Open Access Article
Open Access ArticleAdvances in nanomaterial application in enzyme-based electrochemical biosensors: a review
I. S.
Kucherenko
 *ab,
O. O.
Soldatkin
ac,
D. Yu.
Kucherenko
a,
O. V.
Soldatkina
cd and
S. V.
Dzyadevych
ac
*ab,
O. O.
Soldatkin
ac,
D. Yu.
Kucherenko
a,
O. V.
Soldatkina
cd and
S. V.
Dzyadevych
ac
aDepartment of Biomolecular Electronics, Institute of Molecular Biology and Genetics of the National Academy of Sciences of Ukraine, Zabolotnogo Street 150, Kyiv, 03143, Ukraine. E-mail: kucherenko.i.s@gmail.com
bDepartment of Mechanical Engineering, Iowa State University, Ames, Iowa 50011, USA
cInstitute of High Technologies, Taras Shevchenko National University of Kyiv, Volodymyrska Street 64, Kyiv, 01003, Ukraine
dF. D. Ovcharenko Institute of Biocolloidal Chemistry, Acad. Vernadskoho Blvd. 42, Kyiv, 03142, Ukraine
First published on 31st October 2019
Abstract
Electrochemical enzyme-based biosensors are one of the largest and commercially successful groups of biosensors. Integration of nanomaterials in the biosensors results in significant improvement of biosensor sensitivity, limit of detection, stability, response rate and other analytical characteristics. Thus, new functional nanomaterials are key components of numerous biosensors. However, due to the great variety of available nanomaterials, they should be carefully selected according to the desired effects. The present review covers the recent applications of various types of nanomaterials in electrochemical enzyme-based biosensors for the detection of small biomolecules, environmental pollutants, food contaminants, and clinical biomarkers. Benefits and limitations of using nanomaterials for analytical purposes are discussed. Furthermore, we highlight specific properties of different nanomaterials, which are relevant to electrochemical biosensors. The review is structured according to the types of nanomaterials. We describe the application of inorganic nanomaterials, such as gold nanoparticles (AuNPs), platinum nanoparticles (PtNPs), silver nanoparticles (AgNPs), and palladium nanoparticles (PdNPs), zeolites, inorganic quantum dots, and organic nanomaterials, such as single-walled carbon nanotubes (SWCNTs), multi-walled carbon nanotubes (MWCNTs), carbon and graphene quantum dots, graphene, fullerenes, and calixarenes. Usage of composite nanomaterials is also presented.
1. Introduction
Biosensors are a group of state-of-the-art analytical devices involving a biorecognition material in close contact with a transducer.1 An important benefit of biosensors is their significantly lower cost compared to alternative and commonly used methods. Additionally, biosensors are usually portable and easy-to-use, therefore their development is an urgent task of biotechnology and analytical chemistry.The spectrum of biosensors applications is quite wide. Supposedly, in future biosensors will be widely used in medicine, agriculture, control of various biotechnological processes, environmental monitoring of toxic compounds and other areas. Electrochemical enzyme-based biosensors are one of the most advanced and commercially successful bioanalytical devices because of a high catalytic activity and selectivity of enzymes, as well as commercial availability of purified enzymes. However, traditional enzyme-based biosensors have limited sensitivity, selectivity, and stability, thus different approaches to improvement of the biosensors are considered.
A new promising trend in biosensorics is the use of nanoscale materials of various types, which have unique physical and chemical properties. Nanomaterials (NMs), the substances with the size of structural elements of 1–100 nm, significantly differ from similar macro-scale materials. In biosensors, NMs are used to improve the basic analytical characteristics of biosensors, such as sensitivity, limit of detection (LOD), linear detection range, selectivity, reproducibility, stability, response time, etc.2 Unique properties of NMs, in particular, a high surface-to-volume ratio, ensure significant increase in the sensitive surface of the transducer and more effective enzyme immobilization. Additionally, NMs are characterized by high electrical conductivity, magnetic properties, catalytic activity, etc., which are important for biosensors.3 Moreover, surface of NMs can be easily modified with different chemical groups,4,5 which is essential for the interaction with biomaterial in biosensors and other biotechnological assays.6 Doped NMs also provide a flexible way to obtain highly effective sensors.7 A perspective approach is the synthesis of NMs that form colored complexes with their targets – such complexes can be observed with the naked eye.8 Separation of molecules in complex matrixes can be also achieved using NMs.9–12
By the chemical structure, NMs can be divided into organic and inorganic. Inorganic NMs include metals and their oxides, quantum dots, zeolites, etc.; fullerenes, carbon nanotubes (CNTs), graphene and graphene oxide, calixarenes, etc. are organic NMs.13 Inorganic NMs are characterized by relatively simple synthesis; they catalyze some electrochemical reactions,14,15 and participate in acceleration of electron transfer. Organic NMs are characterized by the properties, which contribute to the amplification of the electrochemical signal and ensure a high degree of biocompatibility.6,16 Both types of NMs are useful in the development of electrochemical sensors.17 More exotic NMs such as semiconductor or composite NMs are also studied.18 General comparison of NMs is presented in Table 1.
