Open Access Article
Open Access ArticleAn automatable platform for genotoxicity testing of nanomaterials based on the fluorometric γ-H2AX assay reveals no genotoxicity of properly surface-shielded cadmium-based quantum dots†
D.
Geißler
 a,
M.
Wegmann
ab,
T.
Jochum
c,
V.
Somma
b,
M.
Sowa
b,
J.
Scholz
b,
E.
Fröhlich
a,
M.
Wegmann
ab,
T.
Jochum
c,
V.
Somma
b,
M.
Sowa
b,
J.
Scholz
b,
E.
Fröhlich
 d,
K.
Hoffmann
d,
K.
Hoffmann
 a,
J.
Niehaus
d,
D.
Roggenbuck
be and
U.
Resch-Genger
a,
J.
Niehaus
d,
D.
Roggenbuck
be and
U.
Resch-Genger
 *a
*a
aBundesanstalt für Materialforschung und -prüfung (BAM), Division 1.2 Biophotonics, Richard-Willstätter-Str. 11, 12489 Berlin, Germany. E-mail: ute.resch@bam.de
bMEDIPAN GmbH, Ludwig-Erhard-Ring 3, 15827 Dahlewitz, Germany
cFraunhofer-Zentrum für Angewandte Nanotechnologie CAN, Grindelallee 117, 20146 Hamburg, Germany
dMedizinische Universität Graz, Zentrum für Medizinische Forschung (ZMF), Stiftingtalstrasse 24, 8010 Graz, Austria
eInstitute of Biotechnology, Faculty Environment and Natural Sciences, Brandenburg University of Technology, Germany
First published on 9th July 2019
Abstract
The large number of nanomaterial-based applications emerging in the materials and life sciences and the foreseeable increasing use of these materials require methods that evaluate and characterize the toxic potential of these nanomaterials to keep safety risks to people and environment as low as possible. As nanomaterial toxicity is influenced by a variety of parameters like size, shape, chemical composition, and surface chemistry, high throughput screening (HTS) platforms are recommended for assessing cytotoxicity. Such platforms are not yet available for genotoxicity testing. Here, we present first results obtained for application-relevant nanomaterials using an automatable genotoxicity platform that relies on the quantification of the phosphorylated histone H2AX (γ-H2AX) for detecting DNA double strand breaks (DSBs) and the automated microscope system AKLIDES® for measuring integral fluorescence intensities at different excitation wavelengths. This platform is used to test the genotoxic potential of 30 nm-sized citrate-stabilized gold nanoparticles (Au-NPs) as well as micellar encapsulated iron oxide nanoparticles (FeOx-NPs) and different cadmium (Cd)-based semiconductor quantum dots (QDs), thereby also searching for positive and negative controls as reference materials. In addition, the influence of the QD shell composition on the genotoxic potential of these Cd-based QDs was studied, using CdSe cores as well as CdSe/CdS core/shell and CdSe/CdS/ZnS core/shell/shell QDs. Our results clearly revealed the genotoxicity of the Au-NPs and its absence in the FeOx-NPs. The genotoxicity of the Cd-QDs correlates with the shielding of their Cd-containing core, with the core/shell/shell architecture preventing genotoxicity risks. The fact that none of these nanomaterials showed cytotoxicity at the chosen particle concentrations in a conventional cell viability assay underlines the importance of genotoxicity studies to assess the hazardous potential of nanomaterials.
Introduction
One of the key technologies of this century is nanotechnology, generating and utilizing inorganic, organic, and hybrid functional nanomaterials with one or more dimensions below 100 nm and specific optical, electrical, magnetic or catalytic characteristics.1–3 Applications for such nanomaterials range from devices for energy generation like solar cells, batteries, and fuel cells, and solid-state lighting like converter materials for light emitting diodes and optically active materials for plasma displays, over nanoelectronics such as printable conductive inks, to the health sector and nanomaterial-containing consumer products.4–7 Examples are metal nanomaterials like gold nanoparticles (Au-NPs) assessed for different diagnostic applications as well as for drug and gene delivery, silver nanoparticles for antibacterial uses in consumer products like clothes, and iron oxide nanoparticles (FeOx-NPs) as contrast agents for nuclear magnetic resonance imaging (MRI) and cancer therapy via hyperthermia. All these applications require biocompatibility and no cytotoxicity.8,9 Moreover, the ongoing increase in the production of unique nanomaterials of different sizes, shapes, chemical compositions, charges, and surface chemistries triggers the need for reliable methods to evaluate and characterize the toxic potential of these nanomaterials to keep safety risks to people and environment as low as possible.10 These methods must also be generally accepted, as for example studies with semiconductor quantum dots (QDs) of similar physicochemical characteristics and other nanomaterials revealed differences in toxicity results. These differences originate from cell type-dependent variations,11,12 NP degradation in certain cellular compartments,13 serum protein-concentration dependent degree of QD agglomeration, different compositions of the cell culture media used,14 NP concentration,15–17 and varying exposure times.This underlines the importance of the awareness on the limitations when assessing the complex interactions of nanoparticles with and their influence on biological entities.18,19 In addition, the nanoparticles used for such toxicity studies must be well characterized with respect to the physicochemical parameters that can potentially affect toxicity like size, size distribution, shape, charge, and surface chemistry.20–22 This also requires control experiments with the applied capping/surface ligand(s), as they not only play an important role in rationally designing and tuning some of the physicochemical properties of functional nanomaterials for e.g. biomedical and optoelectronic applications,23,24 but also affect their interactions with and influence on biological systems. Here, effects such as dilution-induced ligand desorption, which can also affect toxicity, must be considered for NPs with coordinatively bound ligands like QDs composed of potentially hazardous constituents such as heavy metal ions like cadmium and lead.
