Open Access Article
Open Access ArticleDroplet probe: coupling chromatography to the in situ evaluation of the chemistry of nature
Nicholas H.
Oberlies
 *a,
Sonja L.
Knowles
*a,
Sonja L.
Knowles
 a,
Chiraz Soumia M.
Amrine
a,
Chiraz Soumia M.
Amrine
 a,
Diana
Kao
a,
Diana
Kao
 a,
Vilmos
Kertesz
a,
Vilmos
Kertesz
 b and
Huzefa A.
Raja
b and
Huzefa A.
Raja
 a
a
aDepartment of Chemistry & Biochemistry, University of North Carolina at Greensboro, Greensboro, North Carolina, USA. E-mail: nicholas_oberlies@uncg.edu
bMass Spectrometry and Laser Spectroscopy Group, Chemical Sciences Division, Oak Ridge National Laboratory, Oak Ridge, Tennessee 37831, USA
First published on 21st May 2019
Abstract
Covering: up to 2019
The chemistry of nature can be beautiful, inspiring, beneficial and poisonous, depending on perspective. Since the isolation of the first secondary metabolites roughly two centuries ago, much of the chemical research on natural products has been both reductionist and static. Typically, compounds were isolated and characterized from the extract of an entire organism from a single time point. While there could be subtexts to that approach, the general premise has been to determine the chemistry with very little in the way of tools to differentiate spatial and/or temporal changes in secondary metabolite profiles. However, the past decade has seen exponential advances in our ability to observe, measure, and visualize the chemistry of nature in situ. Many of those techniques have been reviewed in this journal, and most are tapping into the power of mass spectrometry to analyze a plethora of sample types. In nearly all of the other techniques used to study chemistry in situ, the element of chromatography has been eliminated, instead using various ionization sources to coax ions of the secondary metabolites directly into the mass spectrometer as a mixture. Much of that science has been driven by the great advances in ambient ionization techniques used with a suite of mass spectrometry platforms, including the alphabet soup from DESI to LAESI to MALDI. This review discusses the one in situ analysis technique that incorporates chromatography, being the droplet-liquid microjunction-surface sampling probe, which is more easily termed “droplet probe”. In addition to comparing and contrasting the droplet probe with other techniques, we provide perspective on why scientists, particularly those steeped in natural products chemistry training, may want to include chromatography in in situ analyses. Moreover, we provide justification for droplet sampling, especially for samples with delicate and/or non-uniform topographies. Furthermore, while the droplet probe has been used the most in the analysis of fungal cultures, we digest a variety of other applications, ranging from cyanobacteria, to plant parts, and even delicate documents, such as herbarium specimens.
1. Introduction
Throughout a scientific career, there are times when a new technology develops that allows one not only to carry out experiments differently, but to imagine experimentation in new ways. Having been fortunate to work with scientists that have been in the natural products field for >50 years,1 I (NHO) have heard stories about disruptive technologies, things that changed the way natural products chemistry was carried out. If you go back far enough, there are countless examples, including 2D NMR and HPLC, both of which are tools essentially ubiquitous to the study of natural products today. How many of us have a natural products chemistry lab without at least one HPLC instrument? Conversely, how many of us have seen, let alone used, a Craig Countercurrent Apparatus,2,3 despite the fact that they were instrumental in the isolation of many iconic natural products, including taxol?4,5In my own time in science, just beyond two and a half decades, I recall the excitement I felt when I saw for the first time a presentation by Dr Pieter Dorrestein that used mass spectrometry imaging to study bacterial cultures. While the idea of examining the chemistry of nature in situ may have been pondered and discussed for years, I had never before seen it executed so eloquently and in such a visual way. Afterwards, I started contemplating both the questions I would ask via such experiments, and more pragmatically, what tools and expertise would be required. In many ways, this review on the droplet-liquid microjunction-surface sampling probe (droplet probe) originates in attempting to carry out similar in situ chemistry experiments but on fungal cultures. However, back to the preceding paragraph, I am not suggesting that the droplet probe will become as ubiquitous as HPLCs are today. Instead, what I firmly believe is that the process of studying the chemistry of nature in situ is here to stay, and most likely, it will only become both more powerful and more accessible in the future. Quite simply, the ability to probe research questions that address the timing and/or spatial distribution of natural products are both too tempting and too important to ignore.
1.1 Ambient ionization and mass spectrometry imaging vs. droplet probe
There are many techniques available today that use ambient ionization and/or mass spectrometry imaging.6–8 This review is not intended to address all of the possibilities and permutations thereof, but instead, compare and contrast some of the more well-known ones with the droplet probe. In my own group,9,10 and indeed in the earlier studies that I noted by Dorrestein's group,11,12 DESI (desorption electrospray ionization) mass spectrometry was used to examine many different types of substrates. When we first started experimenting with this technique on fungal cultures, we were inspired by the work of Kubanek and colleagues, who had used DESI-MS to examine chemical ecology questions between algae and marine fungal pathogens.13,14 Even in the abstract of their paper, they noted that limits in methodology had, until then, impeded the ability to measure and evaluate defense chemicals on native surfaces.14 To begin working with DESI-MS, we initiated a collaboration with Professor R. Graham Cooks at Purdue University, who is one of the authorities on the technique.7,15 Collaboratively, we were able to measure and observe secondary metabolites in fungal cultures,10 noting both temporal and spatial variability. However, there were several challenges using the DESI approach, most notably the spray could induce divots on the cultures, causing us to use imprints of the surface for some of the measurements. Other researchers have used imprints with DESI-MS examinations of fungi too.16,17 In a follow up study, we spent considerable effort working out ways to grow fungi in a manner that would facilitate DESI-MS,9 eventually using autoclaved cardboard to impart a firm/flat surface, which we believed minimized damage to the fungal culture and enhanced the transfer of ions into the source. While that process was successful, it was not something that could be implemented in a routine manner. Of course, DESI is not the only technique tried for examining natural products chemistry in situ, and some other notable examples include MALDI (matrix assisted laser desorption ionization) and LAESI (laser ablation electrospray ionization).7,18–20In all cases, the key difference between any of those techniques and the droplet probe comes down to a single word/concept: chromatography. None of those other techniques use chromatography, while the droplet probe does. Having attended conferences across many different disciplines, I find it interesting to hear how one group's challenge may be considered straight forward to another. In the mass spectrometry community, ambient ionization techniques are often discussed in the context of sample preparation, or more precisely, the lack thereof. It reminds me of what I learned once in a graduate course in the 1990s at Purdue about LC-MS. That is: do you consider the LC the injector for a mass spectrometer, or do you consider the mass spectrometer the detector for the chromatography system? My general feeling is that if experts in mass spectrometry could avoid sample preparation almost entirely, they would be happy to do so, instead focusing their time, talents and passion on the mass spectrometer. Perhaps the converse is true for the natural products chemist, who may find the intricacies and physics of sample analyzers and ionization principles to be onerous. Indeed, one of the driving forces in the growing use of mass spectrometry by so many fields is the fact that those instruments are becoming more user friendly to operate.
The point of this review is not to get into a philosophical debate or to say that one way of thinking is better than the other. Rather, I simply note that people trained in different ways may think of the challenges and opportunities differently. For natural products chemists, chromatography is not usually the challenge, even when working on seemingly difficult purifications. That is not because we are geniuses. Rather, it is because we are very accustomed to working with complex mixtures and may use chromatography on a daily basis. Thus, from my perspective, chromatography is an attribute of droplet probe that drove me toward it. Nature is full of chemical complexity, and if that can be distilled and simplified into distinct peaks using chromatography, then my thought was to embrace it. More precisely, with DESI-MS, or any technique that essentially ionizes the entire sample with no separation, it is not possible to distinguish between isobars, i.e. compounds with the same molecular weight, although tandem mass spectrometry may help further differentiate such compounds. However, as long as those compounds are not enantiomers, they likely can be separated via chromatography, thereby simplifying the mass spectrometric analyses. In addition, if the effluent into the mass spectrometer has been clarified, then it may be possible to dig even deeper into the baseline, perhaps seeing chemical complexity that may be swamped by the compounds in the organism in greatest quantity or with the best ionization properties.
The other factor that drove the adoption of the droplet probe in my lab was the use of the droplet itself. Fungi are morphologically complex, and they grow in a three-dimensional fashion. While we had some success with DESI, particularly when we were collaborating with one of the seminal researchers in this field (Cooks),10 it was not always straight forward. The spray was too strong and could make a divot on the Petri dish, or the spores could literally be blown around the lab. Furthermore, we had to make imprints of the fungal cultures10 or get them to grow on a more sturdy substrate.9 There was too much sample handling and “tricks of the trade” for routine use. In fact, the way we discovered the droplet probe was at an American Society of Mass Spectrometry meeting, where one of my students was presenting on methodology he had developed for DESI-MS on fungal cultures,9 and the team of Kertesz and Van Berkel from Oak Ridge National Laboratory, the inventors of the droplet probe, were presenting a poster nearby. As described below, they had not designed it for the natural products environment, but they could see how it might help circumvent some of the problems my student had worked so diligently to resolve. The rest of this manuscript is a digest of those experiments and expansion into a range of study materials, from fungal to cyanobacterial to plant (and even delicate documents).