| Nanomaterial | High conductance | High adsorption capability | Catalysis of reactions | Other features |
|---|---|---|---|---|
| a AgNPs – silver nanoparticles; AuNPs – gold nanoparticles; CNTs – carbon nanotubes; NADH – reduced nicotinamide adenine dinucleotide; PdNPs – palladium nanoparticles; PtNPs – platinum nanoparticles; QDs – quantum dots. | ||||
| AuNPs | Yes | No | No | Possibility of thiol bonds formation |
| PtNPs | Yes | No | H2O2 decomposition | No |
| AgNPs | Yes | No | No | No |
| PdNPs | Yes | No | H2O2 decomposition | Relatively cheap |
| Zeolites | No | Yes | No | No |
| Inorganic QDs | No | No | No | Semiconductor and optical properties |
| CNTs | Yes | No | No | No |
| Organic QDs | No | No | NADH oxidation | Optical properties |
| Graphene and derivatives | Yes | No | No | No |
| Fullerenes | Yes | No | No | No |
| Calixarenes | No | No | No | Specific binding of small molecules |
An alternative classification of NMs is based on their dimensions.19 NMs are divided into 0D clusters and particles, 1D nanowires and nanotubes, 2D films, and 3D structures. However, in the present review we use the chemical classification of NMs.
To improve sensors' analytical characteristics, nanoparticles can be used in different ways. They can be either co-immobilized with the enzymes or integrated into the transducer surface; some nanoparticles can be used as a selective element of chemosensors.20–24 Thus, depending on the objective it is possible to obtain required parameters of the sensors by appropriate choice of NMs and procedure of their application.
The utilization of nano- and micro-materials to improve the analytical characteristics of biosensors is one of the major trends of analytical biotechnology.
Here, we reviewed the recent applications of various types of NMs in electrochemical enzyme-based biosensors for the detection of environmental pollutants, food contaminants, and clinical biomarkers. Additionally, we discussed the benefits and limitations of using NMs for analytical purposes.
2. Methods of embedding nanomaterials in the enzyme-based biosensors
An electrochemical transducer can be modified with NMs before the biomaterial immobilization, or NMs can be immobilized together with the bioreceptor component (Fig. 1). In the first case, NMs are synthesized directly on the transducer surface by applying constant or variable voltage, and then the enzymes are adsorbed on NMs or immobilized in any other way.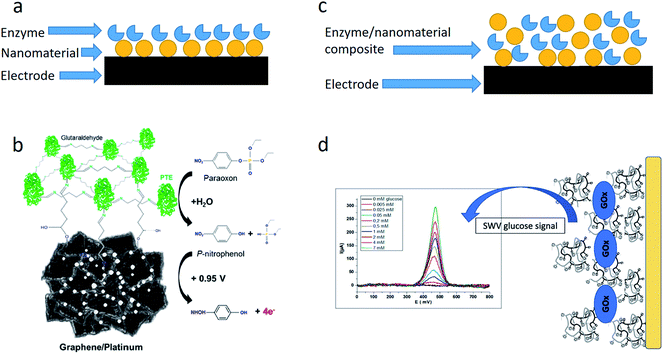 | ||
| Fig. 1 Ways of embedding NMs in the enzyme-based biosensors. (a) Enzyme immobilization on the NM-modified electrode. (b) Schematic of the biosensor based on phosphotriesterase (PTE) immobilized via glutaraldehyde on the graphene surface with platinum nanoparticles. Reprinted with permission from J. A. Hondred, J. C. Breger, N. J. Alves, S. A. Trammell, S. A. Walper, I. L. Medintz and J. C. Claussen, Printed Graphene Electrochemical Biosensors Fabricated by Inkjet Maskless Lithography for Rapid and Sensitive Detection of Organophosphates, ACS Applied Materials & Interfaces, 2018, 10, 11125–11134. Copyright 2018 American Chemical Society.25 (c) Enzyme/NM co-immobilization on the electrode. (d) Schematic of the biosensor based on glucose oxidase encapsulated in a chitosan-kappa-carrageenan bionanocomposite. Reprinted from Material Science and Engineering: C, 95, I. Rassas, M. Braiek, A. Bonhomme, F. Bessueille, G. Rafin, H. Majdoub, and N. Jaffrezic-Renault, Voltammetric glucose biosensor based on glucose oxidase encapsulation in a chitosan-kappa-carrageenan polyelectrolyte complex, 152–159, Copyright (2018), with permission from Elsevier.26 | ||
Enzymes can be easily immobilized directly onto the nanoparticles, since enzymes have many functional groups such as carboxylic (–COOH), amino (–NH2), thiol (–SH), etc. NMs with hydrophobic or charged sites on their surface, which can interact with enzymes, or NMs with chemical groups, which are able to bind to the corresponding enzyme groups, can be the enzymes adsorbents. In the first place, inorganic mesoporous materials are attractive adsorbents since they have a large surface area of the crystal and can carry various chemical groups, such as zeolites and mesoporous silicon spheres.27