Moreover, there are increasing concerns regarding the potential genotoxicity of nanomaterials which are often not addressed in many toxicity studies that focus mainly on cytotoxicity.25–30 This triggered the development of a battery of in vitro assays addressing nanomaterial-induced DNA damage to assess a number of genotoxicity endpoints to minimize costly in vivo testing.31 Genotoxicity can be unveiled using the AMES test (the bacterial reverse mutation test used to determine the mutagenicity of exogenous substances; OECD 471), micronucleus (MN; gross chromosomal damage; MNvit OECD 487), and hypoxanthine guanine phosphoribosyltransferase gene forward mutation assay (HPRT assay; point mutagenicity; OECD 476).25 In addition, the COMET assay, the mouse lymphoma L5178Y cell thymidine kinase gene mutation assay (MLA), and the mammalian chromosomal aberration test can be employed, which detect DNA double strand breaks (DSBs). For the detection of DSBs, the quantification of phosphorylated histone H2AX (γ-H2AX), by ELISA, flow cytometry, western blotting, or microscopy, proved to be suitable because, compared to other methods for the detection of DSBs like the COMET assay, the γ-H2AX assay shows a higher sensitivity and specificity and provides a direct link to the formation of DSBs.32–34 Various high throughput screening (HTS) systems for detecting γ-H2AX have been used for assessing the genotoxic effects of radiation exposure35 and of chemicals such as radiosensitizers, environmental toxicants, and drugs by employing different detection techniques including optical microscopy.36–41 Evaluation by optical microscopy has the huge advantage that each subnuclear focus, i.e. the local accumulation or modification of DNA damage response proteins at the site of a DNA double-strand break,42 is thought to represent a single DSB,43 enabling direct DSB counting. In addition, a direct correlation of DSB results and cell morphology is possible. Meanwhile, assays measuring DSBs have also been employed for assessing the potential genotoxicity of NPs. Examples are HTS platforms for detecting DSBs caused by NPs utilizing the COMET assay.44
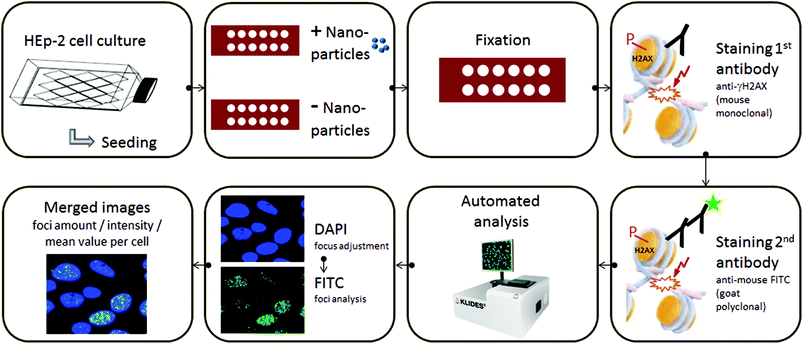
Assessing the large number of already existing nanomaterials together with the huge number of continuously emerging new nanoparticles (NPs) calls for efficient and standardized test procedures to screen these materials not only for their potential cytotoxicity but also for their genotoxicity, preferably in an automatable manner. In this respect and to identify negative and positive controls for assay users, we assessed the feasibility of the automatable fluorometric γ-H2AX assay for the detection of nanomaterial-induced DSBs in exemplarily chosen HEp-2 cells. The automated assessment of γ-H2AX foci has been performed by using several approaches for high-throughput analysis35,45,46 that efficiently provides the quantification of foci size, number, and intensity. In our work, we use the automated fluorescence microscopic detection system AKLIDES® for γ-H2AX foci detection, as schematically shown in Fig. 1.47–49 AKLIDES software, which has been validated by comparison with manual data evaluation using the visual recognition of γ-H2AX foci,47,48 automatically performs a qualitative image analysis to eliminate artefacts, thereby excluding cell aggregates and heavily damaged cells, and detects foci by local thresholds of objects inside the cell nucleus followed by the morphological characterization of each object. The latter allows for a wide range of detectable foci.47 In addition, a fully automated image acquisition based on Haralick's image characterization of objects through a grey-scale transition is done using DAPI as the fluorescent dye for focusing, quality evaluation, and object recognition.
The representative nanomaterials selected for this proof-of-concept study were of similar matching sizes, yet with different chemical compositions and surface chemistries. This included frequently used citrate-stabilized 30 nm Au-NPs as well as FeOx-NPs and three Cd-based QDs having a similar Cd content but differing in their particle architecture and surface passivation, all of which bear a tightly bound shell of an amphiphilic polymer resulting in a hydrodynamic diameter of about 30 nm.50 As a positive control, CdCl2 was used, known to be a potent inducer of genotoxicity. In the following, first results of this genotoxicity study are presented and compared to the cytotoxic potential of these nanomaterials assessed using the conventional MTS assay51,52 for the same cell line under identical conditions.53–55
Experimental
Materials
HEp-2 cells (ATCC® CCL-23) were obtained from ATCC/LGC Standards. Dulbecco's phosphate buffered saline (PBS) was obtained from Biochrom/Merck, protease-free bovine serum albumin (BSA) was purchased from SERION Immunologics, formalin solution was obtained from Carl Roth, CdCl2 and Triton X-100 were purchased from Sigma-Aldrich. Monoclonal mouse anti-phospho-histone H2AX antibodies were purchased from Merck, polyclonal Alexa Fluor 488- (AF488-) labeled goat anti-mouse antibodies were obtained from Dianova, 4,6′-diamidino-2-phenylindole (DAPI)-mounting medium was obtained from the AKLIDES® Cell Damage reagent kit from MEDIPAN GmbH. Dulbecco's modified Eagle medium (DMEM) with GlutaMAX supplement and fetal bovine serum (FBS) were purchased from Gibco. Penicillin/streptomycin was obtained from Life Technologies GmbH/ThermoFisher. The CellTiter 96® AQueous Non-Radioactive Cell Proliferation assay including (3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium (MTS) and phenazine methosulfate (PMS) was obtained from Promega. All solutions and buffers were prepared with Milli-Q water (Millipore). The chemicals used for NP synthesis are described in the respective publications cited (see below).NP synthesis
NP characterization
NP toxicity testing
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 HEp-2 cells were seeded per well and cultured for 24 h prior to the exposure with the samples. Visual checks for changes in morphology and cell density were performed by microscopy after 4 h and 24 h. After dilution with cell culture medium, the NPs were added to the cells in similar concentrations to those used for genotoxicity testing. After incubation with the NPs, the cells were washed with PBS. Positive controls (cells treated with 70% ethanol + 2 μL TritonX-100 for 10 minutes), negative controls (cells treated with cell culture medium), color controls (NP without cells), and solvent controls (water in medium) were included in the cytotoxicity testing. The cell viability was measured using CellTiter 96® AQueous Non-Radioactive Cell Proliferation Assay (Promega). The MTS and PMS solutions were thawed, 100 μL PMS solution was mixed with 2 mL MTS solution, and 20 μL of the combined MTS/PMS solution was added to 100 μL of each well. The plates were then incubated for 2 h at 37 ± 1 °C and 5% CO2 in a cell incubator. During the assay, the tetrazolium compound MTS is reduced by cellular dehydrogenases into a formazan product that is soluble in tissue culture medium and can be quantified photometrically.51,52 Thus, the absorbance at 490 nm was measured with a microplate reader (SPECTRA MAX plus 384, Molecular Devices), and the dehydrogenase activity as an indicator of cell viability was calculated as the ratio of the blank-corrected absorbances of sample and control.
000 HEp-2 cells were seeded per well and cultured for 24 h prior to the exposure with the samples. Visual checks for changes in morphology and cell density were performed by microscopy after 4 h and 24 h. After dilution with cell culture medium, the NPs were added to the cells in similar concentrations to those used for genotoxicity testing. After incubation with the NPs, the cells were washed with PBS. Positive controls (cells treated with 70% ethanol + 2 μL TritonX-100 for 10 minutes), negative controls (cells treated with cell culture medium), color controls (NP without cells), and solvent controls (water in medium) were included in the cytotoxicity testing. The cell viability was measured using CellTiter 96® AQueous Non-Radioactive Cell Proliferation Assay (Promega). The MTS and PMS solutions were thawed, 100 μL PMS solution was mixed with 2 mL MTS solution, and 20 μL of the combined MTS/PMS solution was added to 100 μL of each well. The plates were then incubated for 2 h at 37 ± 1 °C and 5% CO2 in a cell incubator. During the assay, the tetrazolium compound MTS is reduced by cellular dehydrogenases into a formazan product that is soluble in tissue culture medium and can be quantified photometrically.51,52 Thus, the absorbance at 490 nm was measured with a microplate reader (SPECTRA MAX plus 384, Molecular Devices), and the dehydrogenase activity as an indicator of cell viability was calculated as the ratio of the blank-corrected absorbances of sample and control.
