Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Protein deglycosylation can drastically affect the cellular uptake†
Artur
Ghazaryan
a,
Katharina
Landfester
 *a and
Volker
Mailänder
*a and
Volker
Mailänder
 ab
ab
aMax Planck Institute for Polymer Research, Ackermannweg 10, 55128 Mainz, Germany. E-mail: katharina.landfester@mpip-mainz.mpg.de
bDermatology Clinic, University Medical Center Mainz, Langenbeckstraße 1, 55131 Mainz, Germany
First published on 3rd May 2019
Abstract
Targeted drug delivery mediated by nanocarriers is a major issue in modern-day medicine. Upon coming in contact with biological fluids (e.g. blood), nanocarriers are rapidly covered by biomolecules (proteins, lipids, etc.) which results in the formation of a surface layer, widely known as the biomolecular corona. The biomolecular corona subsequently confers a certain biological identity to the corona-covered nanocarriers which can be crucial during their subsequent interactions with cells or other biological entities. In contrast to the proteins of the corona, little is known about the impact of the non-protein constituents of the corona, such as sugars. Here, we investigate the role of protein glycosylation of the corona in cellular uptake. We show that deglycosylation of clusterin (CLU) and apolipoprotein AI (Apo AI) significantly changes (increases and decreases, respectively) the cellular uptake of nanocarriers covered with these proteins.
Introduction
In recent years, nanocarriers have been employed for targeted drug delivery. Biomolecular corona formation is the first event nanocarriers experience upon entering into the bloodstream and coming into contact with its components. The rapidly forming corona consists of proteins, but also of sugars, lipids, and other components of the plasma.The biomolecular corona formed on the surface of nanocarriers confers them a unique identity, which later determines their biological fate within the body, such as cellular uptake, biodistribution, retention, and clearance.1,2
The clearance of nanocarriers from the body is usually carried out by scavenging the activity of immune system cells, such as macrophages. Thus, one of the strategies towards targeted drug delivery is to make nanocarriers “invisible” to the immune system, a phenomenon also known as the stealth effect. This will allow nanocarriers to circulate in the bloodstream for an extended period of time and to reach their target.3
Studies demonstrated that surface functionalization of nanocarriers (e.g. attachment of polyethylene glycol or PEGylation), and the appearance of certain proteins in the corona result in an increased stealth effect.4 Schöttler et al. showed that clusterin confers to the nanoparticles a strong stealth effect.4
Clusterin (also known as apolipoprotein J) is a 70–80 kDa lipoprotein consisting of two polypeptide chains that are connected by up to 5 disulfide bonds. Seven asparagine residues of clusterin are highly glycosylated and the glycosylic groups add up to ca. 30% of the total protein weight5 (UniProt Protein Database, Protein ID P10909).
Other major examples of lipoproteins include high-density lipoprotein (HDL), low-density lipoprotein (LDL) and very low-density lipoprotein (vLDL) complexes of the blood. Similar to clusterin, they consist of glycosylated apolipoproteins.
Scavenger receptors (SR)
The uptake of lipoprotein particles or their components by immune system cells (e.g. macrophages) or tissue cells (e.g. hepatocytes) is mostly carried out via plasma membrane receptors of those cells. Macrophage receptors that are involved in lipoprotein uptake are of great interest in the field of targeted drug delivery.Macrophages possess a number of surface receptors that are capable of mediating the internalization of lipoproteins or their components, the most famous ones being scavenger receptors (SR). Among other receptors that are involved in their metabolism are LDL receptors, β-vLDL receptors, phosphatidylserine receptors, mannose receptors, and LRP1 receptors (also known as Apo E-receptors).6,7
SRs are a ‘superfamily’ of membrane-bound receptors that can bind to a variety of ligands including endogenous and modified host-derived molecules, as well as microbial pathogens. In the 1970s, Brown and Goldstein found that modified LDL (mLDL), but not native LDL, is internalized and degraded by macrophages which ultimately led to the discovery of SRs.8,9 According to a recent classification, there are 10 classes of these receptors, named classes A–J.10
The first scavenger receptors were purified in 1988![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 11 and cloned in 1990
11 and cloned in 1990![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 12,13 by Monty Krieger's group. Nowadays these receptors are known as SR-AI and SR-AII scavenger receptors.
12,13 by Monty Krieger's group. Nowadays these receptors are known as SR-AI and SR-AII scavenger receptors.