Alternatively, NMs are synthesized separately and mixed with the enzyme solution prior to immobilization. This approach can be used for incorporation of NMs, which do not adsorb the enzymes. However, many NMs (especially organic ones) do not dissolve in aqueous solutions at all or easily aggregate in them, thus an addition of surfactants or other stabilizers to the resulting NM/enzyme mixture is often required. The auxiliary compounds should be carefully selected, since they can worsen the enzyme activity.
It is possible to combine both options of NMs incorporation, first immobilizing the enzyme on NM and then attaching the enzyme/NM composite to the electrode.
The application of NMs in biosensors results in:
- Enhancement of the transfer of electrons, which are formed or used in the enzymatic reaction, between the transducer surface and the enzyme;
- An increase of the sensor sensitive surface and thus enables immobilization of a larger amount of enzyme molecules;
- Improvement of enzyme stability;
- Catalysis of additional chemical reactions.28
3. Inorganic nanomaterials in enzyme-based biosensors
The most widespread inorganic NMs are nanosized particles of metals and metal oxides (TiO2, Al2O3, Fe2O3, ZrO2, MoO3, and CeO2), quantum dots and zeolites. In biosensors, metal nanoparticles are commonly used due to their unique physical and chemical properties.29–31There are two groups of methods of simple production of inorganic NMs – physical (fragmentation of the initial material to nanoscaled particles) and chemical (synthesis from precursors). The synthesized nanoparticles usually are more uniform compared to fragmented ones.
The advantages of inorganic NMs are simple production, the possibility of various surface modifications, catalysis of chemical reactions, acceleration of the electron transfer, biocompatibility, and improvement of conditions of enzyme immobilization. It makes NMs promising in the development of electrochemical enzyme-based biosensors.17 Some examples of the application of inorganic NMs are given in Table 2.
| Sensitive element | Analyte | Method of detection | LOD | Real sample | Ref. |
|---|---|---|---|---|---|
| a AuE – gold electrode; CA – chronoamperometry; CNT – carbon nanotubes; CV – cyclic voltammetry; GCE – glassy carbon electrode; ITO – indium tin oxide; N/D – no data; NP – nanoparticle; NT – nanotube; SPE – screen-printed electrode; SWV – square wave voltammetry. | |||||
| Acetylcholine esterase/ZnO/SPE | Paraoxon | CA | 0.035 ppm | N/D | 32 |
| Acetylcholine esterase/Fe3O4NP/CNT/ITO | Malathion, chlorpyrifos, monocrotophos, endosulfan | CV | 0.1 nM | Cabbage, onions, spinach, soil | 33 |
| Cholesterol esterase/cholesterol oxidase/quantum dots CdS/chitosan | Cholesterol and cholesterol esters | CV | 0.47 mM | N/D | 34 |
| Glucose oxidase/CNT/PtNP/GCE | Glucose | CA | 6 μM | N/D | 35 |
| Glucose oxidase/PdNP/CNT | Glucose | CV | 150 μM | N/D | 36 |
| Glutamate oxidase/CNT/AuNP/AuE | Glutamate | CA | 1.6 μM | Human blood serum | 37 |
| Horseradish peroxidase/TiO2NT | Hydrogen peroxide | CA | 0.1 μM | N/D | 38 |
| Horseradish peroxidase/TiO2NT | Hydrogen peroxide | Detection of photocurrent | 0.7 nM | N/D | 39 |
| Laccase/AuNP/AuE | Formetanate | SWV | 0.095 μM | Mango, grapes | 40 |
| Lactate oxidase/AgNP | Lactate | CA | 1 mM | N/D | 41 |
| Tyrosinase/graphene/PtNP/GCE | Chlorpyrifos, profenofos, malathion | CA | 0.2 ppb, 0.8 ppb, 3 ppb | N/D | 42 |
| Tyrosinase/quantum dots CdS/chitosan | Phenol compounds | CA | 0.3 nM | Water | 43 |
3.1. Gold nanoparticles
Gold nanoparticles (AuNPs) are widely utilized in enzyme-based biosensors. AuNPs are highly conductive and biocompatible, and their distinctive feature is the formation of strong thiol bonds between organic substances (i.e., cysteine residues of enzymes) and nanoparticles.28 Thus, the nanoparticles form a suitable microenvironment for the enzyme immobilization. The Enzyme activity can be significantly enhanced by immobilization onto AuNPs.44The researchers proposed a biosensor for the determination of organophosphorus pesticide formetanate, using laccase immobilized onto a gold electrode, modified with electrochemically deposited AuNPs.40 Detection was based on inhibition of the laccase activity by the pesticide. The biosensor appeared to be highly sensitive to the pesticide (LOD – 95 nM), and was successfully used to identify this pesticide in fruits.