Results and discussion
As a proof-of-concept study for the suitability of the automatable fluorometric γ-H2AX assay for the detection of nanomaterial-induced DSBs, we assessed the genotoxicity of five representatively chosen nanomaterials of similar size, yet different chemical compositions and surface chemistries at varying NP concentrations in Hep-2 cells. This included citrate-stabilized 30 nm Au-NPs as well as 4 nm FeOx-NPs and three Cd-based QDs differing in particle architecture and surface passivation, namely CdSe core-only QDs (C-QDs), CdSe/CdS core/shell QDs (CS-QDs), and CdSe/CdS/ZnS core/shell/shell QDs (CSS-QDs). TEM images of the different nanoparticles are shown in Fig. S1 and S2 in the ESI.† Whereas the Au-NPs with their hydrophilic citrate capping ligands are directly dispersible in aqueous media, the FeOx-NPs and Cd-based QDs bear apolar surface ligands directly after NP synthesis. These hydrophobic NPs and QDs were transferred to water using a versatile micellar encapsulation strategy involving polyisoprene–diethylenetriamine (PI–N3) and a polyisoprene-b-polyethylene glycol di-block copolymer (PI-b-PEG) to realize water dispersibility (see Fig. S3 in the ESI†).64,65A micellar encapsulation of the C-QDs, CS-QDs, and CSS-QDs, prepared to have a similar Cd content, was chosen here, because the tightly bound cross-linked polymer shell on the QD surface prevents ligand desorption, as previously revealed by concentration independent fluorescence measurements,50 which underlined the extraordinary fluorescence stability of these NPs in a biological environment. Hence, a concentration-dependent diminution or loss in the QD emission intensity, associated with the ligand loss and release of Cd ions, could be precluded. Pure PI-b-PEG without any NPs forms empty micelles with a hydrodynamic diameter of approximately 30 nm. Thus, although the actual inorganic FeOx-NPs and Cd-based QDs are smaller than 10 nm, the eventually resulting hydrodynamic diameter of the micellar encapsulated particles is about 30 nm, matching the size of the empty PI-b-PEG micelles. This can be seen from the DLS size curves of the different water-dispersible NPs shown in Fig. S4 in the ESI.† This is also the reason why the Au-NPs were synthesized to be of 30 nm in the first place, thereby providing the basis to compare the performance of NPs of similar size in the genotoxicity assays. To ensure that the NP surface chemistry does not account for potential toxic effects, the empty PI-b-PEG micelles were also assessed with regard to their toxicity under identical conditions to those of the NPs. Moreover, to address the potential Cd-related toxicity of the QDs, control experiments with CdCl2 were also performed, which has been classified as a group 1 carcinogen and is known to show positive results in common mammalian in vitro genotoxicity assays.53–55
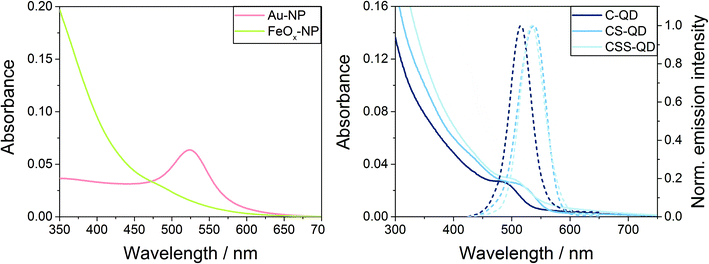
As a preliminary test for the desired automated genotoxicity test with our automated microscopy platform, spectroscopic measurements (absorption spectra; for the luminescent QDs also emission spectra) of the NPs were carried out to assess potential interferences with the fluorescent reporters DAPI and AF488 used in γ-H2AX assay. The 30 nm Au-NPs showed a plasmonic absorption band at around 525 nm, whereas the FeOx-NPs displayed an unstructured absorption spectrum with an onset of absorption at around 600 nm and an increasing absorbance towards the ultraviolet region (see Fig. 2, left). The QDs showed the typical absorption spectra of high quality II/VI semiconductor nanocrystals with a first excitonic absorption peak at 485 nm (for C-QDs) or 505–510 nm (for CS-QDs and CSS-QDs), and a narrow symmetrical emission band at 515 nm (for C-QDs) or 535–540 nm (for CS-QDs and CSS-QDs) with a full-width at half maximum of ca. 40–50 nm (Fig. 2, right). As expected, in comparison with the C-QDs, the additional shell(s) of the CS-QDs and CSS-QDs result in an increased absorption at a wavelength below 450 nm (below the onset of the absorption of the CdS and ZnS shell materials) and a slightly red-shifted emission. Thus, all NPs and QDs absorb at wavelengths where the γ-H2AX assay reporter dyes DAPI (at ca. 360 nm) and AF488 (at ca. 490 nm) also absorb. Hence, both QDs and dyes are excited during the assay readout. Moreover, the QDs also emit in the FITC emission channel (at about 520 nm) used for AF488 detection.
In order to circumvent interferences with the fluorescence signals originating from the dyes used as assay reporters, control experiments with the NPs and the respective cells alone were performed using the microscopic detection system AKLIDES®. Although this revealed slight optical interferences, the signals arising from the QDs were much weaker than those of the reporter dyes. Thus, we can exclude that the fluorescence of the QDs affects the counting of the NP-initiated DSBs by using the AKLIDES® data evaluation software, as the NP signal intensities were below the threshold applied for DSB counting.
In addition to optical characterization, the NPs were also monitored for at least 3 months regarding their long-term colloidal stability using regular absorption spectroscopy and the measurements of photoluminescence spectra, decay kinetics, and luminescence lifetimes, as well as photoluminescence quantum yields. All NP and QD samples displayed an excellent colloidal stability and revealed no significant changes in their absorption and/or emission properties over time. This makes them perfectly suitable for reliable and reproducible toxicity studies. Moreover, this is a very important prerequisite for their applications as reference materials for the automated γ-H2AX genotoxicity assay platform.
In addition, the total amount of protonable/deprotonable surface functional groups was assessed using conductometric acid/base titration.66 The results, summarized in Table S1 in the ESI,† show that the Au-NPs bear a large number of citrate ligands, and thus, functional groups (–COOH) on their surface, as expected for such electrostatically stabilized NPs. The micellar encapsulated FeOx-NPs, C-QDs, CS-QDs, and CSS-QDs have a moderate, and importantly, fairly constant number of functional groups (–NH2) on their surface. Thus, it can be excluded that the surface chemistry and surface functionalities of the micellar encapsulated particles account for differences in genotoxicity between these samples detected using the γ-H2AX assays.