2. History and development of the droplet probe
In 2001, liquid microjunction-surface sampling probes (LMJ-SSPs) employing concentric capillaries to deliver solvent to and from the surface coupled with mass spectrometric analysis were introduced.21,22 Realizing the potential of such a continuous flow liquid extraction system for surface sampling purposes, it was immediately adapted for direct mass spectrometric analysis of thin layer chromatography plates.23 The use of the LMJ-SSP devices was extended to spot sampling and imaging of drugs and metabolites from thin tissue sections24,25 and the analysis of surface deposited and affinity captured proteins.26 In 2013 an implementation of the LMJ-SSP device, named flowProbe, became commercially available.27Another breakthrough was made in 2009 when a new type of sampling mode was implemented using the LMJ-SSP system. This methodology took advantage of first creating, and then breaking, a liquid junction of about 100–300 μm in thickness between the probe and the surface, making the probe-to-surface positioning less critical.28 This “droplet” sampling mode was later implemented on a chip-based robotic nanoelectrospray platform, demonstrating analysis of various sample surface types, including whole-body thin tissue sections from drug dosed mice.29 The success of the method led to commercialization of the liquid extraction surface analysis (LESA) mode on the same device.30
In 2010, the LESA mode was implemented on a commercially available autosampler system.31 The driving force of this improvement was to couple droplet-based liquid microjunction surface sampling with HPLC-MS for spatially resolved surface analysis. The hybrid system had better performance characteristics, especially for the analysis of complex matrix samples. In addition, it provided a greater degree of chemical information from a single spot sample than was possible with direct analysis of an extract. (As a note, in 2013 a continuous flow LMJ-SSP system was also demonstrated to allow for coupling of such a probe with HPLC-MS, enabling extraction, separation and detection of proteins from surfaces in a spatially resolved manner.32 Furthermore, in 2017 the LESA method was also coupled to HPLC-MS).33 Improvements of the autosampler-based system (e.g. incorporation of a laser distance sensor enabling unattended analysis of samples and sample locations of dramatically disparate height; use of an open bed tray system to accommodate samples as large as whole-body rat thin tissue sections and to shorten sampling time to approximately 1 min per sample; camera system for quality control of sampling) resulted in such speed, reliability, sensitivity and selectivity of the autosampler-HPLC-MS combination that it was commercialized in 2015 by the name of dropletProbe.34
In summary, the sampling of surfaces could be envisioned in several different ways, from continuous flow to droplet based. Since the droplet based techniques were amenable to both the nature of the study materials (i.e. non-uniform surfaces, spores, distinct morphological features) and were compatible with tools and techniques that were common to natural products chemistry labs (i.e. HPLC and UHPLC coupled to an array of detectors, including mass spectrometers), the droplet-liquid microjunction-surface sampling probe (droplet probe) seemed well suited for natural products chemistry experimentation.
2.1 Comparing droplet probe with other techniques
There are several other techniques that have been used to analyse the chemistry of nature in situ. This section is not intended to be exhaustive, but instead, used as a way to examine key differences and/or similarities with some of the more prominent techniques. As all of these are coupled to mass spectrometry, they are discussed based on how secondary metabolites are ionized (Table 1).The key step any technique has to accomplish is the ionization of secondary metabolites directly from the organism in situ. MALDI is a mass spectrometry imaging technique that uses a laser to energize a matrix, which aids in desorbing and ionizing secondary metabolites.35,36 The laser is probably the easier thing to visualize, but the matrix, which is often benzoic acid, sinapinic acid, or cinnamic acid,11 is critical for transferring energy to the secondary metabolites. With DESI, the ionization can be thought of as a rebound, where charged solvent droplets from a modified electrospray ionization source cause the secondary metabolites to desorb from the surface into the mass spectrometer.9,18,37 While a matrix is not needed, there is a level of skill required to optimize the spray angle for maximum desorption with minimal damage to the surface20,37,38 of the organism. LAESI can be thought of as a hybrid of the two,7,19 where a mid-IR laser is used to generate gas phase particles, which are then ionized with an ESI source.18,19 With the droplet probe, ionization is decoupled from the organismal sampling, occurring post chromatography. Natural product samples can be analysed directly by MALDI, DESI, LAESI, and droplet probe but with varying amounts of sample preparation. LAESI and droplet probe have the advantage that they do not require intricate sample preparation.9,39 In contrast, MALDI utilizes a matrix that is applied to the surface of the organism.40 Sample preparation could be limited for DESI,9 however, we10 and others17 have found that an imprint of the culture surface may be ideal, at least for fungi.16
Another key point that distinguishes droplet probe from DESI, MALDI and LAESI is the ability to image a surface. Using droplet probe, it is possible to map the surface of a sample, but because of the size of the droplet, it is not possible to image.39 Alternatively, imaging has been performed a great deal with MALDI, LAESI, and DESI.8,11,12 For the applications we have examined with droplet probe, mapping the surface was sufficient to answer the posited questions. However, if a more spatially resolved measurement is needed for true imaging, one of the other three techniques is likely superior.
For temporal measurements, another consideration may be if the sample is damaged during analysis. With the use of a laser in MALDI and LAESI, it is obvious that part of the organism will be destroyed, and the matrix required for MALDI may be the biggest drawback. However, even with DESI, due to the spray that will raster across the surface, this too can hinder the ability to use the same sample over time. The sampling in droplet probe can be directed to a distinct spot, and the droplet itself can be dispersed several times so as to concentrate the sample before injecting into the chromatography system. We find that 3–5 replicates are often sufficient,39 but in some samples where the concentration of secondary metabolites could border on the limit of detection, we have replicated a spot more than a dozen times.41 As discussed below, that examination,41 in particular, was on a delicate substrate, and we used high resolution photographs to show that no visual damage to the sample surface was observed. The only caveat to temporal sampling with the droplet probe could be if the sample becomes contaminated via exposure during analysis, although that is true for all of the techniques.
Out of these four techniques, droplet probe has the broadest scope of substrate sampling capability because it is not limited to polarity, topography, or matrix application (Table 1).9,41,42 All four techniques could include tandem mass spectrometry and any other mass spectrometry driven parameters. However, droplet probe can be coupled with an LC system, which gives the advantage of chromatographic separation of isomers and identification of retention times. It also opens the door to the installation of other detectors post column, including UV-vis or photodiode array, providing spectroscopic data about secondary metabolites that are not possible with other in situ sampling techniques. In addition, quasi-universal detectors, such as evaporative light scattering detectors (ELSD)43,44 and charged aerosol detectors (CAD),45,46 may facilitate quantitative (or at least semi-quantitative) studies with the droplet probe; such applications are currently under development.
3. Use of droplet probe with fungi
3.1 Initial pilot studies and dereplication
For natural products chemists working toward the discovery of drug leads, irrespective of the source material and targets, a common challenge is preventing the rediscovery of already described compounds, commonly referred to as “dereplication”.39,47–49 We have developed several strategies to address this for studying fungal cultures, particularly with respect to the elimination of mycotoxins in the context of searching for anticancer drug leads.48,50,51 While our approach has evolved, those processes were developed for analysing an extract of an entire fungal culture from a single time point. Thus, when we first started to adapt the droplet probe for studying fungi, one of our early goals was to test its ability to dereplicate samples in situ from the Petri dish. This was initially proposed as a way to speed the dereplication process via interrogating the fungi much earlier in their growth phase (Fig. 1). However, as noted throughout this review, the framework of that approach has been applied nearly universally when using the droplet probe.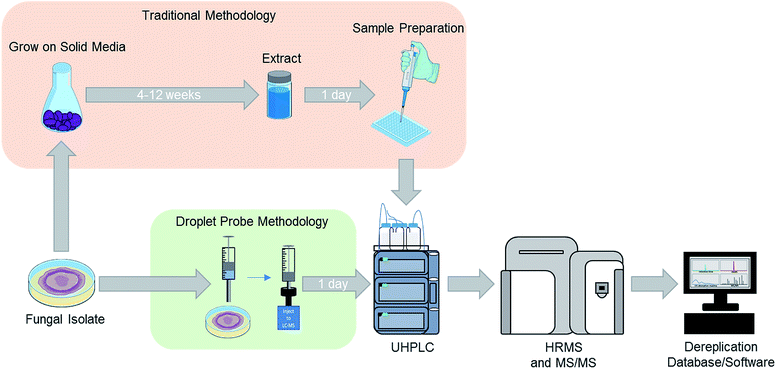 | ||
| Fig. 1 For traditional screening of fungal cultures for drug discovery purposes, the initial analysis may occur after growing a fungal culture for approximately 6–12 weeks (top). Using the droplet probe, the chemistry of cultures is evaluated much sooner and directly from the Petri dish (bottom), opening the door for examining a suite of different growth conditions both readily and rapidly. Adapted from Sica et al.39 | ||
Cogent to the point of embracing chromatography in the in situ analysis, we had developed a dereplication protocol based upon retention time, UV, HRMS, and MS/MS data in a database that now includes over 500 fungal metabolites.48,51 Thus, that protocol was adapted for studying fungal cultures in situ, and it became the foundation upon which many of our later studies were built. By incorporating the droplet probe to screen fungal cultures directly from the Petri dish, the dereplication process could begin before even extracting a fungus (Fig. 1). To test this, we first spotted pure fungal metabolites onto Teflon-coated slides. After calibrating the retention time data based on a longer inlet (due to the tubing for the droplet probe), we found that the same dereplication database48,51 could be utilized. We then demonstrated the practicality of this approach with in situ analysis of fungal cultures, rapidly dereplicating living cultures.39
While dereplication was a driver in that study, there were four major goals, which were (1) to eliminate the need to extract the fungal sample, (2) to conduct the analysis directly from the Petri dish, (3) to avoid optimizing growth conditions to facilitate ambient ionization, and (4) to include the acquisition of mutually supportive data.39 Of all those, goal 3 was a major challenge when we worked with DESI-MS.9 Once we realized that goal 3 was attainable, plans for the breadth of experimentation discussed later in this review began to be formalized.
An additional finding in that initial study was verification that the droplet probe could be used to distinguish isomers and adducts. We analysed a culture that biosynthesized sets of isomeric resorcylic acid lactones. Since those isomers had different retention times, they could readily be distinguished (e.g. 4.03 vs. 4.21 min; Fig. 2). Such information could be used to observe differences in relative abundance of distinct isobars. In addition, the chromatographic separation facilitated the recognition of multiple adducts, such as assigning [M + H]+, [M − H2O + H]+, and [M + Na]+ to a compound. One example was noting the loss of H2O via two separate signals that originated from a single compound (m/z 399.1204 and m/z 381.1098). Since we could observe those signals arising as a peak eluted from the UHPLC and into the mass spectrometer, we could correlate them to a single compound, with the latter simply being the loss of H2O from the former. In general, such assignments are not as straight forward with imaging techniques that infuse secondary metabolite ions as a mixture from the organism directly into the mass spectrometer. For example, those resorcylic acid lactone isobars would be indistinguishable, and the loss of H2O from a peak could be construed as a separate compound, if we had been using other ambient ionization techniques.
