum., 1999, 70, 3967–3969 CrossRef CAS.
- I. L. Ivanovska, R. Miranda, J. L. Carrascosa, G. J. Wuite and C. F. Schmidt, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 12611–12616 CrossRef CAS PubMed.
- W. L. DeLano, The Pymol Molecular Graphics System, San Carlos, CA, USA, 2002. URL: http://www.pymol.org Search PubMed.
- W. Humphrey, A. Dalke and K. Schulten, J. Mol. Graphics, 1996, 14, 33–38 CrossRef CAS PubMed.
- G. Vriend, J. Mol. Graphics, 1990, 8, 52–56 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9nr01935a |
| This journal is © The Royal Society of Chemistry 2019 |