To establish the Au-NPs and FeOx-NPs as γ-H2AX assay-specific positive and negative control standards for NP genotoxicity testing and to assess the genotoxicity of the Cd-based QDs with different shell compositions, the established AKLIDES® assay protocol was adapted for NP testing (see Fig. 1 and the procedure described in the Materials and methods section). AKLIDES® Cell Damage software is capable of measuring and analyzing various sample parameters automatically, such as the number of cells in total, number of foci per cell, foci diameter, foci mean intensity, and percentage of positive cells. To identify the most suitable parameter for the automated NP-induced DSB detection and quantification with the γ-H2AX assay, different concentrations of the potential reference materials Au-NPs and FeOx-NPs were studied in HEp-2 cells. Although other sample parameters yielded similar trends, the most reliable and reproducible results were obtained for the detection of foci mean number. Hence, this parameter was applied to assess NP and QD genotoxicity in this work (see Fig. 3). Interestingly, the Au-NPs displayed a considerable genotoxicity already at a very low particle concentration of 50 pM and reached the genotoxicity values of the positive control (CdCl2) at a NP concentration of 500 pM. In contrast, the FeOx-NPs showed low genotoxicity that exceeds the values of the cell culture medium used here as a negative control only at a relatively high particle concentration of 1 nM and did not reach the genotoxicity value of the positive control even at 500 nM, i.e. at a 1000-fold higher concentration, compared to the Au-NPs. The detected genotoxicity of the FeOx-NPs observed at high nM concentrations is probably not due to the iron oxide material but is most likely governed by the genotoxicity of the PI-b-PEG micelles itself. This becomes obvious from a comparison of the measured γ-H2AX foci for the FeOx-NPs and the empty micelles in Fig. 3 (left). When using an intermediate concentration between 100 pM and 500 pM, the Au-NPs display high genotoxicity, whereas the FeOx-NPs show no genotoxicity.
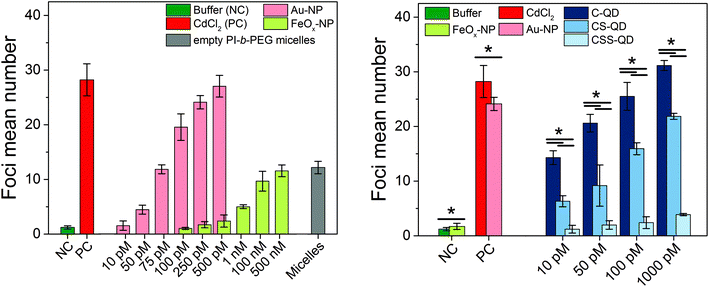 | ||
| Fig. 3 Genotoxicity studies of the Au-NPs and FeOx-NPs (left) as well as the Cd-based quantum dots (right) with different shell compositions (C-QD = CdSe core-only quantum dot, CS-QD = CdSe/CdS core/shell quantum dot, CSS-QD = CdSe/CdS/ZnS core/shell/shell quantum dot). Genotoxicity was measured in HEp-2 cells in comparison to established toxicity standards (cell culture medium as the negative control and 25 μM CdCl2 in DMEM as the positive control). For the micellar encapsulated FeOx-NPs (left), the empty PI-b-PEG micelles (concentration ca. 500 nM) were measured as an additional control to investigate the origin of the slight genotoxicity at large particle concentrations. For the Cd-based QDs (right), the potential reference materials Au-NPs and FeOx-NPs (concentration 250 pM) were additionally measured to demonstrate their use as NP-based positive and negative genotoxicity controls, respectively. Error bars represent the standard deviation derived from three independent replicates. The foci mean numbers of Au-NP and FeOx-NP concentrations (left) were significantly different (ANOVA, P < 0.001). Post-hoc analysis of the data revealed significant differences for the comparison of the foci mean numbers obtained for an Au-NP concentration of 10 pM and the results derived for measurements with Au-NP concentrations of 75, 100, 250, and 500 pM. For the FeOx-NPs, the foci mean numbers observed for incubation with 100 pM FeOx-NPs differed from those observed for 1 nM, 100 nM, and 500 nM. The P values for the comparison of two foci mean numbers (right) are marked with asterisks (* P < 0.001). For more details, see Fig. S6 and S7 in the ESI.† | ||
To evaluate and compare the quality of the results of the γ-H2AX assays, the so-called Z′ factors (screening window coefficients) were calculated, which are common quality metrics for all kinds of bioassays, and particularly for HTS assays.67,68 The Z′ factors were obtained using the established negative and positive controls (buffer and CdCl2, respectively) as well as the potential NP-based controls (FeOx-NPs and Au-NPs, respectively) identified in this study. The resulting Z′ factors were 0.64 for the established controls and 0.65 for the NP controls (particle concentration range 100–500 pM), indicating excellent assays with sufficient assay resolution in both cases. This underlines that these well-characterized Au-NPs and FeOx-NPs, which can be synthesized with a high reproducibility, are well suited as positive and negative NP-based control standards for the determination of NP genotoxicity with the γ-H2AX assay, as demonstrated in Fig. S5 in the ESI.† A standardization of the assay is important as γ-H2AX staining background differs considerably between cell lines.32
As a proof of principle, Cd-based QDs with three different particle architectures and surface passivation were tested for their genotoxicity with the γ-H2AX assay using the exact same assay conditions (incubation time, washing steps, detection parameters) as those for the Au-NPs and FeOx-NPs. These QDs varied in the number and material of the inorganic shells, ranging from CdSe core-only (C-) QDs over CdSe/CdS core/shell (CS-) QDs to CdSe/CdS/ZnS core/shell/shell (CSS-) QDs, but were all encapsulated using PI-N3/PI-b-PEG micelles. The results of the γ-H2AX assays for these QDs, shown in Fig. 3 (right), demonstrate that the shell composition strongly influences the genotoxicity of these NPs, which are typically considered as inherently toxic due to their Cd content. As expected, the CdSe core-only QDs without any protective shell displayed high genotoxicity even at the lowest particle concentration of 10 pM. The CdSe/CdS core/shell QDs, in contrast, showed a reduced genotoxicity as indicated by a reduction of the foci mean number by a factor of 2. The CdSe/CdS/ZnS core/shell/shell QDs, which contain the same Cd amount as the C- and CS-QDs, revealed no significant genotoxicity even at the highest tested QD concentration of 1 nM. This underlines the efficient shielding of the CdSe core by the shell/shell architecture and the beneficial impact of the outer ZnS shell. These results demonstrate that the genotoxicity of Cd-based QDs, which are typically considered as toxic, can be reduced or even completely suppressed by an appropriate shell design together with optimized and reproducible synthetic procedures, and an efficient and versatile phase transfer strategy utilizing micellar encapsulation.