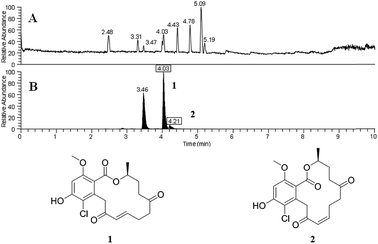 | ||
| Fig. 2 Panel (A) shows the base peak chromatogram for sampling a fungal culture by droplet probe. Panel (B) shows the XIC of m/z 381.1099 (±5 ppm), where the boxed retention times corresponded to resorcylic acid lactones 1 and 2, respectively, and the peak at 3.46 min represents a possible regioisomer of those compounds. Without chromatographic separation afforded by the droplet probe, the variance of the relative amounts of all three compounds would not be discernible. Adapted from Sica et al.39 | ||
In that initial study, there were a few other interesting applications that arose. When working on the taxonomy of a fungal culture, we observed that the inoculum seemed to contain two different fungi. This presented a significant challenge to the future publication52 of those results, as we needed to assign the chemistry to one of those cultures. Using the droplet probe, we could grow both of these fungi, separately, on Petri dishes and then analyse their chemistry in situ; the turnaround time for doing so was rapid, compared to the weeks to months required to grow the fungus for traditional natural products chemistry studies (Fig. 1). In doing so, we were quickly able to determine the culture that required taxonomic analysis, which was accomplished using both morphological and molecular (i.e. DNA-based) methods.53
There were several other pragmatic details that were piloted. For droplet retention on the syringe of the droplet probe, it is important to include some H2O, and after testing a suite of conditions, we settled on a droplet of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 MeOH–H2O. Alternatively, if the chromatography conditions utilized an acetonitrile gradient, then 1
1 MeOH–H2O. Alternatively, if the chromatography conditions utilized an acetonitrile gradient, then 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 CH3CN–H2O also worked. Additionally, the same droplet could be dispensed three to five times on the same spot. This served to concentrate the secondary metabolites, thereby improving our ability to detect them. In examining scores of fungal cultures, we have found that the droplet probe works on both the mycelium and the agar medium. In the rare instances where the mycelium was too absorbent, we found that pre-wetting with a droplet that absorbed into the mycelium, and then sampling with a second droplet on the same spot, circumvented this problem. We also found that droplet probe could sample distinct fungal morphologies, such as stroma and guttates. The latter was of interest biologically, as we studied a fungus that produced an herbicidal compound, termed mevalocidin, and its concentration in guttates may explain how this saprobic fungus interacts with its environment (see Section 3.4.1).54 In short, many future investigations benefited from the piloting and optimization experiments that were carried out in this initial study.39
1 CH3CN–H2O also worked. Additionally, the same droplet could be dispensed three to five times on the same spot. This served to concentrate the secondary metabolites, thereby improving our ability to detect them. In examining scores of fungal cultures, we have found that the droplet probe works on both the mycelium and the agar medium. In the rare instances where the mycelium was too absorbent, we found that pre-wetting with a droplet that absorbed into the mycelium, and then sampling with a second droplet on the same spot, circumvented this problem. We also found that droplet probe could sample distinct fungal morphologies, such as stroma and guttates. The latter was of interest biologically, as we studied a fungus that produced an herbicidal compound, termed mevalocidin, and its concentration in guttates may explain how this saprobic fungus interacts with its environment (see Section 3.4.1).54 In short, many future investigations benefited from the piloting and optimization experiments that were carried out in this initial study.39
3.2 Identifying unique residues and disputing artefacts
Peptaibols are prominent fungal metabolites that have been evaluated extensively,55–57 likely due to interesting biological activities, including those in the realm of antibacterial activity.58 There are more than 1000 of these non-ribosomal biosynthesized peptides (NRPs) reported in the literature,59 and they typically consist of 5–20 amino acid residues. From a structure elucidation standpoint, and due to the linear nature of the assemblage of those molecules, there is a great deal that can be determined directly by mass spectrometry. For example, in source fragmentation often occurs around proline residues, the N-terminus is typically acetylated, and the C-terminus amino acid is reduced to the alcohol.60 As such, fungal cultures that biosynthesize peptaibols are well positioned for in situ analysis by the droplet probe.It is rare to discover peptaibols that include a Thr residue (∼25 out of more than 1000).59 We found this surprising, as one could hypothesize that the secondary alcohol side chain of Thr could impart favourable properties to peptaibols, both biologically and physically; peptaibols with such residues have antibiotic61 and anthelmintic62 activities. Using the droplet probe, Sica et al.60 profiled in situ a strain of Nectriopsis sp., identifying four new peptaibols that included a Thr residue in the tenth position of 11-mer peptaibols. Importantly, the in situ measurements by droplet probe were validated by scaling up the cultures and isolating and characterizing the four new compounds, which were assigned the sequential trivial names necthreonin A through D. In summary, via this one study,60 the number of Thr-containing peptaibols in the literature was increased by about 20%, and in situ analysis paved the way for prioritizing this fungal culture as biosynthesizing new chemical diversity.
In another peptaibol-related study, three new peptaibols were identified, and the droplet probe helped to establish that they were true natural products.63 The question of artefacts often arises when studying secondary metabolites.64 This question was relevant, since the new compounds were isolated in extremely low yield, essentially as side fractions while scaling up the production of alamethicin F50 (3). In addition, these new analogues could be envisioned as simple derivatives of the more prominent compounds. If any conclusions were to be drawn on their potential biosynthesis, it was important to either prove their natural origin or admit that they were artefacts.
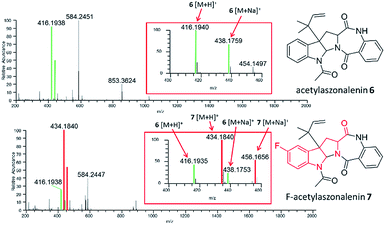
Specifically, one of the new peptaibols was a methyl ester analogue of alamethicin F50 (Fig. 3), with the ester occurring at residue 18 in compound 3, where Gln18 was converted to Glu-OMe. Of the over 1000 reported peptaibols, there are only four other compounds that had such a structural feature, and all were reported in a single manuscript.65 In this case, the droplet probe was not being used initially, as the scale up was performed using traditional natural products chemistry techniques. However, the droplet probe was implemented to analyse a living culture of the source organism, Trichoderma arundinaceum, and in doing so, several key ions were identified to support the conclusion that this new compound was an authentic secondary metabolite.63
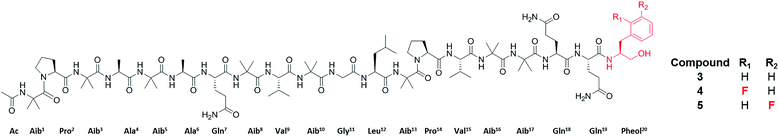 | ||
| Fig. 3 Structures of the peptaibols, alamethicin F50 (3), ortho-F-pheol alamethicin F50 (4), and meta-F-pheol alamethicin F50 (5). The incorporation of the fluorinated derivatives of Phe at residue 20 via precursor-directed biosynthesis was monitored and optimized by in situ evaluation by droplet probe. The structures of these non-natural natural products were then verified by scaled up isolation and structure elucidation,66 including Marfey's analysis, which confirmed incorporation of the L-enantiomers of F-Phe. | ||
In a separate study discussed later (Section 3.4.1), the droplet probe was used to probe for the presence of mevalocidin and its lactone form in situ. Prior to that, it was unknown whether the fungal strains were biosynthesizing both compounds, or if the lactone was formed due to the use of acid during the isolation process. When sampling fungal cultures of both strains, mevalocidin and its lactone form were observed directly from the fungal cultures, thereby establishing them as secondary metabolites.54
3.3 Biosynthesis of non-natural natural products
Synthetic biology approaches can be used to generate new secondary metabolites by introducing unnatural building blocks into parent compounds, essentially biosynthesizing non-natural natural products.67,68 Our own interest was to biosynthesize fungal metabolites that incorporated a fluorine atom, as fluorinated compounds make up at least 25% of all FDA approved drugs.69,70 Moreover, there are no fungal metabolites reported in the literature that include a fluorine atom naturally, with only a few reported based on precursor-directed biosynthesis.71,72There were many aspects of this study that were ideally suited to in situ examination by the droplet probe. Based on the aforementioned studies on peptaibols, we had a good grasp of the chemistry of these compounds,60,63,73,74 particularly from a mass spectrometry perspective. Moreover, the linear biosynthetic logic75 for the assembly of these compounds suggested that a precursor-directed biosynthetic approach could be used to incorporate non-canonical amino acids. Fortuitously, we had identified a robust producer of the peptaibol, alamethicin F50 (3), where the C-terminal amino acid is a reduced form of Phe (i.e. pheol). Fluorinated versions of Phe, with the fluorine in the ortho, meta, or para positions, are readily available, both as racemic mixtures and as pure enantiomers. Thus, the goal of this study was to use the droplet probe to pilot various ways to incorporate the fluorinated metabolites into alamethicin F50 (Fig. 3). This included testing the biosynthetic incorporation of all three fluorinated regioisomers, first as racemic mixtures, and then re-testing as pure enantiomers, to validate the hypothesized preferential incorporation of L-isomers. The studies were followed in two different fungi, both harvested from the wild, and validated by scaled up isolation and structure elucidation.