Class A SRs are glycoproteins (ca. 400–500 amino acids), which comprise a homotrimer. They are membrane-spanning proteins with a short N-terminal cytoplasmic domain followed by a transmembrane region and a large extracellular domain. The latter one, responsible for ligand binding, usually consists of an α-helical coiled-coil domain, a collagen-like domain and a C-terminal cysteine-rich domain.14 One of the intriguing features of SRs is their ability to recognise multiple ligands e.g. natural/endogenous, modified host, microbial (bacterial, viral, fungal, and parasitic), environmental, soluble, or particulate ligands by endocytosis and phagocytosis. The chemical nature of SR ligands varies greatly, but generally depends on the regions of net negative charge.15 For instance, mLDL, cholesterol, phosphatidylserine, dextran sulfate, apolipoprotein AI, and apolipoprotein E serve as ligands for SR-A1 receptors.16
Other major representatives of scavenger receptors include class B scavenger receptors of about 450–500 amino acids (SR-B1, SR-B2, and SR-B3). The characteristic feature of these receptors is the presence of two transmembrane domains and the N-, C-termini being located in the cytoplasm. The extracellular domain of these receptors is heavily N-linked glycosylated, providing protection against proteases.10 As with SR-A receptors, class B receptors also recognise diverse types of ligands. SR-B1 has a major role in HDL internalization.10,14 There are also other SRs involved in the lipoprotein metabolism, such as SR-E1, also known as lectin-type oxidized-LDL receptor 1 (LOX-1-receptor). This receptor belongs to the C-type lectin superfamily, for which Ca2+ ions are required for their binding activity.
Various compounds, such as dextran sulfate, are often used as inhibitors for blocking SR-A receptors. Dextran sulfate, a polyanionic derivative of dextran, is a specific ligand for SR-A1, which can bind to the receptor and competitively inhibit cellular internalization.17,18
Protein glycosylation
One of the key post-translational modifications is glycosylation of proteins, including many apolipoproteins. The sugar chains, which are called glycans, play an important role in structural stability, enzymatic activity, molecular recognition, and receptor binding among other biological processes. N- and O-glycans are the two most common types in eukaryotic systems. N-Glycans are attached to the asparagine residues of proteins, while O-glycans connect to proteins via serine or threonine residues. The carbohydrate chains are usually branched and contain versatile sugars, such as mannose, galactose, fucose, N-acetylglucosamine, N-acetylgalactosamine, and sialic acid. Various enzymes, called deglycosylases, can entirely or partly remove the sugar moieties from the proteins. PNGase F (Peptide:N-glycosidase F) is an enzyme that can fully deglycosylate the proteins that consist of N-glycans by cutting between the asparagine molecule and the carbohydrate chains. Due to its specificity, PNGase F does not affect O-glycans.19 However, clusterin is not attached to O-glycans.Here, we investigate the non-protein, particularly the sugar parts of HDL and (v)LDL (a mixture of LDL and vLDL), as well as of clusterin. For elucidating the role of glycans in murine macrophage cellular uptake, deglycosylation of apolipoproteins was applied. We found that deglycosylation of the proteins of the corona can significantly change the cellular uptake of nanocarriers.
Materials and methods
Materials
Human citrate plasmas out of ten donors were pooled into one batch, aliquoted and stored at −80 °C. Before the use, the citrate plasma was thawed and centrifuged (4 °C, 20.000g, 1 h) to remove any aggregated compounds.
For the synthesis of TW192 nanoparticles (D = 162 nm) the NHS-chemistry was applied as described by Schöttler et al.4 The number of polyethylene glycol (PEG) chains, with a size of 5000 g mol−1, was 2700 per nanoparticle.
All the experiments were performed on KK132a nanoparticles unless mentioned additionally.
Methods
The corona preparation was as follows: 30 μL nanoparticles were incubated with 1 mL of sample. In the case of clusterin, apolipoprotein AI (Apo AI) or apolipoprotein B100 (Apo B100), the protein amount was 50 μg. In the case of HDL, (v)LDL fractions 1 mL plasma equivalent was taken. The incubation was carried out at 37 °C for 1 hour. After the incubation in order to remove unbound compounds, the nanoparticles were centrifuged (20.000g, 4 °C, 1 h). The pelleted nanoparticles were subsequently given to cells (75 μg mL−1) in 1 mL DMEM (without FBS) and incubated for 2 h.
After 2 h incubation was over, the cells were washed twice with PBS (1 mL), detached with trypsin–EDTA (0.25%, Thermo Fisher Scientific), centrifuged (500g, room temperature, 5 minutes) and resuspended in 1 mL PBS.
Flow cytometry measurements were carried out on an Attune NxT cytometer (Thermo Fisher Scientific, USA) with a 488 nm blue laser to excite the nanoparticles labeled with Bodipy (523/535 nm) and a 530 nm pass filter to detect the emission (BL1-H channel). Analysis was performed by FCS Express v4 (DeNovo Software, USA). Cells that were selected/gated on a forward-scatter/sideward-scatter plot were analyzed in the BL1-H channel. The median fluorescence intensity (MFI) was determined from a 1D histogram and the data measurements with three technical replicates are presented as mean ± standard deviation.
The ζ-potentials of the abovementioned coronas were measured using again a Nano Z Zetasizer (Malvern Instruments GmbH, Herrenberg, Germany). Triplicates were used for every corona sample, each of them represented an average of 12 measurements.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) protein = 1
protein = 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50, w/w). Afterwards, the created peptides were mixed with formic acid (final concentration of 0.1% (v/v)) and were spiked with Hi3 E. coli Standard (final concentration of 20 fmol μL−1) (Waters Corporation) for absolute quantification.