A conductometric biosensor was developed for the hydrogen peroxide determination based on horseradish peroxidase immobilized in a chitosan film with and without 11 nm AuNPs.45 Interestingly, the addition of nanoparticles resulted in a decrease of the biosensor response, which, due to the authors, occurred because of the difference in the oxidative states of AuNPs and the active center of horseradish peroxidase.
A potentiometric biosensor for the determination of pesticide glyphosate was proposed; it was based on urease immobilized with AuNPs 2.54 nm in diameter.46 Glyphosate inhibited urease, which led to a decrease in the response of ammonium-sensitive ion-selective electrode. The linear range of the glyphosate detection was 0.5–50 ppm (3–300 μM), LOD – 0.5 ppm (3 μM).
The biosensor, recently proposed for determination of the protein kinase A activity, was based on horseradish peroxidase, antibodies (IgG) and AuNPs.47 The use of nanoparticles reduced the working potential to 0.08 V.
The change in the diameter of AuNPs due to their aggregation or growth during synthesis causes the variation in optical properties of their suspension (an adsorption of light with a wavelength from 400 to 700 nm increases accompanied by a change in color). This phenomenon can be used to visualize a certain reaction. In particular, a biosensor for the colorimetric determination of acetylcholinesterase inhibitors was proposed.48 In the absence of inhibitors, the enzyme catalyzed the cleavage of acetylthiocholine to acetic acid and thiocholine. The latter reduced AuCl4−, which led to an increase in the AuNPs diameter and the change in the suspension color from a weakly pink to purple. In the presence of acetylcholinesterase inhibitors in the sample, neither the growth of 2–3 nm AuNPs nor the corresponding change of solution color was observed.
Other platinum nanostructures can be also used in enzyme-based biosensors for effective H2O2 detection. For example, fractal nanoplatinum structures were synthesized by four different methods and used for the glucose oxidase (GOx) immobilization.49 The developed amperometric biosensor demonstrated very high sensitivity to hydrogen peroxide (3335 ± 305 μA cm2 mM−1), but sensitivity to glucose was notably lower (155 ± 25 μA cm2 mM−1). The biosensor response to glucose was rapid (2.0 ± 0.6 s). According to the authors, in general the analytical characteristics were not worse or even exceeded those of other glucose biosensors based on GOx and NMs.
3.2. Platinum nanoparticles
Platinum nanoparticles (PtNPs) differ from nanoparticles of other metals by the ability to catalyze decomposition of hydrogen peroxide (H2O2), a common product of oxidative reactions. An oxidation or reduction of hydrogen peroxide results in the formation or adsorption of electrons, which can be registered by amperometric methods. Basically, hydrogen peroxide decomposes spontaneously, but the catalyst significantly accelerates this reaction and increases the biosensor response. Though conventional platinum electrodes have the same catalytic activity, usage of platinum nanoparticles considerably increases the surface area and number of catalytic sites. The electrode modification with platinum nanoparticles can be carried out by chemical reduction, electrochemical and photochemical deposition of platinum compounds (usually PtCl6−). The choice of method for the nanoparticles synthesis defines their chemical inertness, electrical resistance, background current, and catalytic properties.50–52Numerous amperometric biosensors contain oxidases and platinum NMs. For example, the biosensors for the determination of glucose based on GOx and diamond microfibers were compared with the analogues which differed in the addition of PtNPs; the latter demonstrated a significantly higher sensitivity.53
An amperometric biosensor based on GOx, co-immobilized with PtNP/SnS2 composite is described in.54 Due to the authors, the nanocomposite improved the direct electron transfer between the enzyme and the surface of the glassy carbon electrode, and allowed the potential −0.4 V vs. saturated calomel electrode to be used. The biosensor had two linear ranges of glucose determination (0.1–1.0 mM and 1.0–12 mM), LOD was 2.5 μM.