In situ chemical data, derived from droplet probe analysis, was instrumental in this project, rapidly going from pilot studies to scaled up isolation, characterization, and biological analysis of fluorinated analogues.66 Evaluating how the incorporation of various precursors manipulate biosynthesis is executed more rapidly via in situ monitoring in a Petri dish, drastically reducing time and resources needed to process unsuccessful experiments with traditional natural products chemistry techniques (Fig. 1). Using the fungus, Trichoderma arundinaceum, in situ analysis of various growth conditions showed a signal of m/z 1963.1313 ([M + H]+; monoisotopic precursor ion), which was characteristic of the presence of the parent compound, alamethicin F50 (Fig. 3 and 4). In the meta- and ortho-F-Phe supplemented cultures, an additional peak was observed at m/z 1981.1241 ([M + H]+) resulting from the addition of 17.99 amu, which indicated the addition of fluorine to the targeted peptaibol. Interestingly, probing the para-F-Phe supplemented culture did not show incorporation of the fluorinated amino acid (Fig. 4).66,76 This preliminary in situ analysis of the supplemented fungal cultures with non-canonical building blocks facilitated prioritization of the ortho- and meta-fluorinated analogues. Moreover, this experiment gave insight into the selectivity of the building blocks by the fungal biosynthetic machinery. Ortho and meta F-pheol alamethicin F50 analogues (4 and 5) were isolated and tested in vitro against a panel of human cancer cell lines. The results showed comparable potency to that of the non-fluorinated parent compound.66
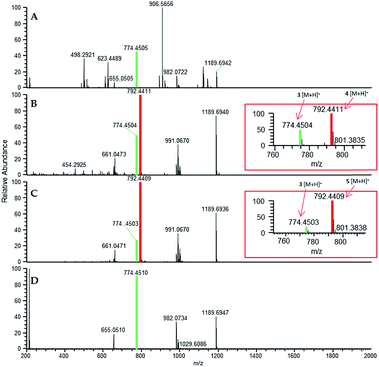 | ||
| Fig. 4 (A) Full-scan MS data of a Trichoderma arundinaceum (strain MSX70741) grown on PDA medium (control). (B) Full-scan MS data of MSX70741 grown on PDA supplemented with a racemic mixture of ortho-F-DL-Phe. (C) Full-scan MS data of MSX70741 grown on PDA supplemented with a racemic mixture of meta-F-DL-Phe. (D) Full-scan MS data of MSX70741 grown on PDA supplemented with a racemic mixture of para-F-DL-Phe. All cultures were sampled in situ using the droplet probe. In panels (B, C), the peaks corresponding to the fragment b7+ (m/z 792.4411 and 792.4409 for 4 and 5, respectively), indicating the incorporation of fluorine (19F), are boxed in red. In all panels, the green indicates ions for 3, whereas red indicates ions that support the incorporation of fluorinated building blocks (i.e.4 in (B) and 5 in (C)). Adapted from Rivera-Chávez et al.66 | ||
This general approach has been used successfully with precursor-directed biosynthesis of other non-canonical amino acids into different fungal metabolites. For example, fluorinated Trp was incorporated into growth media for Aspergillus fischeri. In situ analysis was employed for rapid processing of the conditions that yielded the best incorporation (Fig. 5).
3.4 Optimized production of fungal metabolites on the lab scale
It has long been known that media studies can be used to optimize the production of fungal metabolites, sometimes codified as an OSMAC (one strain, many cultures) approach.77,78 However, how one goes about that can be quite variable, and we have found that evaluating the chemistry of fungal cultures in situ via droplet probe enables scouting growth conditions rapidly, especially when spatial and temporal studies are taken into consideration.The droplet probe facilitated in situ sampling of the fungal chemistry on the surface of the cultures (mycelium, guttates, and surrounding agar), which would be challenging to accomplish via traditional natural product extraction methods (Fig. 1). The use of droplet probe was important in understanding that both strains of Coniolariella sp. (strains MSX56446 and MSX92917) released mevalocidin into their surroundings via guttates (exudates or liquid droplets), where the highest concentration of this secondary metabolite was mapped. Similarly, mevalocidin was also detected in the surrounding agar but not on the surface of the mycelium. Since the fungi that biosynthesize it are both saprobes (i.e. decomposers of dead organic matter), it was fascinating to map that the fungi seem to concentrate mevalocidin in this fashion, so as to exude the phytotoxic compound into their surroundings.54 Extrapolating, we hypothesized that this imparts an advantage to the fungus, potentially killing (or at least weakening) surrounding plant material, so that the fungus can utilize the dead organic matter for growth and reproduction. Such information could not be obtained without instruments that sampled chemistry in situ in this manner.
This general approach has been used several times, where it has been interesting to see where fungal secondary metabolites are concentrated.81,82 Based on additional literature,83–86 we suspect that guttates may be a common place for such compounds to be localized.
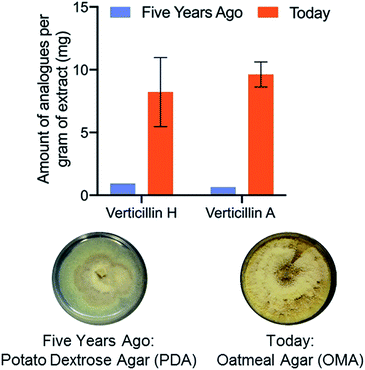
Fungal cultures that produced verticillins were examined in situ via droplet probe on a suite of different media (Fig. 6), ranging from both rich (i.e. those based on extracts of potatoes, yeast, etc.) and defined (i.e. those that are made via a specific recipe).87 We were pleased that fermentation on oatmeal-based medium seemed to produce the highest yield of verticillins, a fact that could only be discerned empirically. This observation was then tested in scaled up cultures (including three biological replicates), using a traditional natural products approach (Fig. 1) to validate the measurements from in situ analysis. Not only did the in situ results translate well to the pragmatic need for isolating more of the verticillins, but also, a timing study noted their peak production from 7 to 22 days, meaning that cultures could be processed on a weekly to biweekly basis.87 All told, we went from a challenging provision of a few mg of verticillins to a steady state production of 50–100 mg monthly (Fig. 6). Importantly, those materials are now being used in a suite of further studies, including in vivo pharmacology, in vivo pharmacokinetics, and semi-synthesis; none of those would be possible without the enhanced supply.
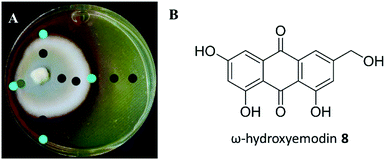
A suite of media types were explored, and this included varying pH. Among these, P. restrictum seemed to upregulate biosynthesis of the polyhydroxyanthraquinones on Sabouraud dextrose agar (SDA), where a colour change was noted from light yellow in young cultures to the diffusion of red into the agar after about 2–3 weeks of growth (Fig. 7). Analysis with droplet probe showed enhanced production of the target compound, ω-hydroxyemodin (8), with SDA. Similar to studies with mevalocidin, very little compound was detected on the surface of the mycelium, as most of the ω-hydroxyemodin (8) was exuded into the surrounding agar (Fig. 7).
4. Cyanobacteria
As part of a project to identify anticancer drug leads from a range of study materials, some of our collaborators work with cyanobacteria collected from fresh water habitats.107 As noted previously, dereplication is a key component for natural products drug discovery studies, irrespective of the source material. For our colleagues, scaling up the production of cyanobacteria is resource and time intensive, requiring lighted chambers and as many as 4–6 months to go from strain isolation to an 8 L culture. Obviously, it is a great disappointment if, at the end of that process, the cyanobacterium then yields known or uninteresting chemistry.In an attempt to improve this process, the droplet probe was used to examine the chemistry of cyanobacterial cultures in situ.108 In a pilot study, about 25 different cyanobacterial cultures were grown on solid Z medium, a process that takes about 2–3 weeks. These were then examined by droplet probe, and one culture, identified as a Calothrix sp. (strain UIC 10520), revealed two compounds of interest via m/z values of 517.3975 and 515.3975. Each of these signals was attributed to sodiated molecular ions, and the molecular formulae for the cyanobacterial metabolites were computed as C29H52N2O4 and C29H54N2O4, respectively. Since these formulae did not match any known secondary metabolites, this culture was targeted for scale up, whereupon two new compounds were isolated and elucidated using a suite of NMR and MS techniques.108
Given the aforementioned studies of fungi, it may not be surprising that in situ analysis of secondary metabolite profiles by droplet probe can also be applied to cyanobacteria. However, at the time, it was not clear if cyanobacterial cultures grown on solid phase media would recapitulate the chemistry observed in liquid cultures. Indeed, of the 25 strains that were examined, not all of them yielded valuable chemical information, for reasons that are unknown at this time. However, of the ones that did (representing about 70% of the strains), it was possible to either rule them out based on dereplication or prioritize them for scaled up isolation and structure elucidation. Given that this analysis can be completed within 2–3 weeks of plating a culture, vs. months for scale up, the investment in carrying out such in situ analyses seems worthwhile. We hypothesize that natural product researchers will be able to efficiently seek out and prioritize unique compounds from many different kinds of natural resources using the droplet probe.
5. Plant studies
5.1 Spatial mapping of acetogenins in Asimina triloba
Plants of the Annonaceae have been the subject of intense phytochemical studies for over 30 years due to the biological activity of their secondary metabolites, termed acetogenins.109,110 These plants typically biosynthesize a suite of structurally related acetogenins, and like peptides, they often fragment in predictable patterns, helping to establish the position of each hydroxy, the length of the hydrocarbon chains, and the position of THF rings (Fig. 8).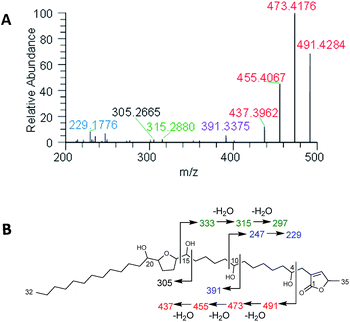 | ||
| Fig. 8 (A) In situ analysis of seeds of Asimina triloba shows the fragmentation pattern of annonacin. (B) Structure and key fragments for annonacin. Diagnostic signals are shown in the same colour in both panels. Reproduced from Sica et al.111 with permission from the Royal Society of Chemistry. | ||
One of our initial goals was to test the limits of the droplet probe to sample a range of botanical specimens. Much of the literature on acetogenins from Asimina triloba (paw paw) has been from the seeds,112,113 twigs114,115 and leaves.116 The flowers of this plant have not been explored, likely due to the difficulty in obtaining them, and acetogenins have not been reported from the flowers of any plant in the Annonaceae. This presented an interesting test case for the droplet probe, as the flowers were a plant organ that was not amenable to traditional natural products procedures, and thus, in situ chemistry could answer a question that had never been probed. Acetogenins were detected via in situ analysis of the seeds, fruit pulp, twigs, leaves, flower petals, and ovaries, and interestingly, the ovaries had the most extensive list of acetogenins,111 suggesting that the plant may be sequestering the secondary metabolites there to protect its progeny. Two pragmatic details were also uncovered, which were to strip any waxy layer from plant tissue by rubbing with CHCl3 prior to in situ analysis or implementing cryotome cross-sectioning to sample internal plant tissue.