50, w/w). Afterwards, the created peptides were mixed with formic acid (final concentration of 0.1% (v/v)) and were spiked with Hi3 E. coli Standard (final concentration of 20 fmol μL−1) (Waters Corporation) for absolute quantification.
Quantitative analysis of proteins was carried out using a nanoACQUITY UPLC system coupled with a Synapt G2-Si mass spectrometer (Waters Corporation). Tryptic-digested peptides were separated on the nanoACQUITY system equipped with a C18 nanoACQUITY Trap Column (5 μm, 180 μm × 20 mm, Waters Corporation) and a C18 analytical reversed-phase column (1.7 μm, 75 μm × 150 mm, Waters Corporation). Chromatographic separation was performed with two mobile phases: A [0.1% (v/v) formic acid in water] and B [acetonitrile with 0.1% (v/v) formic acid]. The sample flow rate was set at 0.3 μL min−1 using a gradient of 2–40% mobile phase B for 70 minutes. As a lock-mass reference compound 150 fmol μL−1 Glu-Fibrinopeptide was infused at a flow rate of 0.8 μL min−1. Data-independent acquisition (MSE) experiments were performed on the Synapt G2-Si operated in resolution mode. Electrospray Ionization was performed in positive ion mode using a NanoLockSpray source. Data were acquired over a range of m/z 50–2000 Da with a scan time of 0.5 s and a total acquisition time of 80 min. All samples were analysed in two technical replicates and averaged. Data acquisition and processing was carried out using MassLynx v4.1, and Progenesis QI for Proteomics v2.0 software was used to process data and to identify the proteins. The generated peptide masses were searched against reviewed human or appropriate animal protein sequence databases downloaded from UniProt.
Ethics
This study was performed in strict accordance with the DFG guidelines for the use of human material. The study was approved by the Landesärztekammer Rheinland-Pfalz (Bearbeitungsnummer: 837-439-12 (8540-F)).Results
Previous studies showed that clusterin (CLU) possesses a strong anti-uptake (stealth) property, when covering the surface of polystyrene nanoparticles. To date, little is known about what constitutes the stealth properties of clusterin. To gain a deeper understanding, clusterin was studied by focusing on its non-protein, namely sugar composition. Clusterin belongs to the family of lipoproteins, where the apoprotein part is heavily N-glycosylated (Fig. S-1†). Seven asparagine residues of clusterin are covalently bound to various oligosaccharides, which are usually branched. Besides the CLU, other classical representatives of lipoproteins, namely HDL, LDL, and vLDL particles, were investigated, since their apolipoproteins are also glycosylated. HDL, LDL, and vLDL particles were obtained by purification from human plasma.Purification of HDL and (v)LDL particles
We wanted to check the deglycosylation effect on the uptake of nanocarriers covered with lipoprotein coronas. Therefore, we purified HDL, LDL and vLDL from human plasma. The purification kit used in this study allowed us to separate two different lipoprotein fractions: HDL and (v)LDL, the latter one being a mixture of LDL and vLDL complexes.The purified fractions were subsequently run on SDS-PAGE to check the purification quality. As shown in Fig. 1, the HDL fraction has a major band at around 28 kDa height which corresponds to Apo AI, the major protein constituent of HDL particles. A few bands at the low molecular weight region most probably correspond to apolipoprotein C proteins (C1, C2 and C3). The purification cannot avoid the occurrence of unspecific proteins. The band slightly above 62 kDa is most probably human serum albumin, which is not part of HDL particles but rather a purification unspecific outcome. However, a full list of the proteome of these fractions has been published by us elsewhere.23
In the case of the (v)LDL fraction, the band of apolipoprotein B100 (Apo B100) is visible (above 198 kDa). The high molecular weight protein with an approximate size of 500 kDa is the major protein constituent of (v)LDL.
Overall, Fig. 1 shows that the purified HDL and (v)LDL fractions are of good purity and can be used for further downstream applications.