In the γ-H2AX assay, NP-induced DNA damage and the subsequent activation of the cell's repair mechanisms are used as an indicator of genotoxic action. However, DNA DSBs and fragmentation can also be caused by apoptosis because of the cytotoxic action of the NPs, and phosphorylation of the H2AX histone occurs in both genotoxicity and cytotoxicity cases.69 Thus, in vitro cytotoxicity screening, which is a routine step in the evaluation of biological effects, is also needed for the correct interpretation of the genotoxicity testing results shown above.70–72 When testing NPs that need active uptake to cause genotoxicity, recommended concentrations should range from nontoxic to around 80% cell viability to exclude DNA DSBs as a secondary effect of cytotoxicity.73 Moreover, the exposure time must be sufficiently long (18–24 h) to allow nanoparticle uptake, and appropriate controls are recommended to correct for, e.g., NP absorption at the detection wavelength. Hence, to validate that the detected DNA double strand breaks are in fact due to NP-induced genotoxicity, NPs from the same batch were assessed for their potential cytotoxicity using exactly the same concentration ranges, exposure time and cell line as for genotoxicity testing. To detect potential NP cytotoxicity, colorimetric MTS assay was used to measure cell viability after incubation with the NP samples. To correct for NP absorption, color controls (nanoparticles without cells) were included in the testing and showed only 0–3.6% of the absorbance of unexposed cells, meaning negligible. The cell viability of the cells in the positive control decreased to 5–8%. Viability values of the cells exposed to solvent controls ranged from 78% to 114% of the untreated cells. The lower viability of the solvent controls is due to the dilution of the nutrients in the cell culture medium by the addition of water. The results of the MTS cytotoxicity assays with the NP and QD samples are displayed in Fig. 4. From the results of the MTS assays shown in Fig. 4 it becomes obvious that for none of the NP or QD samples the viability of the Hep2 cells decreased to <90%. According to the classification used for the testing of medical devices, decrease of viability to <70% of the non-exposed cells is interpreted as cytotoxicity.74 This implies that the NPs and QDs assessed did not show cytotoxicity in the concentration range used for the genotoxicity testing. Since the tested concentrations were already unrealistically high, we deliberately did not use higher NP concentrations/doses for this proof-of-concept study. To confirm that the events caused by the NPs are DSBs and hence a critical kind of DNA damage, which can also cause cytotoxicity, one could also include higher doses in the NP testing and study whether a dose-related increase in both γ-H2AX foci and cytotoxicity is observed as for conventional molecular compounds.40 However, we demonstrated that the genotoxicity studies with the Au-NPs, FeOx-NPs and Cd-based QDs using the fluorometric γ-H2AX assay in combination with the microscopic detection system AKLIDES® can be applied as a fast and easy to use, automatable detection method for genotoxicity testing of various nanomaterial classes when using the standardized test procedure developed in this work.
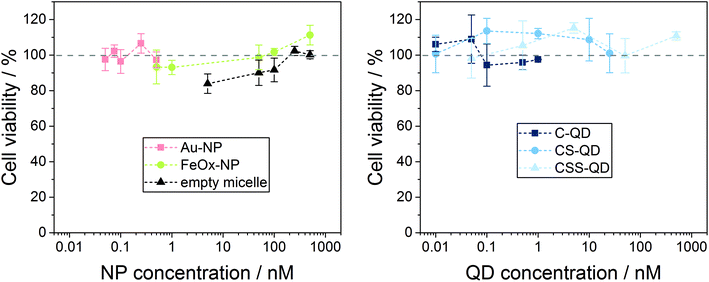 | ||
| Fig. 4 Results of cytotoxicity tests using the MTS assay for the different NP and QD samples as well as the empty micelles. Values were normalized to the viability of untreated cells as 100%. | ||
Conclusion and outlook
We developed a new detection method for genotoxicity screening of nanomaterials, based on the fluorometric γ-H2AX assay and the automated microscopic platform AKLIDES®. The comparison of the results of our genotoxicity assays using this assay platform revealed genotoxicity for 30 nm-sized citrate-stabilized gold nanoparticles (Au-NP) and 30 nm-sized micellar encapsulated CdSe core and CdSe/CdS core/shell semiconductor quantum dots (QDs).The observed genotoxicity induced by 30 nm Au-NPs in our study may be unexpected because in general gold NPs are regarded as biocompatible and genotoxicity of Au-NPs with sizes >20 nm has been rarely reported. Au-NPs <20 nm are small enough to penetrate the nucleus and accumulate there, with the generation of reactive oxygen species (ROS) being reported as the main mode of action. The proposed mechanisms for ROS-induced damage in the nucleus are the generation of DNA–protein crosslinks, damage of the deoxyribose phosphate backbone, specific modifications of purine and pyrimidine bases, and damage of DNA repair proteins.75,76 In contrast, very small gold particles of 1.4 nm can directly interact with nuclear DNA.77
A genotoxicity study of larger Au-NPs with sizes of 30, 50, and 90 nm has been reported only recently based on the Somatic Mutation and Recombination Test (SMART). Here, Au-NPs up to 90 nm induced genotoxicity, for example, in Drosophila cells and DNA double strand breaks in human HepG2 and HL-60 cells. The oxidation of pyrimidines and purines was identified as an important mode of action.78 Other studies suggest that citrate-coating can also increase the genotoxicity of Au-NPs.79 The controversial findings for the genotoxicity of Au-NPs may be due to the fact that a repair of DNA damage may also occur after Au-NP exposure.80 We can only speculate on the mechanism of the genotoxicity induced by the 30 nm-sized AuNPs in this study. Although our 30 nm-sized citrate-stabilized Au NPs are too large to penetrate the nucleus, they may nevertheless be able to generate ROS in the cytoplasm which could then induce oxidative damage in the nucleus.
Cytotoxicity studies using the conventional MTS cell viability assay, however, revealed no indication of cytotoxicity for all nanomaterials at the assessed particle concentrations. This suggests that the DNA damage caused by low concentrations of certain nanomaterials does not necessarily cause cell death but may convey a genotoxic risk as has been reported in a few studies on the genotoxicity of Cd-based nanoparticles, which also did not lead to cytotoxic effects.70–72 The results of our study underline the importance of genotoxicity studies for the safe use of nanomaterials, which can straightforwardly be addressed by the HTS system AKLIDES®, characterized by its high sensitivity for genotoxicity.
Our results can also be employed to establish nanomaterials with well-characterized physicochemical properties including surface chemistry as positive and negative controls or standards for the regular performance validation for users of this fluorometric genotoxicity assay. This enables the setting up of the assay for inexperienced users and day-to-day performance control. In this context, the potential of the tested Au-NPs and core and core–shell QDs as positive controls and the potential of the micellar encapsulated iron oxide nanoparticles (FeOx-NPs) and the micellar encapsulated CdSe/CdS/ZnS core/shell/shell QDs as negative controls is currently being studied, also in combination with the long-term stability of these nanomaterials. Supported by the positive and negative controls identified in this study and the in parallel ongoing development of reference materials for instrument performance validation, like micrometer-sized fluorescent calibration beads matching the reporters used in the γ-H2AX assay run on the platform AKLIDES®, this automated assay platform is expected to provide the basis for better comparable and reliable results of genotoxicity studies and the speeding up of such measurements.
