High speed atomic force microscopy to investigate the interactions between toxic Aβ1-42 peptides and model membranes in real time: impact of the membrane composition†
Abstract
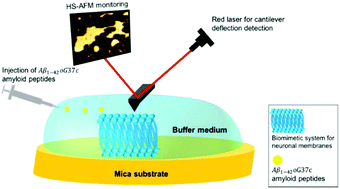
Due to an aging population, neurodegenerative diseases have become a major health issue, the most common being Alzheimer's disease. The mechanisms leading to neuronal loss still remain unclear but recent studies suggest that soluble Aβ oligomers have deleterious effects on neuronal membranes. Here, high-speed atomic force microscopy was used to assess the effect of oligomeric species of a variant of Aβ1-42 amyloid peptide on model membranes with various lipid compositions. Results showed that the peptide does not interact with membranes composed of phosphatidylcholine and sphingomyelin. Ganglioside GM1, but not cholesterol, is required for the peptide to interact with the membrane. Interestingly, when they are both present, a fast disruption of the membrane was observed. It suggests that the presence of ganglioside GM1 and cholesterol in membranes promotes the interaction of the oligomeric Aβ1-42 peptide with the membrane. This interaction leads to the membrane's destruction in a few seconds. This study highlights the power of high-speed atomic force microscopy to explore lipid–protein interactions with high spatio-temporal resolution.
- This article is part of the themed collection: Editor’s Choice: Recent breakthroughs in nanobiotechnology


 Please wait while we load your content...
Please wait while we load your content...