Deglycosylation
In order to investigate the role of glycans in the stealth effect, full deglycosylation of glycoproteins was carried out. As proof that the sugar molecules were completely removed from the protein clusterin upon PNGase F treatment, SDS-PAGE and subsequent glycoprotein staining were performed as depicted in Fig. 2. On the left side of the Coomassie-stained gel (blue gel), the clusterin band shifted from ca. 62 kDa to ca. 49 kDa upon PNGase F treatment. The absence of a higher molecular weight band clearly indicates that all the sugar molecules were successfully removed and that the 49 kDa band corresponds to sugarless clusterin. This fact is additionally confirmed by the glyco-staining procedure (Fig. 2, pink gel): the clusterin band is stained in pink when not treated with PNGase F and no pink bands could be observed anymore when clusterin was treated with PNGase F. This indicates that clusterin underwent complete deglycosylation. Besides clusterin, other major glycoproteins were also subjected to PNGase F treatment, namely Apo AI as the major protein of HDL and Apo B100 as the major protein of (v)LDL. Unlike clusterin, SDS-PAGE and subsequent glycostaining did not confirm the (complete) removal of sugar molecules from Apo AI (Fig. S-2†). The reason is that this protein has a mixed (N-type and O-type) glycosylation pattern. Deglycosylation of Apo B100 was not checked, because no uptake effect had been observed upon PNGase F treatment (Fig. 3), hence not of great interest for us.Cellular uptake of nanoparticles with various coronas
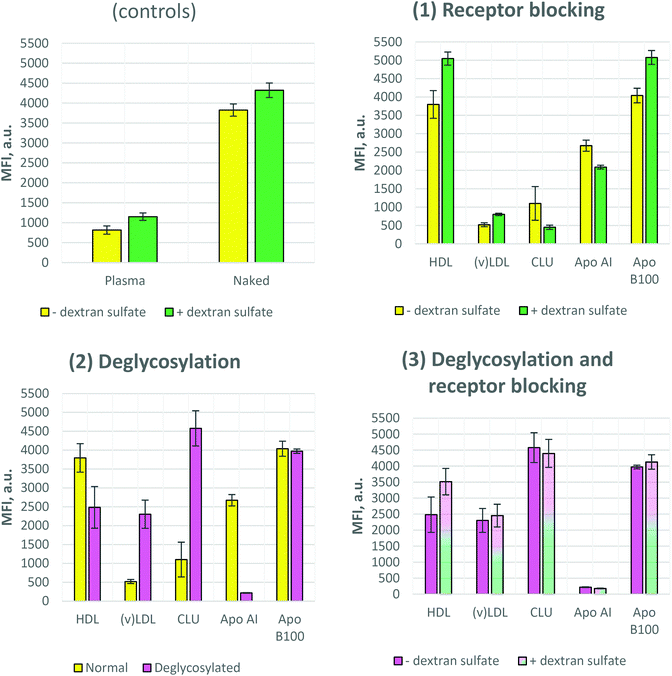
It was previously reported that polystyrene nanoparticles bearing no corona (so called “naked” nanoparticles) trigger a high uptake, while the nanoparticles covered by plasma or serum (Fig. S-3†) proteins show a significantly reduced uptake (Fig. 3 controls).23 These are classical controls always applied along the experiments. The main reason for this reduced uptake of plasma-covered or serum-covered nanoparticles is the adsorption of clusterin which is the most dominant protein in the plasma corona of polystyrene particles.4 Clearly this may be different for other materials and should always be tested first. The fact that clusterin inhibits the cellular uptake of our nanoparticles is also shown in Fig. 3-1. As in the case of plasma- and CLU-coronas, a similar uptake rate was detected for (v)LDL-corona, i.e. a very low uptake by mouse macrophages. In contrast to this, the uptake of particles with a HDL-corona was similar to that of the naked ones. We covered the nanoparticles with the major single proteins of lipoprotein fractions. This is Apo AI for HDL and Apo B100 for (v)LDL. Both Apo AI- and Apo B100-covered nanoparticles showed similar uptake compared to that of naked nanoparticles.Dextran sulfate blocking
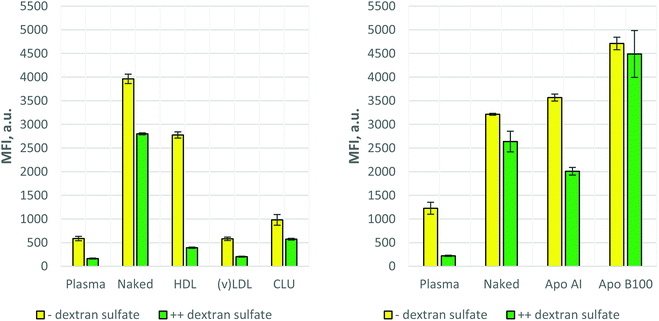
Two types of dextran sulfate blocking experiments were carried out as described in Materials and Methods. In the first case (Fig. 3) cell surface receptors were only blocked by dextran sulfate but during the uptake process (cell–nanoparticle incubation) no dextran sulfate was present in the incubation media. In the second case (Fig. 4) after blocking the cell surface receptors, dextran sulfate remained in the incubation environment during the uptake process. As Fig. 3 (controls and 1) shows, dextran sulfate blocking results in slightly increased uptake of the control samples, as well as HDL, (v)LDL, and Apo B100 coronas, while for the Apo AI and CLU coronas the inhibition of the uptake was observed.The results differed when the dextran sulfate was not washed away from the incubation media after receptor blocking. Fig. 4 shows that except for Apo B100, all other coronas resulted in further uptake inhibition, especially for plasma and HDL. Together with the (v)LDL corona, plasma and HDL exhibited an extremely low cellular uptake. The fluorescence signal of these nanoparticles was comparable to that of the auto-fluorescence of nanoparticle-untreated cells.