An additional goal was to elucidate the structures of the acetogenins based on comparisons to the rich literature on these compounds.109,110,112,117 However, much of that was developed at a time when electron impact and fast atom bombardment were used for ionizing metabolites. Unfortunately, under typical electrospray ionization conditions, which is how the eluent from the column is infused into the mass spectrometer using droplet probe (Fig. 1), acetogenins do not form prominent product ions, and this confounds the use of tandem mass spectrometry for structure elucidation. However, recent studies have shown that the infusion of lithium ions, post column, enhances the fragmentation of acetogenins.118 Thus, Sica et al.111 infused a 2 mM solution of LiF (dissolved in MeOH), and this greatly facilitated structure elucidation efforts based on tandem mass spectrometry fragmentation patterns (Fig. 8), even when analysing samples in situ. Again, the ability to add detectors, or in this case, infuse a counter ion post column, represents an advantage of the modular set up of droplet probe (Fig. 1).
5.2 Mapping of phytochemicals on herbarium specimens
Herbarium voucher specimens are used most often for taxonomic purposes. However, those specimens also hold a record of the metabolic profile of a plant, at least at the time of sampling, and possibly at the time of collection. The setup of the droplet probe is quite versatile, making it straight forward to sample materials of various shapes and sizes. Thus, the droplet probe was tested for the analysis of phytochemicals on a herbarium voucher specimen.41 Importantly, a major goal was to do so in a manner that does not mar or destroy the integrity of the voucher, such that its appearance remained intact.A voucher of Garcinia mangostana (mangosteen) was analysed by droplet probe for xanthones (Fig. 9).41 Similar to acetogenins, there are many analogues and isomers of the xanthones, and thus, chromatographic separation was a key element to enhance detection and elucidation via in situ studies. There were two interesting modifications that grew out of this study. First, due to the fact that xanthones were poorly soluble in MeOH and CH3CN, the droplet solution was modified to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 DMSO
1 DMSO![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O. In addition, due to the low concentration of the xanthones in the herbarium voucher, the droplet was replicated as many as a dozen times on a single spot, so as to concentrate the secondary metabolites. While it was easier to measure the concentration of some metabolites over others, based on their relative abundance, the droplet probe was able to discern the chemistry of the herbarium voucher specimen in a manner that did not damage its appearance. While the direct application to herbarium vouchers could be considered niche, we believe this study is proof-of-concept that the droplet probe could be used to analyse the chemistry of delicate documents and other artifacts.
H2O. In addition, due to the low concentration of the xanthones in the herbarium voucher, the droplet was replicated as many as a dozen times on a single spot, so as to concentrate the secondary metabolites. While it was easier to measure the concentration of some metabolites over others, based on their relative abundance, the droplet probe was able to discern the chemistry of the herbarium voucher specimen in a manner that did not damage its appearance. While the direct application to herbarium vouchers could be considered niche, we believe this study is proof-of-concept that the droplet probe could be used to analyse the chemistry of delicate documents and other artifacts.
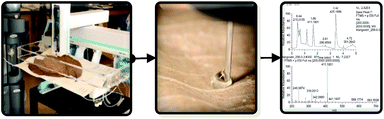 | ||
| Fig. 9 An example of sampling a herbarium voucher specimen of Garcinia mangostana with droplet probe. The left shows the herbarium voucher being analysed by droplet probe, the middle shows how the droplet interacts with the surface of the voucher, and the right shows the chromatographic and spectrometric data that are acquired. Adapted from Kao et al.41 | ||
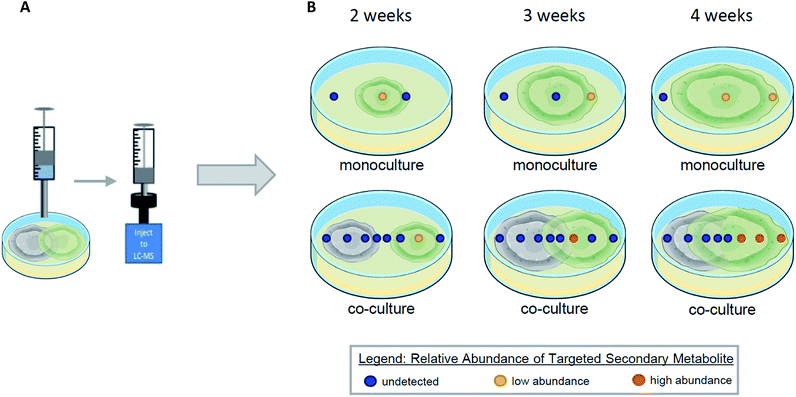
6. Interspecific interactions (i.e. co-culturing)
Co-culturing is a way to model and exploit the natural conditions in which microorganisms have evolved to survive, by growing two organisms within the same environment under laboratory conditions.119–123 When microorganisms are co-cultured, interspecific interactions between them leads to the activation of previously silenced secondary metabolites and/or an up/down regulation in the biosynthesis of secondary metabolites.101,124,125 Using droplet probe for in situ analysis during co-cultures has several advantages that are lost through ex situ techniques. The droplet probe analysis of a co-culture can shed light on the temporal, spatial, and relative abundance of secondary metabolites (Fig. 10). By screening the co-culture chemistry, droplet probe has the ability to provide chemical ecology information. By sampling two fungi and their surroundings directly, droplet probe serves to map the location of the fungal compounds produced during interspecific interactions.126,127 This provides a deeper understanding of why the fungi are producing compounds differently in monoculture versus co-culture conditions and how they are spatially distributed.39 Determining whether a compound is produced for defence, communication, attraction, or other purposes, can be explored via these mapping experiments. By obtaining temporal information during co-culture experiments, droplet probe can shed light on the time frame when organisms begin to produce secondary metabolites during interspecific interactions (Fig. 10).When Xylaria cubensis and Penicillium restrictum were co-cultured and analysed via droplet probe in a temporal study, the secondary metabolite activation and distribution changed based on how long the fungi were growing towards each other.127 Griseofulvin, which is produced by X. cubensis, was observed mainly toward the growing edge of the mycelium. In addition, the concentration of griseofulvin in guttates was over two magnitudes greater than that of a stroma (both base and tip) and about half a magnitude greater than on the mycelium.127 Given the fungistatic properties of griseofulvin,127 it is easy to suggest the protective role these adaptations play.
In a recent study, Knowles et al.128 showed that when X. cubensis was co-cultured with a mycotoxin producing strain, Aspergillus fischeri, a junction (conflict zone) was formed at the site where the two fungal strains interact with each other. Droplet probe analysis showed that X. cubensis marshalled its antifungals (griseofulvin and dechlorogriseofulvin) to the site of interaction between the two fungi. Interestingly, griseofulvin was not present on the side of the fungus away from A. fischeri.
Droplet probe analysis can compare the relative abundance of secondary metabolites in the co-culture to that of the monoculture to determine if the biosynthesis of secondary metabolites has been up/down regulated (Fig. 10). For example, when X. cubensis and A. fischeri were grown in co-culture, X. cubensis stunted the growth of A. fischeri. Not surprisingly, when analysed using traditional techniques, diminished growth corresponded with lower abundance of the secondary metabolites from A. fischeri. However, when performing the same analysis via droplet probe, i.e. monoculture to co-culture, the relative production of secondary metabolites by A. fischeri had increased significantly due to interspecific interactions in co-culture.128 In summary, the visual appearance of the co-culture experiment indicated that the growth of A. fischeri was stunted. However, based on in situ measurements, the relative production of secondary metabolites actually increased, a point that would not be discernible without droplet probe analysis.
Another advantage of using droplet probe in co-culturing experiments is the ability to quickly assess if the interspecific interactions lead to the activation of biosynthesis of secondary metabolites, especially when coupling the measurements to dereplication protocols.48,51 Altogether droplet probe analysis of co-culturing can help to glean information about the temporal, spatial, and up/down regulation of secondary metabolites, which are traditionally lost through ex situ techniques. The droplet probe technique is advantageous, as it allows us to understand how fungal species interactions may lead to changes in secondary metabolite profiles between two or more species, rapidly analysing the chemistry of the cultures in situ.127,128
7. Conclusions and future directions
In general, the advent of ambient ionization mass spectrometry techniques has made an enormous impact on natural products research, since organisms can be examined in situ with limited sample preparation.38 Key references for many of these techniques are noted in the preamble. When it comes to the droplet probe, and with the caveat that we may be biased based on our mycological research, fungi represent ideal candidates for in situ mass spectrometry mapping experiments. Compared to plants, microorganisms respond quickly to changes in environment (i.e. media)129,130 or by the introduction of other organisms (i.e. co-cultures),77,131 thus creating unique profiles of their biosynthesized secondary metabolites. Furthermore, as opposed to many bacteria, fungi are morphologically diverse and often develop unique physical characteristics as the culture grows. The presence of stroma (finger-like projections),132,133 guttates or liquid exudates10,83–85 and mycelium color changes/gradients give rise to several questions about the spatial distribution of the metabolites associated with such features. While, as we noted above, those distinct characteristics were problematic for using DESI-MS, they seemed to open up opportunities when working with droplet probe. Therefore, measuring chemistry in situ presents an avenue for profiling fungal cultures to examine the spatial and temporal distribution patterns of secondary metabolites that may not be possible via traditional natural products extraction methods.Measuring the chemical diversity of fungal cultures in situ can help address numerous biological questions, allowing us to delve deeper and wider into how fungal hyphae respond to experimental perturbations along space and time. In a concerted fashion, in situ techniques can measure and map the changing chemistry of fungi in situ during growth and development. The history of research on fungal secondary metabolites has taken a reductionist approach.134–136 Historically, the development and growth of multicellular hyphal cells (and their associated secondary metabolite chemistry) were examined in cultures at a defined time point and with zero regard to spatial distribution. Moreover, such studies did not use mass spectrometry mapping tools to specify where in the hyphal cells or nutrient media surroundings the secondary metabolites were localized during these interactions (e.g. guttates, aerial mycelium, vegetative hyphae or surrounding agar). However, in situ techniques can examine hyphal interactions in a fluid manner, using spatial (space) and temporal (time) sampling to measure and map the chemical diversity of fungal cultures. In situ methods can also examine how co-culturing affects chemical diversity by measuring and mapping how the secondary metabolites change/evolve under competition.
