DNA-free directed assembly in single-molecule cut-and-paste†
Abstract
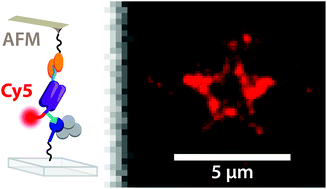
Single-molecule cut-and-paste facilitates bottom-up directed assembly of nanoscale biomolecular networks in defined geometries and enables analysis with spatio-temporal resolution. However, arrangement of diverse molecules of interest requires versatile handling systems. The novel DNA-free, genetically encodable scheme described here utilises an orthogonal handling strategy to promote arrangement of enzymes and enzyme networks.
- This article is part of the themed collections: Editor’s Choice: Recent breakthroughs in nanobiotechnology and 2018 Nanoscale HOT Article Collection


 Please wait while we load your content...
Please wait while we load your content...