Effect of deglycosylation on uptake
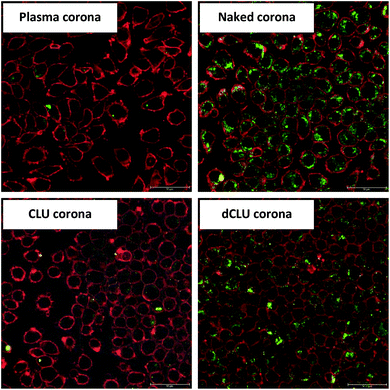
To address the question whether deglycosylation can affect the cellular uptake of the nanoparticles, we performed protein deglycosylation with PNGase F. Fig. 3-2 shows that deglycosylation resulted in an increase of cellular uptake for (v)LDL and CLU coronas, while the opposite effect, i.e. decreased uptake upon PNGase F treatment, was observed for HDL and Apo AI coronas. Moreover, the effect was more prominent for Apo AI and CLU coronas. PNGase F treatment did not have an effect on the uptake of the Apo B100 corona. Interestingly, when comparing the uptake of deglycosylated coronas, no significant differences were observed between normal and dextran sulfate-blocked cells (Fig. 3-3).To prove that the increased uptake upon deglycosylation is indeed due to the uptake and not due to nanoparticle aggregation on the cell surface, confocal laser scanning microscopy (cLSM) images were obtained. For visualization, the cell membrane was stained with a red dye (CellMask Orange) and the nanoparticles contained a green-fluorescing dye (Bodipy 523/535). Fig. 5 confirms the flow cytometry data of the controls: low uptake for the plasma corona and high uptake for naked nanoparticles, as expected. In addition, these results showed that the uptake of the CLU corona drastically increased upon clusterin deglycosylation.
To show that deglycosylation does not cause a change in the size or charge of the corona, which could affect the uptake, dynamic light scattering and ζ-potential measurements were performed for both glycosylated and deglycosylated clusterin and apolipoprotein AI coronas. As can be seen in Fig. S-4† deglycosylation of clusterin or Apo AI does not alter the ζ-potential. As for the size, only for the Apo AI corona a very little (ca. 10%) size decrease could be observed upon PNGase F treatment. However, this is not the case for clusterin deglycosylation.
To show that the amount of total protein (or clusterin) which is adsorbed on the surface of nanoparticles has no effect on the uptake, total protein (or clusterin) quantification was carried out. As Fig. S-5† shows, the amount of total protein on the surface of nanoparticles does not change upon deglycosylation. Moreover, according to the MS data, the clusterin amount is the same in both the CLU and dCLU coronas.
Discussion
The aim of the current work was to investigate the non-protein, namely sugar parts of apolipoproteins and their role in the uptake of nanocarriers by mammalian immune cells. Therefore, we chose clusterin, as well as HDL and (v)LDL. Particularly, protein deglycosylation was carried out in order to elucidate the role of glycans in cellular uptake. Moreover, the class A scavenger receptors were blocked with dextran sulfate to investigate the uptake under receptor blockage conditions.Several biological replicates confirmed that the HDL-corona decorated nanoparticle uptake is higher (similar to that of naked nanoparticles), while the (v)LDL-corona uptake is highly inhibited (similar to that of the plasma-corona). However, when we analysed the uptake of their main protein constituents, Apo AI and Apo B100, we observed a similar uptake to that for naked nanoparticles (slightly less for Apo AI and slightly higher for Apo B100). Thus, the question is where the huge uptake inhibition of (v)LDL stems from, when its main protein constituent (Apo B100) has ca. 8-fold higher uptake (Fig. 3-1).
The important role of deglycosylation has been elucidated previously. Furbish et al. and Blakey et al. demonstrated in the 1980s that the sugar molecules of ricin and glucocerebrosidase are important for receptor binding in mouse and rat livers, respectively, and deglycosylation results in altered uptake by liver cells.24,25 Colaço and his colleagues reported that deglycosylation had an influence on gelonin uptake: it reduces the uptake by a third in the liver, but does not affect the uptake by the kidney.26 The deglycosylation impact on the uptake by macrophages is known as well.27 Moreover, deglycosylation effects have been demonstrated in the past, which involve nanoparticle coronas.28 According to these authors deglycosylated nanoparticle–protein complexes led to an increase in nanoparticle uptake by macrophages.28
(v)LDL and other coronas mentioned above consist not only of proteins but also of non-proteins, such as carbohydrates and lipids. To elucidate the role of sugars in the stealth effect, N-glycans attached to the proteins were enzymatically removed. As we found in Fig. 3-2, the (v)LDL fraction treated with PNGase F deglycosylase exhibited a ca. 4.4-fold increase in cellular uptake. However, this increased uptake was less than that of naked nanoparticles. This means that glycosylation is an important part of the stealth effect for (v)LDL, while also other components like lipids may contribute to uptake inhibition. Interestingly, PNGase F treatment had no influence on Apo B100, the main protein of (v)LDL, although Apo B100 has 19 glycosylated sites, according to the UniProt Protein Database (Protein ID P04114). Two possibilities could be the reason for the lack of this effect: either these N-glycans have no role in nanoparticle internalization or no deglycosylation occurred upon PNGase F treatment, because the sugar groups are internally located and the enzyme has no access to them. Since (v)LDL, besides Apo B100, contains other glycosylated apolipoproteins as well, the partial effect of increased uptake upon deglycosylation could stem from these apolipoproteins.