Evaluating chemistry in situ has numerous implications for fungal biology studies, and we have tried to conjure a few examples. In situ mass spectrometry can be used for probing plant surfaces for mycotoxins. This could be expanded to evaluating fungal pathogens in culture, cataloging differences and similarities in mycotoxin profiles between pathogens in artificial vs. natural environments. As global climate change alters our environment and ecosystems, we need to prepare for risks and challenges posed by fungal mycotoxins.137 Indoor air pollution by molds is a growing problem,138 estimated to occur in about 30–40% of households in the USA.139 We envisage that droplet probe can be used to evaluate the toxin profile of mold growing on surfaces (such as drywall). Moreover, for obligate pathogens that cannot be isolated in pure culture, these techniques could provide culture-independent chemical profiles.
Pragmatically, the design of the droplet probe is rather straight forward, as it essentially makes a microextraction on the surface of a substrate. Droplet sampling is amenable to a wide range of sample types, include those with distinct topography. Numerous examples of fungal cultures have been provided, since these organisms grow in a complex fashion and have distinct parts, such as mycelium, guttates, stroma, and spores. Additionally, it has been shown to work on cyanobacterial cultures, a range of plant materials, and delicate documents. Thus, a scientist's imagination may be the only limit to what types of substrates can be sampled.
Once the droplet probe has sampled the surface of an organism, that extract can then be interfaced with a wide range of analytical tools that are common to natural products laboratories. Chromatographic separation is the most unique aspect, as that is how it differs from the range of ambient ionization techniques that are also used to study the chemistry of nature in situ. In turn, chromatographic resolution of the extract from in situ sampling via droplet probe serves to enhance the measurements from any detectors added post column, including UV-vis and HRMS/tandem mass spectrometry. In one example, LiF was infused post column to enhance the ionization and fragmentation of a class of compounds (acetogenins), which made their structure elucidation more straight forward. The system works well with dereplication protocols, and thus, the droplet probe can be used in a variety of ways, examining secondary metabolites in both targeted and untargeted protocols. We have found that it is a great tool for determining ways to enhance the production of secondary metabolites, to probe conditions for the biosynthesis of non-natural natural products, and to examine the generation of new chemical diversity via co-culturing. Opportunities to study the chemistry of a range of materials in situ abound, and only time will tell what other questions can be analysed using the droplet probe.
8. Conflicts of interest
VK reports that he collects royalties for the copyrighted dropletProbe Premium software used by some of the research reported in the enclosed paper.9. Acknowledgements
SLK and DK were supported by the National Center for Complementary and Integrative Health/National Institutes of Health (NIH), under award numbers T32 AT008938 and F31 AT009264, respectively. VK was supported by the U. S. Department of Energy, Office of Science, Basic Energy Sciences, Chemical Sciences, Geosciences, and Biosciences Division. Research on bioactive fungal metabolites in NHO's lab is supported by the National Cancer Institute/NIH under grant P01 CA125066.10. References
- N. H. Oberlies and D. J. Kroll, J. Nat. Prod., 2004, 67, 129–135 CrossRef CAS PubMed.
- G. F. Pauli, S. M. Pro and J. B. Friesen, J. Nat. Prod., 2008, 71, 1489–1508 CrossRef CAS PubMed.
- J. B. Friesen, J. B. McAlpine, S.-N. Chen and G. F. Pauli, J. Nat. Prod., 2015, 78, 1765–1796 CrossRef CAS PubMed.
- M. C. Wani, H. L. Taylor, M. E. Wall, P. Coggon and A. T. McPhail, J. Am. Chem. Soc., 1971, 93, 2325–2327 CrossRef CAS PubMed.
- M. C. Wani and J. B. McAlpine, Personal Communications.
- J. M. Wiseman, D. R. Ifa, A. Venter and R. G. Cooks, Nat. Protoc., 2008, 3, 517 CrossRef CAS PubMed.
- C. Wu, A. L. Dill, L. S. Eberlin, R. G. Cooks and D. R. Ifa, Mass Spectrom. Rev., 2013, 32, 218–243 CrossRef CAS PubMed.
- J. D. Watrous, T. Alexandrov and P. C. Dorrestein, J. Mass Spectrom., 2011, 46, 209–222 CrossRef CAS PubMed.
- V. P. Sica, H. A. Raja, T. El-Elimat and N. H. Oberlies, RSC Adv., 2014, 4, 63221–63227 RSC.
- M. Figueroa, A. K. Jarmusch, H. A. Raja, T. El-Elimat, J. S. Kavanaugh, A. R. Horswill, R. G. Cooks, N. B. Cech and N. H. Oberlies, J. Nat. Prod., 2014, 77, 1351–1358 CrossRef CAS PubMed.
- E. Esquenazi, Y. L. Yang, J. Watrous, W. H. Gerwick and P. C. Dorrestein, Nat. Prod. Rep., 2009, 26, 1521–1534 RSC.
- J. D. Watrous and P. C. Dorrestein, Nat. Rev. Microbiol., 2011, 9, 683–694 CrossRef CAS PubMed.
- L. Nyadong, E. G. Hohenstein, A. Galhena, A. L. Lane, J. Kubanek, C. D. Sherrill and F. M. Fernández, Anal. Bioanal. Chem., 2009, 394, 245–254 CrossRef CAS PubMed.
- A. L. Lane, L. Nyadong, A. S. Galhena, T. L. Shearer, E. P. Stout, R. M. Parry, M. Kwasnik, M. D. Wang, M. E. Hay and F. M. Fernandez, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 7314–7319 CrossRef CAS PubMed.
- A. K. Badu-Tawiah, L. S. Eberlin, Z. Ouyang and R. G. Cooks, Annu. Rev. Phys. Chem., 2013, 64, 481–505 CrossRef CAS PubMed.
- A. Tata, C. J. Perez, M. O. Ore, D. Lostun, A. Passas, S. Morin and D. R. Ifa, RSC Adv., 2015, 5, 75458–75464 RSC.
- A. Tata, C. Perez, M. L. Campos, M. A. Bayfield, M. N. Eberlin and D. R. Ifa, Anal. Chem., 2015, 87, 12298–12305 CrossRef CAS PubMed.
- M.-Z. Huang, S.-C. Cheng, Y.-T. Cho and J. Shiea, Anal. Chim. Acta, 2011, 702, 1–15 CrossRef CAS PubMed.
- P. Nemes and A. Vertes, Anal. Chem., 2007, 79, 8098–8106 CrossRef CAS PubMed.
- A. K. Jarmusch and R. G. Cooks, Nat. Prod. Rep., 2014, 31, 730–738 RSC.
- T. Wachs and J. Henion, Anal. Chem., 2001, 73, 632–638 CrossRef CAS PubMed.
- A. D. Modestov, S. Srebnik, O. Lev and J. Gun, Anal. Chem., 2001, 73, 4229–4240 CrossRef CAS PubMed.
- G. J. Van Berkel, A. D. Sanchez and J. M. E. Quirke, Anal. Chem., 2002, 74, 6216–6223 CrossRef CAS PubMed.
- G. J. Van Berkel, V. Kertesz, K. A. Koeplinger, M. Vavrek and A. N. T. Kong, J. Mass Spectrom., 2008, 43, 500–508 CrossRef CAS PubMed.
- E. Q. Blatherwick, G. J. Van Berkel, K. Pickup, M. K. Johansson, M.-E. Beaudoin, R. O. Cole, J. M. Day, S. Iverson, I. D. Wilson and J. H. Scrivens, Xenobiotica, 2011, 41, 720–734 CrossRef CAS PubMed.
- G. J. Van Berkel, M. J. Ford, M. J. Doktycz and S. J. Kennel, Rapid Commun. Mass Spectrom., 2006, 20, 1144–1152 CrossRef CAS PubMed.
- http://https://prosolia.com/products/flowprobe/, January 1, 2019.
- G. J. Van Berkel, V. Kertesz and R. C. King, Anal. Chem., 2009, 81, 7096–7101 CrossRef CAS PubMed.
- V. Kertesz and G. J. Van Berkel, J. Mass Spectrom., 2010, 45, 252–260 CrossRef CAS PubMed.
- http://https://advion.com/products/triversa-nanomate/mode-3-liquid-extraction-surface-analysis-lesa/, January 1, 2019.
- V. Kertesz and G. J. Van Berkel, Anal. Chem., 2010, 82, 5917–5921 CrossRef CAS PubMed.
- G. J. Van Berkel and V. Kertesz, Rapid Commun. Mass Spectrom., 2013, 27, 1329–1334 CrossRef CAS PubMed.
- https://advion.com/rsc-product-note/lesa-plus-liquid-extraction-surface-analysis-plus-lc-separation/, January 1, 2019.
- http://www.htximaging.com/htx-sepquant-dropletprobe/, January 1, 2019.
- M. Karas and R. Krüger, Chem. Rev., 2003, 103, 427–440 CrossRef CAS PubMed.
- R. Knochenmuss, Analyst, 2006, 131, 966–986 RSC.