In the case of the different QDs, the genotoxicity data presented also underline the importance of surface passivation and protection for nanomaterials that consist of hazardous constituents such as heavy metal ions, and the potential of a suitable surface chemistry to prevent or at least strongly reduce such health and environmental risks. For example, for our micellar encapsulated QDs, we could demonstrate the transformation of initially genotoxic CdSe cores into non-genotoxic QDs by growing a ZnS passivation shell on its surface. This highlights the considerable importance of surface passivation and underlines that information on the chemical composition of a nanomaterial alone does not enable the estimation of its cytotoxic and genotoxic potential.
Conflicts of interest
D. G., K. H., U. R. G., and E. F. declare no competing financial interest. M. W., V. S., and J. S. are employees and D. R. is chief executive officer of MEDIPAN GmbH which is the manufacturer and provider of the AKLIDES® microscopic system and AKLIDES® diagnostic kits. T. J. and J. N. are employees of the Fraunhofer-Zentrum für Angewandte Nanotechnologie CAN which might benefit from a possible commercial launch of the NP-based negative and positive standards for γ-H2AX genotoxicity assays.Acknowledgements
The authors acknowledge the financial support from the Federal Ministry of Education and Research (BMBF) via the “KMU-innovativ: Nanotechnologie” (NanoChance) project 03XP0018C “NanoGenotox”.References
- W. J. Stark, P. R. Stoessel, W. Wohlleben and A. Hafner, Chem. Soc. Rev., 2015, 44, 5793–5805 RSC.
- R. Ghosh Chaudhuri and S. Paria, Chem. Rev., 2011, 112, 2373–2433 CrossRef PubMed.
- F. P. Zamborini, L. Bao and R. Dasari, Anal. Chem., 2012, 84, 541–576 CrossRef CAS PubMed.
- J. J. Giner-Casares, M. Henriksen-Lacey, M. Coronado-Puchau and L. M. Liz-Marzan, Mater. Today, 2016, 19, 19–28 CrossRef CAS.
- D. Kim, J. Kim, Y. I. Park, N. Lee and T. Hyeon, ACS Cent. Sci., 2018, 4, 324–336 CrossRef CAS PubMed.
- B. Pelaz, C. H. Alexiou, R. A. Alvarez-Puebla, F. Alves, A. M. Andrews, S. Ashraf, L. P. Balogh, L. Ballerini, A. Bestetti, C. Brendel, S. Bosi, M. Carril, W. C. W. Chan, C. Y. Chen, X. D. Chen, X. Y. Chen, Z. Cheng, D. X. Cui, J. Z. Du, C. Dullin, A. Escudero, N. Feliu, M. Y. Gao, M. George, Y. Gogotsi, A. Grunweller, Z. W. Gu, N. J. Halas, N. Hampp, R. K. Hartmann, M. C. Hersam, P. Hunziker, J. Jian, X. Y. Jiang, P. Jungebluth, P. Kadhiresan, K. Kataoka, A. Khademhosseini, J. Kopecek, N. A. Kotov, H. F. Krug, D. S. Lee, C. M. Lehr, K. W. Leong, X. J. Liang, M. L. Lim, L. M. Liz-Marzan, X. M. Ma, P. Macchiarini, H. Meng, H. Mohwald, P. Mulvaney, A. E. Nel, S. M. Nie, P. Nordlander, T. Okano, J. Oliveira, T. H. Park, R. M. Penner, M. Prato, V. Puntes, V. M. Rotello, A. Samarakoon, R. E. Schaak, Y. Q. Shen, S. Sjoqvist, A. G. Skirtach, M. G. Soliman, M. M. Stevens, H. W. Sung, B. Z. Tang, R. Tietze, B. N. Udugama, J. S. VanEpps, T. Weil, P. S. Weiss, I. Willner, Y. Z. Wu, L. L. Yang, Z. Yue, Q. Zhang, Q. Zhang, X. E. Zhang, Y. L. Zhao, X. Zhou and W. J. Parak, ACS Nano, 2017, 11, 2313–2381 CrossRef CAS PubMed.
- D. Bobo, K. J. Robinson, J. Islam, K. J. Thurecht and S. R. Corrie, Pharm. Res., 2016, 33, 2373–2387 CrossRef CAS PubMed.
- P. Rivera-Gil, D. J. De Aberasturi, V. Wulf, B. Pelaz, P. Del Pino, Y. Y. Zhao, J. M. De La Fuente, I. R. De Larramendi, T. Rojo, X. J. Liang and W. J. Parak, Acc. Chem. Res., 2013, 46, 743–749 CrossRef CAS PubMed.
- N. Lewinski, V. Colvin and R. Drezek, Small, 2008, 4, 26–49 CrossRef CAS PubMed.
- A. Gnach, T. Lipinski, A. Bednarkiewicz, J. Rybka and J. A. Capobianco, Chem. Soc. Rev., 2015, 44, 1561–1584 RSC.
- B. B. Manshian, S. J. Soenen, A. Al-Ali, A. Brown, N. Hondow, J. Wills, G. J. S. Jenkins and S. H. Doak, Toxicol. Sci., 2015, 144, 246–258 CrossRef CAS PubMed.
- F. Joris, D. Valdeperez, B. Pelaz, T. Q. Wang, S. H. Doak, B. B. Manshian, S. J. Soenen, W. J. Parak, S. C. De Smedt and K. Raemdonck, Acta Biomater., 2017, 55, 204–213 CrossRef CAS PubMed.
- S. J. Soenen, W. J. Parak, J. Rejman and B. Manshian, Chem. Rev., 2015, 115, 2109–2135 CrossRef CAS PubMed.
- F. Joris, B. B. Manshian, K. Peynshaert, S. C. De Smedt, K. Braeckmans and S. J. Soenen, Chem. Soc. Rev., 2013, 42, 8339–8359 RSC.
- B. B. Manshian, S. Munck, P. Agostinis, U. Himmelreich and S. J. Soenen, Sci. Rep., 2015, 5, 13890 CrossRef PubMed.
- B. B. Manshian, A. M. Abdelmonem, K. Kantner, B. Pelaz, M. Klapper, C. Nardi Tironi, W. J. Parak, U. Himmelreich and S. J. Soenen, Nanotoxicology, 2016, 10, 1318–1328 CrossRef CAS PubMed.
- S. J. Soenen, S. Abe, B. B. Manshian, T. Aubert, Z. Hens, S. C. De Smedt and K. Braeckmans, J. Biomed. Nanotechnol., 2015, 11, 631–643 CrossRef CAS PubMed.
- B. B. Manshian, U. Himmelreich and S. J. Soenen, Chem. Res. Toxicol., 2017, 30, 595–603 Search PubMed.
- M. Henriksen-Lacey, S. Carregal-Romero and L. M. Liz-Marzan, Bioconjugate Chem., 2017, 28, 212–221 CrossRef CAS PubMed.
- B. B. Manshian, C. Pfeiffer, B. Pelaz, T. Heimerl, M. Gallego, M. Moller, P. del Pino, U. Himmelreich, W. J. Parak and S. J. Soenen, ACS Nano, 2015, 9, 10431–10444 CrossRef CAS PubMed.