A similar effect, i.e. increased uptake upon deglycosylation, was observed with clusterin (around 4.1-times increase). Moreover, deglycosylation by PNGase F could be easily achieved even at room temperature (data not shown). To confirm the phenomenon of increased uptake, the same experiments were conducted using another polystyrene–PEG nanoparticle (TW192, Fig. 6).
Increased uptake upon deglycosylation of (v)LDL and clusterin can be explained by involvement of other receptors. For (v)LDL, deglycosylation can be considered a modification of the particle, thus creating a so called modified LDL (mLDL) which could be internalized by SR-A1 receptors. However, the internalization via this receptor is less probable, because SR-A1 dextran sulfate-blockage did not affect the uptake of deglycosylated (v)LDL (Fig. 3-3). It means that other receptors are involved in the uptake of deglycosylated (v)LDL, which are insensitive to dextran sulfate-blockage. LOX-1, abbreviated as lectin-type oxidized-LDL receptor 1, is a type of scavenger receptor expressed on the surface of macrophages. In addition to recognizing oxidized-LDL, LOX-1 recognizes a variety of other ligands, including modified lipoproteins.7 Thus, this receptor could potentially serve as an uptake way for deglycosylated (v)LDL.
The same hypothesis holds true for the uptake of clusterin. There are several ways and receptors, which are able to recognize clusterin and internalize it. Clusterin is a ligand for LRP (low-density lipoprotein receptor-related protein) family receptors, such as LRP1, LRP2, LRP8, and VLDLR.29–32
In contrast to (v)LDL and clusterin, HDL and Apo AI showed an opposite effect. Upon PNGase F treatment, the uptake slightly decreased in the case of the HDL corona and dramatically decreased for the Apo AI corona (Fig. 3-2). The fact that the two showed similar uptake efficiencies is not surprising, because Apo AI is the major protein of HDL. Interestingly, using the UniProt Protein Database (Protein ID P02647) showed that Apo AI should be N-glycosylated at Lys-263. In addition, Cubedo et al. demonstrated that Apo AI is indeed N-glycosylated and PNGase F treatable.33 Thus, a dramatic decrease of cellular uptake strongly supports deglycosylation. However, the confirmation of deglycosylation of Apo AI upon PNGase F treatment was not achieved using the glycostaining kit (Fig. S-2†). This is due to the mixed (N-type and O-type) glycosylation pattern of ApoAI33 and therefore PNGase F cannot remove all the sugar molecules. The dramatic decrease of Apo AI uptake shows that glycans are crucial for receptor binding. Hence, removal of these groups can cause a failure of receptor binding and thus inhibit the uptake. This is partially true also for HDL deglycosylation.
In summary, the inhibition of uptake can be divided into three categories: strong (plasma, HDL), intermediate (naked, (v)LDL, Apo AI, clusterin), and none (Apo B100). For the strong uptake inhibition, one can assume that mostly A-type SRs are responsible for the binding and internalization. However, for the intermediate ones, most probably also alternative (other than class A SR) ways exist which allow the nanoparticles to bypass the class A scavenger receptor blockage. Thus, a certain particle has more than one way to get inside and vice versa as mentioned before, i.e. a defined receptor can bind more than one ligand. As regards to the third category, i.e. no change of uptake upon dextran sulfate blockage, internalization most probably is carried out via receptors that do not get blocked by dextran sulfate.
The most dramatic influence on cellular uptake was seen for the deglycosylation of Apo AI and clusterin as single proteins, and (v)LDL as a lipoprotein complex.
Conclusions
• Deglycosylation dramatically increases the cellular uptake of (v)LDL and CLU coronas by mouse macrophages.• Deglycosylation dramatically decreases the cellular uptake of the Apo AI corona and the HDL corona.
• We detected three types of uptake upon dextran sulfate blocking: strong inhibition, intermediate inhibition and no inhibition.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors are thankful to Katja Klein and Dr Thomas Wolf for the synthesis of nanoparticles, to Raweewan Thiramanas and Jonas Reinholz for their help with cLSM pictures, and to Stefan Schuhmacher for creation of the lipoprotein image. Dr Lorna Moll is acknowledged for editing the manuscript. We also gratefully acknowledge the support by the DFG (Deutsche Forschungsgemeinschaft, SFB 1066) and the Open Access funding provided by the Max Planck Society.References
- C. D. Walkey and W. C. Chan, Understanding and controlling the interaction of nanomaterials with proteins in a physiological environment, Chem. Soc. Rev., 2012, 41(7), 2780–2799 RSC.
- E. Hellstrand, I. Lynch, A. Andersson, T. Drakenberg, B. Dahlback, K. A. Dawson, S. Linse and T. Cedervall, Complete high-density lipoproteins in nanoparticle corona, FEBS J., 2009, 276(12), 3372–3381 CrossRef CAS PubMed.