- Z. Takats, J. M. Wiseman, B. Gologan and R. G. Cooks, Science, 2004, 306, 471–473 CrossRef CAS PubMed.
- R. G. Cooks, Z. Ouyang, Z. Takats and J. M. Wiseman, Science, 2006, 311, 1566–1570 CrossRef CAS PubMed.
- V. P. Sica, H. A. Raja, T. El-Elimat, V. Kertesz, G. J. Van Berkel, C. J. Pearce and N. H. Oberlies, J. Nat. Prod., 2015, 78, 1926–1936 CrossRef CAS PubMed.
- E. R. A. van Hove, D. F. Smith and R. M. Heeren, J. Chromatogr. A, 2010, 1217, 3946–3954 CrossRef PubMed.
- D. Kao, J. M. Henkin, D. D. Soejarto, A. D. Kinghorn and N. H. Oberlies, Phytochem. Lett., 2018, 28, 124–129 CrossRef CAS PubMed.
- Y. Dong, B. Li, S. Malitsky, I. Rogachev, A. Aharoni, F. Kaftan, A. Svatoš and P. Franceschi, Front. Plant Sci., 2016, 7, 60 Search PubMed.
- R. Lucena, S. Cardenas and M. Valcarcel, Anal. Bioanal. Chem., 2007, 388, 1663–1672 CrossRef CAS PubMed.
- N. C. Megoulas and M. A. Koupparis, Crit. Rev. Anal. Chem., 2005, 35, 301–316 CrossRef CAS.
- M. Ligor, S. Studzińska, A. Horna and B. Buszewski, Crit. Rev. Anal. Chem., 2013, 43, 64–78 CrossRef CAS.
- T. Vehovec and A. Obreza, J. Chromatogr. A, 2010, 1217, 1549–1556 CrossRef CAS PubMed.
- S. P. Gaudêncio and F. Pereira, Nat. Prod. Rep., 2015, 32, 779–810 RSC.
- T. El-Elimat, M. Figueroa, B. M. Ehrmann, N. B. Cech, C. J. Pearce and N. H. Oberlies, J. Nat. Prod., 2013, 76, 1709–1716 CrossRef CAS PubMed.
- T. Ito and M. Masubuchi, J. Antibiot., 2014, 67, 353 CrossRef CAS PubMed.
- A. A. Sy-Cordero, T. N. Graf, M. C. Wani, D. J. Kroll, C. J. Pearce and N. H. Oberlies, J. Antibiot., 2010, 63, 539 CrossRef CAS PubMed.
- N. D. Paguigan, T. El-Elimat, D. Kao, H. A. Raja, C. J. Pearce and N. H. Oberlies, J. Antibiot., 2017, 70, 553 CrossRef CAS PubMed.
- A. Kaur, H. A. Raja, B. A. Darveaux, W.-L. Chen, S. M. Swanson, C. J. Pearce and N. H. Oberlies, Magn. Reson. Chem., 2015, 53, 616 CrossRef CAS PubMed.
- H. A. Raja, A. N. Miller, C. J. Pearce and N. H. Oberlies, J. Nat. Prod., 2017, 80, 756–770 CrossRef CAS PubMed.
- V. P. Sica, M. Figueroa, H. A. Raja, T. El-Elimat, B. A. Darveaux, C. J. Pearce and N. H. Oberlies, J. Ind. Microbiol. Biotechnol., 2016, 43, 1149–1157 CrossRef CAS PubMed.
- C. Toniolo and H. Brückner, Chem. Biodiversity, 2007, 4, 1021–1022 CrossRef CAS.
- H. Bruckner, J. Pept. Sci., 2003, 9, 659 CrossRef.
- C. Toniolo and H. Brückner, Peptaibiotics: Fungal Peptides Containing α-dialkyl α-amino Acids, Wiley VCH Zurich, 2009 Search PubMed.
- P. Zhao, Y. Xue, X. Li, J. Li, Z. Zhao, C. Quan, W. Gao, X. Zu, X. Bai and S. Feng, Peptides, 2019, 113, 52–65 CrossRef CAS PubMed.
- N. K. Neumann, N. Stoppacher, S. Zeilinger, T. Degenkolb, H. Brückner and R. Schuhmacher, Chem. Biodiversity, 2015, 12, 743–751 CrossRef CAS PubMed.
- V. P. Sica, E. R. Rees, H. A. Raja, J. Rivera-Chavez, J. E. Burdette, C. J. Pearce and N. H. Oberlies, Phytochemistry, 2017, 143, 45–53 CrossRef CAS PubMed.
- K. L. Rinehart Jr, L. A. Gaudioso, M. L. Moore, R. C. Pandey, J. C. Cook Jr, M. Barber, R. D. Sedgwick, R. S. Bordoli, A. N. Tyler and B. N. Green, J. Am. Chem. Soc., 1981, 103, 6517–6520 CrossRef.
- M. Schiell, J. Hofmann, M. Kurz, F. R. Schmidt, L. Vériest, M. Vogel, J. Wink and G. Seibert, J. Antibiot., 2001, 54, 220–233 CrossRef CAS.
- J. Rivera-Chávez, H. A. Raja, T. N. Graf, J. M. Gallagher, P. Metri, D. Xue, C. J. Pearce and N. H. Oberlies, RSC Adv., 2017, 7, 45733–45741 RSC.
- R. J. Capon, Nat. Prod. Rep., 2019 10.1039/C9NP00013E.
- I. Panizel, O. Yarden, M. Ilan and S. Carmeli, Mar. Drugs, 2013, 11, 4937–4960 CrossRef CAS PubMed.
- J. Rivera-Chávez, H. A. Raja, T. N. Graf, J. E. Burdette, C. J. Pearce and N. H. Oberlies, J. Nat. Prod., 2017, 80, 1883–1892 CrossRef PubMed.
- M. C. Walker and M. C. Chang, Chem. Soc. Rev., 2014, 43, 6527–6536 RSC.
- M. C. Walker, B. W. Thuronyi, L. K. Charkoudian, B. Lowry, C. Khosla and M. C. Chang, Science, 2013, 341, 1089–1094 CrossRef CAS PubMed.
- I. Ojima, Fluorine in medicinal chemistry and chemical biology, John Wiley & Sons Ltd, Hoboken, NJ, 2009 Search PubMed.
- Y. Zhou, J. Wang, Z. Gu, S. Wang, W. Zhu, J. L. Aceña, V. A. Soloshonok, K. Izawa and H. Liu, Chem. Rev., 2016, 116, 422–518 CrossRef CAS PubMed.
- Y. Xu, J. Zhan, E. K. Wijeratne, A. M. Burns, A. L. Gunatilaka and I. Molnár, J. Nat. Prod., 2007, 70, 1467–1471 CrossRef CAS PubMed.
- R. Thiericke and J. Rohr, Nat. Prod. Rep., 1993, 10, 265–289 RSC.
- M. Figueroa, H. Raja, J. O. Falkinham III, A. F. Adcock, D. J. Kroll, M. C. Wani, C. J. Pearce and N. H. Oberlies, J. Nat. Prod., 2013, 76, 1007–1015 CrossRef CAS PubMed.
- S. Ayers, B. M. Ehrmann, A. F. Adcock, D. J. Kroll, E. J. Carcache de Blanco, Q. Shen, S. M. Swanson, J. O. Falkinham III, M. C. Wani and S. M. Mitchell, J. Pept. Sci., 2012, 18, 500–510 CrossRef CAS PubMed.
- R. D. Süssmuth and A. Mainz, Angew. Chem., Int. Ed. Engl., 2017, 56, 3770–3821 CrossRef PubMed.
- M. Winn, J. Fyans, Y. Zhuo and J. Micklefield, Nat. Prod. Rep., 2016, 33, 317–347 RSC.
- R. T. Hewage, T. Aree, C. Mahidol, S. Ruchirawat and P. Kittakoop, Phytochemistry, 2014, 108, 87–94 CrossRef CAS PubMed.
- C. F. P. Hemphill, P. Sureechatchaiyan, M. U. Kassack, R. S. Orfali, W. Lin, G. Daletos and P. Proksch, J. Antibiot., 2017, 70, 726 CrossRef CAS PubMed.
- B. C. Gerwick, W. K. Brewster, S. C. Fields, P. R. Graupner, D. R. Hahn, C. J. Pearce, P. R. Schmitzer and J. D. Webster, J. Chem. Ecol., 2013, 39, 253–261 CrossRef CAS PubMed.
- B. C. Gerwick III, P. R. Graupner, S. C. Fields, P. R. Schmitzer and W. K. Brewster, Methylidene mevalonates and their use as herbicides. US 7,393,812 B2, 2008.
- N. D. Paguigan, H. A. Raja, C. S. Day and N. H. Oberlies, Phytochemistry, 2016, 126, 59–65 CrossRef CAS.
- H. A. Raja, N. D. Paguigan, J. Fournier and N. H. Oberlies, Mycol. Prog., 2017, 16, 535–552 CrossRef.
- A. Koulman, G. A. Lane, M. J. Christensen, K. Fraser and B. A. Tapper, Phytochemistry, 2007, 68, 355–360 CrossRef CAS PubMed.
- X. Wang, J. G. Sena Filho, A. R. Hoover, J. B. King, T. K. Ellis, D. R. Powell and R. H. Cichewicz, J. Nat. Prod., 2010, 73, 942–948 CrossRef CAS PubMed.
- S. Hutwimmer, H. Wang, H. Strasser and W. Burgstaller, Mycologia, 2010, 102, 1–10 CrossRef CAS PubMed.
- M. Gardes and T. D. Bruns, Mol. Ecol., 1993, 2, 113–118 CrossRef CAS PubMed.
- C. S. M. Amrine, H. A. Raja, B. A. Darveaux, C. J. Pearce and N. H. Oberlies, J. Ind. Microbiol. Biotechnol., 2018, 45, 1053–1065 CrossRef CAS PubMed.
- K. Katagiri, K. Sato, S. Hayakawa, T. Matsushima and H. Minato, J. Antibiot., 1970, 23, 420–422 CrossRef CAS.