- P. del Pino, F. Yang, B. Pelaz, Q. Zhang, K. Kantner, R. Hartmann, N. M. de Baroja, M. Gallego, M. Moller, B. B. Manshian, S. J. Soenen, R. Riedel, N. Hampp and W. J. Parak, Angew. Chem., Int. Ed., 2016, 55, 5483–5487 CrossRef CAS PubMed.
- H. Oliveira, A. Bednarkiewicz, A. Falk, E. Fröhlich, D. Lisjak, A. Prina-Mello, S. Resch, C. Schimpel, I. V. Vrček, E. Wysokińska and H. H. Gorris, Adv. Healthcare Mater., 2019, 8, 1801233 Search PubMed.
- M. A. Boles, D. Ling, T. Hyeon and D. V. Talapin, Nat. Mater., 2016, 15, 141–153 CrossRef CAS PubMed.
- J. Hühn, C. Carrillo-Carrion, M. G. Soliman, C. Pfeiffer, D. Valdeperez, A. Masood, I. Chakraborty, L. Zhu, M. Gallego, Z. Yue, M. Carril, N. Feliu, A. Escudero, A. M. Alkilany, B. Pelaz, P. del Pino and W. J. Parak, Chem. Mater., 2017, 29, 399–461 CrossRef.
- S. H. Doak, B. Manshian, G. J. S. Jenkins and N. Singh, Mutat. Res., Genet. Toxicol. Environ. Mutagen., 2012, 745, 104–111 CrossRef CAS PubMed.
- B. B. Manshian, S. J. Soenen, A. Brown, N. Hondow, J. Wills, G. J. S. Jenkins and S. H. Doak, Mutagenesis, 2016, 31, 97–106 CAS.
- R. Landsiedel, M. D. Kapp, M. Schulz, K. Wiench and F. Oesch, Mutat. Res., Rev. Mutat. Res., 2009, 681, 241–258 CrossRef CAS PubMed.
- S. J. Evans, M. J. D. Clift, N. Singh, J. D. Mallia, M. Burgum, J. W. Wills, T. S. Wilkinson, G. J. S. Jenkins and S. H. Doak, Mutagenesis, 2017, 32, 233–241 CrossRef CAS PubMed.
- W. K. B. Khalil, E. Girgis, A. N. Emam, M. B. Mohamed and K. V. Rao, Chem. Res. Toxicol., 2011, 24, 640–650 Search PubMed.
- E. Demir and V. Castranova, Sci. Pages Nanotechnol., 2017, 1, 1–19 Search PubMed.
- M. Dusinska, E. Mariussen, E. Runden-Pran, A. M. Hudecova, E. Elje, A. Kazimirova, N. El Yamani, N. Dommershausen, J. Tharmann, D. Fieblinger, F. Herzberg, A. Luch and A. Haase, Methods Mol. Biol., 2019, 1894, 83–122 CrossRef PubMed.
- B. C. Nelson, C. W. Wright, Y. Ibuki, M. Moreno-Villanueva, H. L. Karlsson, G. Hendriks, C. M. Sims, N. Singh and S. H. Doak, Mutagenesis, 2017, 32, 215–232 CrossRef CAS PubMed.
- R. Wan, Y. Mo, R. Tong, M. Gao and Q. Zhang, Methods Mol. Biol., 2019, 1894, 145–159 CrossRef PubMed.
- A. Reddig, D. Roggenbuck and D. Reinhold, J. Lab. Precis. Med., 2018, 3, 80 CrossRef.
- Y. N. Hou, A. Lavaf, D. Huang, S. Peters, R. Huq, V. Friedrich, B. S. Rosenstein and J. Kao, Radiat. Res., 2009, 171, 360–367 CrossRef CAS PubMed.
- S. B. Fu, Y. Yang, D. Tirtha, Y. Yen, B. S. Zhou, M. M. Zhou, M. Ohlmeyer, E. C. Ko, R. Cagan, B. S. Rosenstein, S. H. Chen and J. Kao, PLoS One, 2012, 7, e38465 CrossRef CAS PubMed.
- C. Garcia-Canton, A. Anadon and C. Meredith, Mutat. Res., Genet. Toxicol. Environ. Mutagen., 2013, 757, 158–166 CrossRef CAS PubMed.
- G. Harris, T. Palosaari, Z. Magdolenova, M. Mennecozzi, J. M. Gineste, L. Saavedra, A. Milcamps, A. Huk, A. R. Collins, M. Dusinska and M. Whelan, Nanotoxicology, 2015, 9(Suppl 1), 87–94 CrossRef CAS PubMed.
- B. Kopp, L. Hegarat, D. Zalko and M. Audebert, Environ. Mol. Mutagen., 2017, 58, S71–S72 Search PubMed.
- T. Nikolova, M. Dvorak, F. Jung, I. Adam, E. Kramer, A. Gerhold-Ay and B. Kaina, Toxicol. Sci., 2014, 140, 103–117 CrossRef CAS PubMed.
- K. W. Yip, M. Cuddy, C. Pinila, M. Giulanotti, S. Heynen-Genel, S. I. Matsuzawa and J. C. Reed, J. Biomol. Screening, 2011, 16, 251–258 CrossRef CAS PubMed.
- K. Rothkamm, S. Barnard, J. Moquet, M. Ellender, Z. Rana and S. Burdak-Rothkamm, Environ. Mol. Mutagen., 2015, 56, 491–504 CrossRef CAS PubMed.
- M. Podhorecka, A. Skladanowski and P. Bozko, J. Nucleic Acids, 2010, 2010, 920161 Search PubMed.
- A. Azqueta, I. C. Costa-Amaral and A. R. Collins, Issues Toxicol., 2017, 30, 67–92 Search PubMed.
- W. Boker and G. Iliakis, Radiat. Res., 2006, 165, 113–124 CrossRef.
- S. V. Costes, A. Boissiere, S. Ravani, R. Romano, B. Parvin and M. H. Barcellos-Hoff, Radiat. Res., 2006, 165, 505–515 CrossRef CAS PubMed.
- A. Willitzki, S. Lorenz, R. Hiemann, K. Guttek, A. Goihl, R. Hartig, K. Conrad, E. Feist, U. Sack, P. Schierack, L. Heiserich, C. Eberle, V. Peters, D. Roggenbuck and D. Reinhold, Cytometry, Part A, 2013, 83, 1017–1026 CrossRef PubMed.
- R. Runge, R. Hiemann, M. Wendisch, U. Kasten-Pisula, K. Storch, K. Zoephel, C. Fritz, D. Roggenbuck, G. Wunderlich, K. Conrad and J. Kotzerke, Int. J. Radiat. Biol., 2012, 88, 439–447 CrossRef CAS PubMed.
- A. Reddig, C. E. Rübe, S. Rödiger, P. Schierack, D. Reinhold and D. Roggenbuck, J. Lab. Precis. Med., 2018, 3, 31 CrossRef.
- E. Pöselt, C. Schmidtke, S. Fischer, K. Peldschus, J. Salamon, H. Kloust, H. Tran, A. Pietsch, M. Heine, G. Adam, U. Schumacher, C. Wagener, S. Forster and H. Weller, ACS Nano, 2012, 6, 3346–3355 CrossRef PubMed.