- B. Kang, P. Okwieka, S. Schöttler, S. Winzen, J. Langhanki, K. Mohr, T. Opatz, V. Mailander, K. Landfester and F. R. Wurm, Carbohydrate-Based Nanocarriers Exhibiting Specific Cell Targeting with Minimum Influence from the Protein Corona, Angew. Chem., Int. Ed., 2015, 54(25), 7436–7440 CrossRef CAS PubMed.
- S. Schöttler, G. Becker, S. Winzen, T. Steinbach, K. Mohr, K. Landfester, V. Mailander and F. R. Wurm, Protein adsorption is required for stealth effect of poly(ethylene glycol)- and poly(phosphoester)-coated nanocarriers, Nat. Nanotechnol., 2016, 11(4), 372–377 CrossRef PubMed.
- P. Rohne, H. Prochnow, S. Wolf, B. Renner and C. Koch-Brandt, The chaperone activity of clusterin is dependent on glycosylation and redox environment, Cell. Physiol. Biochem., 2014, 34(5), 1626–1639 CrossRef CAS PubMed.
- A. M. Fogelman, B. J. Vanlenten, C. Warden, M. E. Haberland and P. A. Edwards, Macrophage Lipoprotein Receptors, J. Cell Sci., 1988, 135–149 CrossRef CAS.
- P. R. Taylor, L. Martinez-Pomares, M. Stacey, H. H. Lin, G. D. Brown and S. Gordon, Macrophage receptors and immune recognition, Annu. Rev. Immunol., 2005, 23, 901–944 CrossRef CAS PubMed.
- M. S. Brown and J. L. Goldstein, Lipoprotein Metabolism in the Macrophage - Implications for Cholesterol Deposition in Atherosclerosis, Annu. Rev. Biochem., 1983, 52, 223–261 CrossRef CAS PubMed.
- J. L. Goldstein, Y. K. Ho, S. K. Basu and M. S. Brown, Binding site on macrophages that mediates uptake and degradation of acetylated low density lipoprotein, producing massive cholesterol deposition, Proc. Natl. Acad. Sci. U. S. A., 1979, 76(1), 333–337 CrossRef CAS PubMed.
- M. PrabhuDas, D. Bowdish, K. Drickamer, M. Febbraio, J. Herz, L. Kobzik, M. Krieger, J. Loike, T. K. Means, S. K. Moestrup, S. Post, T. Sawamura, S. Silverstein, X. Y. Wang and J. El Khoury, Standardizing Scavenger Receptor Nomenclature, J. Immunol., 2014, 192(5), 1997–2006 CrossRef CAS PubMed.
- T. Kodama, P. Reddy, C. Kishimoto and M. Krieger, Purification and Characterization of a Bovine Acetyl Low-Density Lipoprotein Receptor, Proc. Natl. Acad. Sci. U. S. A., 1988, 85(23), 9238–9242 CrossRef CAS PubMed.
- T. Kodama, M. Freeman, L. Rohrer, J. Zabrecky, P. Matsudaira and M. Krieger, Type-I Macrophage Scavenger Receptor Contains Alpha-Helical and Collagen-Like Coiled Coils, Nature, 1990, 343(6258), 531–535 CrossRef CAS PubMed.
- L. Rohrer, M. Freeman, T. Kodama, M. Penman and M. Krieger, Coiled-Coil Fibrous Domains Mediate Ligand-Binding by Macrophage Scavenger Receptor Type-Ii, Nature, 1990, 343(6258), 570–572 CrossRef CAS PubMed.
- I. A. Zani, S. L. Stephen, N. A. Mughal, D. Russell, S. Homer-Vanniasinkam, S. B. Wheatcroft and S. Ponnambalam, Scavenger receptor structure and function in health and disease, Cells, 2015, 4(2), 178–201 CrossRef PubMed.
- S. Mukhopadhyay and S. Gordon, The role of scavenger receptors in pathogen recognition and innate immunity, Immunobiology, 2004, 209(1–2), 39–49 CrossRef CAS PubMed.
- J. Ben, X. Zhu, H. Zhang and Q. Chen, Class A1 scavenger receptors in cardiovascular diseases, Br. J. Pharmacol., 2015, 172(23), 5523–5530 CrossRef CAS PubMed.
- Y. Chao, M. Makale, P. P. Karmali, Y. Sharikov, I. Tsigelny, S. Merkulov, S. Kesari, W. Wrasidlo, E. Ruoslahti and D. Simberg, Recognition of Dextran-Superparamagnetic Iron Oxide Nanoparticle Conjugates (Feridex) via Macrophage Scavenger Receptor Charged Domains, Bioconjugate Chem., 2012, 23(5), 1003–1009 CrossRef CAS PubMed.
- T. Laumonier, A. J. Walpen, C. F. Maurus, P. J. Mohacsi, K. M. Matozan, E. Y. Korchagina, N. V. Bovin, B. Vanhove, J. D. Seebach and R. Rieben, Dextran sulfate acts as an endothelial cell protectant and inhibits human complement and natural killer cell-mediated cytotoxicity against porcine cells, Transplantation, 2003, 76(5), 838–843 CrossRef CAS PubMed.