- N. Boyer, K. C. Morrison, J. Kim, P. J. Hergenrother and M. Movassaghi, Chem. Sci., 2013, 4, 1646–1657 RSC.
- J. Kim, J. A. Ashenhurst and M. Movassaghi, Science, 2009, 324, 238–241 CrossRef CAS PubMed.
- J. Kim and M. Movassaghi, Acc. Chem. Res., 2015, 48, 1159–1171 CrossRef CAS PubMed.
- M. Figueroa, T. N. Graf, S. Ayers, A. F. Adcock, D. J. Kroll, J. Yang, S. M. Swanson, U. Munoz-Acuna, E. J. C. De Blanco and R. Agrawal, J. Antibiot., 2012, 65, 559 CrossRef CAS PubMed.
- F. Liu, Q. Liu, D. Yang, W. B. Bollag, K. Robertson, P. Wu and K. Liu, Cancer Res., 2011, 71, 6807–6816 CrossRef CAS PubMed.
- C. Lu, D. Yang, M. E. Sabbatini, A. H. Colby, M. W. Grinstaff, N. H. Oberlies, C. Pearce and K. Liu, BMC Cancer, 2018, 18, 149 CrossRef PubMed.
- C. Lu, A. V. Paschall, H. Shi, N. Savage, J. L. Waller, M. E. Sabbatini, N. H. Oberlies, C. Pearce and K. Liu, J. Natl. Cancer Inst., 2017, 109 DOI:10.1093/jnci/djw283.
- A. V. Paschall, D. Yang, C. Lu, J.-H. Choi, X. Li, F. Liu, M. Figueroa, N. H. Oberlies, C. Pearce and W. B. Bollag, J. Immunol., 2015, 195, 1868–1882 CrossRef CAS PubMed.
- J. Wang, X. Zhu, S. Kolli, H. Wang, C. J. Pearce, N. H. Oberlies and M. A. Phelps, J. Pharm. Biomed. Anal., 2017, 139, 187–192 CrossRef CAS PubMed.
- A. Zewdu, G. Lopez, D. Braggio, C. Kenny, D. Constantino, H. Bid, K. Batte, O. Iwenofu, N. Oberlies and C. Pearce, Clin. Exp. Pharmacol., 2016, 6, 221 Search PubMed.
- S. M. Daly, B. O. Elmore, J. S. Kavanaugh, K. D. Triplett, M. Figueroa, H. A. Raja, T. El-Elimat, H. A. Crosby, J. K. Femling, N. B. Cech, A. R. Horswill, N. H. Oberlies and P. R. Hall, Antimicrob. Agents Chemother., 2015, 59, 2223–2235 CrossRef CAS.
- H. A. Raja, A. Kaur, T. El-Elimat, M. Figueroa, R. Kumar, G. Deep, R. Agarwal, S. H. Faeth, N. B. Cech and N. H. Oberlies, Mycology, 2015, 6, 8–27 CrossRef CAS PubMed.
- P. Spiteller, Nat. Prod. Rep., 2015, 32, 971–993 RSC.
- S. Kusari and M. Spiteller, Nat. Prod. Rep., 2011, 28, 1203–1207 RSC.
- T. Sudhakar, S. Dash, R. Rao, R. Srinivasan, S. Zacharia, M. Atmanand, B. Subramaniam and S. Nayak, Curr. Sci., 2013, 104, 178 Search PubMed.
- G. L. Challis, J. Med. Chem., 2008, 51, 2618–2628 CrossRef CAS PubMed.
- N. P. Keller, Nat. Rev. Microbiol., 2018, 1 Search PubMed.
- N. P. Keller, G. Turner and J. W. Bennett, Nat. Rev. Microbiol., 2005, 3, 937 CrossRef CAS PubMed.
- J. Orjala, N. H. Oberlies, C. Pearce, S. Swanson and A. D. Kinghorn, Bioactive compounds from natural sources: natural products as lead compounds in drug discovery, in Discovery of Potential Anticancer Agents from Aquatic Cyanobacteria, Filamentous Fungi, and Tropical Plants, ed. C. Tringali, CRC Press, 2011, pp. 37–64 Search PubMed.
- C. M. Crnkovic, A. Krunic, D. S. May, T. A. Wilson, D. Kao, J. E. Burdette, J. R. Fuchs, N. H. Oberlies and J. Orjala, J. Nat. Prod., 2018, 81, 2083–2090 CrossRef CAS PubMed.
- F. Q. Alali, X.-X. Liu and J. L. McLaughlin, J. Nat. Prod., 1999, 62, 504–540 CrossRef CAS PubMed.
- J. L. McLaughlin, J. Nat. Prod., 2008, 71, 1311–1321 CrossRef CAS PubMed.
- V. Sica, T. El-Elimat and N. Oberlies, Anal. Methods, 2016, 8, 6143–6149 RSC.
- M.-H. Woo, D.-H. Kim and J. L. McLaughlin, Phytochemistry, 1999, 50, 1033–1040 CrossRef CAS.
- M. H. Woo, K. Y. Cho, Y. Zhang, L. Zeng, Z.-M. Gu and J. L. McLaughlin, J. Nat. Prod., 1995, 58, 1533–1542 CrossRef CAS.
- K. He, G.-X. Zhao, G. Shi, L. Zeng, J.-F. Chao and J. L. McLaughlin, Bioorg. Med. Chem., 1997, 5, 501–506 CrossRef CAS PubMed.
- G.-X. Zhao, M. Rieser, Y.-H. Hui, L. Miesbauer, D. Smith and J. McLaughlin, Phytochemistry, 1993, 33, 1065–1073 CrossRef CAS.
- S. Ratnayake, K. J. Rupprecht, W. M. Potter and J. L. McLaughlin, J. Econ. Entomol., 1992, 85, 2353–2356 CrossRef CAS.
- M.-H. Woo, S.-O. Chung and D.-H. Kim, Bioorg. Med. Chem., 2000, 8, 285–290 CrossRef CAS.
- J. Le Ven, I. Schmitz-Afonso, G. Lewin, O. Laprévote, A. Brunelle, D. Touboul and P. Champy, J. Mass Spectrom., 2012, 47, 1500–1509 CrossRef CAS PubMed.
- K. Scherlach and C. Hertweck, Org. Biomol. Chem., 2009, 7, 1753–1760 RSC.
- S. Bertrand, N. Bohni, S. Schnee, O. Schumpp, K. Gindro and J.-L. Wolfender, Biotechnol. Adv., 2014, 32, 1180–1204 CrossRef CAS PubMed.
- J. C. Frisvad, B. Andersen and U. Thrane, Mycol. Res., 2008, 112, 231–240 CrossRef CAS PubMed.
- J. O'Brien and G. D. Wright, Curr. Opin. Biotechnol., 2011, 22, 552–558 CrossRef PubMed.
- G. F. Bills and J. B. Gloer, Microbiol. Spectrum, 2016, 4 DOI:10.1128/microbiolspec.FUNK-0009-2016.
- S. Huang, W. Ding, C. Li and D. G. Cox, Pharmacogn. Mag., 2014, 10, 410–414 CrossRef.
- H.-W. Nützmann, V. Schroeckh and A. A. Brakhage, Regulatory cross talk and microbial induction of fungal secondary metabolite gene clusters, in Methods in enzymology, Elsevier, 2012, vol. 517, pp. 325–341 Search PubMed.
- J. Hiscox, J. O'Leary and L. Boddy, Stud. Mycol., 2018, 89, 117–124 CrossRef CAS.
- V. P. Sica, E. R. Rees, E. Tchegnon, R. H. Bardsley, H. A. Raja and N. H. Oberlies, Front. Microbiol., 2016, 7, 544 Search PubMed.
- S. L. Knowles, H. A. Raja, A. J. Wright, A. M. L. Lee, L. K. Caesar, N. B. Cech, M. E. Mead, J. L. Steenwyk, L. Ries, G. H. Goldman, A. Rokas and N. H. Oberlies, Front. Microbiol., 2019, 10, 285 CrossRef PubMed.
- K. M. VanderMolen, B. A. Darveaux, W.-L. Chen, S. M. Swanson, C. J. Pearce and N. H. Oberlies, RSC Adv., 2014, 4, 18329–18335 RSC.
- G. F. Bills, G. Platas, A. Fillola, M. R. Jiménez, J. Collado, F. Vicente, J. Martín, A. González, J. Bur-Zimmermann, J. R. Tormo and F. Peláez, J. Appl. Microbiol., 2008, 104, 1644–1658 CrossRef CAS PubMed.
- H. Wei, Z. Lin, D. Li, Q. Gu and T. Zhu, Acta Microbiol. Sin., 2010, 50, 701–709 CAS.
- E. Kuhnert, S. Heitkämper, J. Fournier, F. Surup and M. Stadler, Fungal Biol., 2014, 118, 242–252 CrossRef CAS.
- M. Stadler, Curr. Res. Environ. Appl. Mycol., 2011, 1, 75–133 CrossRef.
- K. M. Zuck, S. Shipley and D. J. Newman, J. Nat. Prod., 2011, 74, 1653–1657 CrossRef CAS PubMed.
- S. Bertrand, O. Schumpp, N. Bohni, M. Monod, K. Gindro and J.-L. Wolfender, J. Nat. Prod., 2013, 76, 1157–1165 CrossRef CAS PubMed.
- A. Marmann, A. H. Aly, W. Lin, B. Wang and P. Proksch, Mar. Drugs, 2014, 12, 1043–1065 CrossRef PubMed.
- J. W. Bennett and M. Klich, Clin. Microbiol. Rev., 2003, 16, 497–516 CrossRef CAS PubMed.
- A. S. Amend, K. A. Seifert, R. Samson and T. D. Bruns, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 13748–13753 CrossRef CAS PubMed.
- R. L. Górny, T. Reponen, K. Willeke, D. Schmechel, E. Robine, M. Boissier and S. A. Grinshpun, Appl. Environ. Microbiol., 2002, 68, 3522–3531 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2019 |