- D. A. Scudiero, R. H. Shoemaker, K. D. Paull, A. Monks, S. Tierney, T. H. Nofziger, M. J. Currens, D. Seniff and M. R. Boyd, Cancer Res., 1988, 48, 4827–4833 CAS.
- A. H. Cory, T. C. Owen, J. A. Barltrop and J. G. Cory, Cancer Commun., 1991, 3, 207–212 CrossRef CAS PubMed.
- F. Trabelsi, R. Khlifi, D. Goux, M. Guillamin, A. Hamza-Chaffai and F. Sichel, Environ. Sci. Pollut. Res., 2016, 23, 16127–16136 CrossRef CAS PubMed.
- A. Hartwig, BioMetals, 2010, 23, 951–960 CrossRef CAS PubMed.
- D. Kirkland, P. Kasper, H. J. Martus, L. Muller, J. van Benthem, F. Madia and R. Corvi, Mutat. Res., Genet. Toxicol. Environ. Mutagen., 2016, 795, 7–30 CrossRef CAS PubMed.
- N. G. Bastús, J. Comenge and V. Puntes, Langmuir, 2011, 27, 11098–11105 CrossRef PubMed.
- D. S. Ling and T. Hyeon, Small, 2013, 9, 1450–1466 CrossRef CAS PubMed.
- T. Jochum, J. Niehaus and H. Weller, SID Symp. Dig. Tech. Pap., 2016, 47, 347–349 CrossRef.
- I. Mekis, D. V. Talapin, A. Kornowski, M. Haase and H. Weller, J. Phys. Chem. B, 2003, 107, 7454–7462 CrossRef CAS.
- D. V. Talapin, I. Mekis, S. Gotzinger, A. Kornowski, O. Benson and H. Weller, J. Phys. Chem. B, 2004, 108, 18826–18831 CrossRef CAS.
- U. Resch-Genger and P. C. DeRose, Pure Appl. Chem., 2012, 84, 1815–1835 CAS.
- R. Hiemann, N. Hilger, J. Michel, J. Nitschke, A. Bohm, U. Anderer, M. Weigert and U. Sack, Ann. N. Y. Acad. Sci., 2007, 1109, 358–371 CrossRef PubMed.
- R. Hiemann, T. Buttner, T. Krieger, D. Roggenbuc, U. Sack and K. Conrad, Autoimmun. Rev., 2009, 9, 17–22 CrossRef CAS PubMed.
- J. Ostermann, C. Schmidtke, C. Wolter, J. P. Merkl, H. Kloust and H. Weller, Beilstein J. Nanotechnol., 2015, 6, 232–242 CrossRef PubMed.
- C. Schmidtke, E. Poselt, J. Ostermann, A. Pietsch, H. Kloust, H. Tran, T. Schotten, N. G. Bastús, R. Eggers and H. Weller, Nanoscale, 2013, 5, 7433–7444 RSC.
- M. Moser, N. Nirmalananthan, T. Behnke, D. Geissler and U. Resch-Genger, Anal. Chem., 2018, 90, 5887–5895 CrossRef CAS PubMed.
- A. Birmingham, L. M. Selfors, T. Forster, D. Wrobel, C. J. Kennedy, E. Shanks, J. Santoyo-Lopez, D. J. Dunican, A. Long, D. Kelleher, Q. Smith, R. L. Beijersbergen, P. Ghazal and C. E. Shamu, Nat. Methods, 2009, 6, 569–575 CrossRef CAS PubMed.
- J. H. Zhang, T. D. Y. Chung and K. R. Oldenburg, J. Biomol. Screening, 1999, 4, 67–73 CrossRef PubMed.
- E. P. Rogakou, W. Nieves-Neira, C. Boon, Y. Pommier and W. M. Bonner, J. Biol. Chem., 2000, 275, 9390–9395 CrossRef CAS PubMed.
- L. P. Wang, J. Zhang, Y. F. Zheng, J. Yang, Q. W. Zhang and X. Q. Zhu, J. Nanosci. Nanotechnol., 2010, 10, 8591–8596 CrossRef CAS PubMed.
- L. Ju, G. L. Zhang, C. Zhang, L. Sun, Y. Jiang, C. L. Yan, P. J. Duerksen-Hughes, X. Zhang, X. Q. Zhu, F. F. Chen and J. Yang, Mutat. Res., Genet. Toxicol. Environ. Mutagen., 2013, 753, 54–64 CrossRef CAS PubMed.
- M. Munari, J. Sturve, G. Frenzilli, M. B. Sanders, A. Brunelli, A. Marcomini, M. Nigro and B. P. Lyons, Mutat. Res., Genet. Toxicol. Environ. Mutagen., 2014, 775, 89–93 CrossRef PubMed.
- A. Huk, A. R. Collins, N. El Yamani, C. Porredon, A. Azqueta, J. de Lapuente and M. Dusinska, Mutagenesis, 2015, 30, 85–88 CrossRef CAS PubMed.
- ISO 10993-5:2009 Biological evaluation of medical devices – Part 5: Tests for in vitro cytotoxicity, International Organization for Standardization (ISO), 2009.
- M. Dizdaroglu, Free Radicals Biol. Med., 1991, 10, 225–242 CrossRef CAS.
- J. J. Li, S. L. Lo, C. T. Ng, R. L. Gurung, D. Hartono, M. P. Hande, C. N. Ong, B. H. Bay and L. Y. L. Yung, Biomaterials, 2011, 32, 5515–5523 CrossRef CAS PubMed.
- M. Tsoli, H. Kuhn, W. Brandau, H. Esche and G. Schmid, Small, 2005, 1, 841–844 CrossRef CAS PubMed.
- A. Avalos, A. I. Haza, D. Mateo and P. Morales, Food Chem. Toxicol., 2018, 120, 81–88 CrossRef CAS PubMed.
- S. Fraga, H. Faria, M. E. Soares, J. A. Duarte, L. Soares, E. Pereira, C. Costa-Pereira, J. P. Teixeira, M. D. Bastos and H. Carmo, J. Appl. Toxicol., 2013, 33, 1111–1119 CrossRef CAS PubMed.
- S. May, C. Hirsch, A. Rippl, N. Bohmer, J. P. Kaiser, L. Diener, A. Wichser, A. Burkle and P. Wick, Nanoscale, 2018, 10, 15723–15735 RSC.
Footnote |
| † Electronic supplementary information (ESI) available: TEM images of the Au-NPs and FeOx-NPs (Fig. S1), TEM images of the Cd-based QDs (Fig. S2), scheme of the micellar encapsulation strategy (Fig. S3), number-weighted DLS size distributions (Fig. S4), total amount of surface functional groups measured by conductometry (Table S1), comparison of the genotoxicity at 100 pM (Fig. S5), detailed statistical analysis of genotoxicity of FeOx-NPs (negative control) (Fig. S6), and detailed statistical analysis of genotoxicity of Au-NPs (positive control) (Fig. S7). See DOI: 10.1039/c9nr01021a |
| This journal is © The Royal Society of Chemistry 2019 |