- K. W. Moremen, M. Tiemeyer and A. V. Nairn, Vertebrate protein glycosylation: diversity, synthesis and function, Nat. Rev. Mol. Cell Biol., 2012, 13(7), 448–462 CrossRef CAS PubMed.
- K. Landfester, N. Bechthold, F. Tiarks and M. Antonietti, Miniemulsion polymerization with cationic and nonionic surfactants: A very efficient use of surfactants for heterophase polymerization, Macromolecules, 1999, 32(8), 2679–2683 CrossRef CAS.
- L. L. Hecht, A. Schoth, R. Munoz-Espi, A. Javadi, K. Kohler, R. Miller, K. Landfester and H. P. Schuchmann, Determination of the Ideal Surfactant Concentration in Miniemulsion Polymerization, Macromol. Chem. Phys., 2013, 214(7), 812–823 CrossRef CAS.
- I. García-Moreno, A. Costela, L. Campo, R. Sastre, F. Amat-Guerri, M. Liras, F. L. Arbeloa, J. B. Prieto and I. L. Arbeloa, 8-phenyl-substituted dipyrromethene center dot BF2 complexes as highly efficient and photostable laser dyes, J. Phys. Chem. A, 2004, 108(16), 3315–3323 CrossRef.
- J. Muller, D. Prozeller, A. Ghazaryan, M. Kokkinopoulou, V. Mailander, S. Morsbach and K. Landfester, Beyond the protein corona - lipids matter for biological response of nanocarriers, Acta Biomater., 2018, 71, 420–431 CrossRef PubMed.
- F. S. Furbish, C. J. Steer, N. L. Krett and J. A. Barranger, Uptake and distribution of placental glucocerebrosidase in rat hepatic cells and effects of sequential deglycosylation, Biochim. Biophys. Acta, 1981, 673(4), 425–434 CrossRef CAS.
- D. C. Blakey, D. N. Skilleter, R. J. Price and P. E. Thorpe, Uptake of native and deglycosylated ricin A-chain immunotoxins by mouse liver parenchymal and non-parenchymal cells in vitro and in vivo, Biochim. Biophys. Acta, 1988, 968(2), 172–178 CrossRef CAS.
- M. Colaço, S. Misquith, M. M. Bapat, S. Wattiaux-De Coninck and R. Wattiaux, A comparative study of the subcellular distribution of native and deglycosylated gelonin in rat liver and kidney, Biochem. Biophys. Res. Commun., 2004, 319(4), 1299–1306 CrossRef PubMed.
- D. Kappala, R. Sarkhel, S. K. Dixit, Lalsangpuii, M. Mahawar, M. Singh, S. Ramakrishnan and T. K. Goswami, Role of different receptors and actin filaments on Salmonella Typhimurium invasion in chicken macrophages, Immunobiology, 2018, 223(67), 501–507 CrossRef CAS PubMed.
- S. Wan, P. M. Kelly, E. Mahon, H. Stockmann, P. M. Rudd, F. Caruso, K. A. Dawson, Y. Yan and M. P. Monopoli, The “sweet” side of the protein corona: effects of glycosylation on nanoparticle-cell interactions, ACS Nano, 2015, 9(2), 2157–2166 CrossRef CAS PubMed.
- A. P. Lillis, S. C. Muratoglu, D. T. Au, M. Migliorini, M. J. Lee, S. K. Fried, I. Mikhailenko and D. K. Strickland, LDL Receptor-Related Protein-1 (LRP1) Regulates Cholesterol Accumulation in Macrophages, PLoS One, 2015, 10(6), e0128903 CrossRef PubMed.
- L. Konrad, A. Hackethal, F. Oehmke, E. Berkes, J. Engel and H. R. Tinneberg, Analysis of Clusterin and Clusterin Receptors in the Endometrium and Clusterin Levels in Cervical Mucus of Endometriosis, Reprod. Sci., 2016, 23(10), 1371–1380 CrossRef CAS PubMed.
- C. Leeb, C. Eresheim and J. Nimpf, Clusterin is a ligand for apolipoprotein E receptor 2 (ApoER2) and very low density lipoprotein receptor (VLDLR) and signals via the Reelin-signaling pathway, J. Biol. Chem., 2014, 289(7), 4161–4172 CrossRef CAS PubMed.
- C. Spuch, S. Ortolano and C. Navarro, LRP-1 and LRP-2 receptors function in the membrane neuron. Trafficking mechanisms and proteolytic processing in Alzheimer's disease, Front. Physiol., 2012, 3, 269 Search PubMed.
- J. Cubedo, T. Padro and L. Badimon, Glycoproteome of human apolipoprotein A-I: N- and O-glycosylated forms are increased in patients with acute myocardial infarction, Transl. Res., 2014, 164(3), 209–222 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8nr08305c |
| This journal is © The Royal Society of Chemistry 2019 |