Although PtNPs are useful in the first place for amperometry, PtNPs were integrated in the potentiometric biosensor for the sulfites determination based on sulfite oxidase, immobilized in the polypyrrole film with PtNPs (Fig. 2).55 The biosensor was very sensitive to the substrate; the LOD was 12.4 nM, the linear range 0.75–65 μM, the response time 3–5 s.
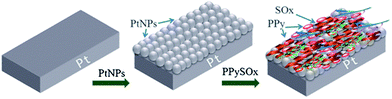 | ||
| Fig. 2 Preparation of the potentiometric sulfite biosensor: modification of the working electrode surface with PtNPs followed by the immobilization of sulfite oxidase (SOx) in the polypyrrole film. Reprinted by permission from: Springer-Verlag Wien, Microchimica Acta, (Potentiometric sulfite biosensor based on entrapment of sulfite oxidase in a polypyrrole film on a platinum electrode modified with platinum nanoparticles, S. B. Adeloju and S. Hussain), © (2016).55 | ||
3.3. Silver nanoparticles
The benefits of using silver nanoparticles (AgNPs) in biosensors are similar to those of other metal nanoparticles – enhancement of the matrix conductivity and amplification of the electrochemical signal.An amperometric biosensor developed for the urea determination was based on urease immobilized in a composite matrix containing polyaniline, polyvinyl acetate and AgNPs stabilized in polyvinyl alcohol.56 For comparison, the characteristics of the biosensor signal were studied without AgNPs. It was found that addition of AgNPs changed the shape of cyclic voltammograms – the cathodic peak (caused by urea addition) became more pronounced, which resulted in more accurate determination of urea concentration.
An amperometric biosensor with a flexible electrode was developed using lactate oxidase and AgNPs.41 The biosensor was intended for lactate determination in sweat. Linear range of detection was 1–25 mM, which coincided with the normal concentrations of lactate in human sweat. Noteworthy, the biosensor operated at a sufficiently high working potential (0.65 V vs. Ag/AgCl reference electrode), and the effect of anionic interfering substances was eliminated by the deposition of an additional negatively charged Nafion® membrane.
On the other hand, when developing the amperometric laccase-based biosensor, it was shown that an addition of AgNPs in high concentrations resulted in a 2390-fold decrease of the sensitivity.57 Such negative experience, though rarely reported, indicates that the amount of nanoparticles should be optimized.
3.4. Palladium nanoparticles
Palladium nanoparticles (PdNPs) have attracted more and more attention not only due to their conductive properties and catalytic activity, but also because of lower cost compared to AgNPs or PtNPs.13 Additionally, PdNPs can catalyze the reactions involving hydrogen peroxide (but probably at a slower rate than PtNPs).It has been recently shown that a glassy carbon electrode, modified with 9 nm PdNPs, effectively reduces hydrogen peroxide at a potential of −0.12 V, which may be useful in the development of amperometric biosensors based on oxidases.58 The limit of hydrogen peroxide detection was 0.34 μM.
The effects of nanoparticles of various metals (gold, platinum, rhodium and palladium) on the performance of superoxide dismutase-based biosensor for determination of aluminum ions were compared.59 The highest sensitivity was observed with PdNPs, LOD was 2 μM.
PdNPs are often used in combination with other NMs. For example, the glucose biosensor in ref. 60 was based on GOx, PdNPs, and multi-walled carbon nanotubes (MWCNTs). The biosensor could operate in the working potential range from −0.2 to −0.5 V. Furthermore, PdNPs improved the biosensor stability. The biosensor was used for glucose determination in honey and blood serum. In a similar biosensor PdNPs were used along with graphene and chitosan.61 Additionally, the sensor based on a reduced graphene oxide (rGO), tert-nonyl mercaptan (TNM), and PdNPs was proposed for hydrogen peroxide determination at the potential from −0.6 to +0.8 V (Fig. 3).62 Linear range of H2O2 detection was up to 12 mM and LOD was 2.5 μM.
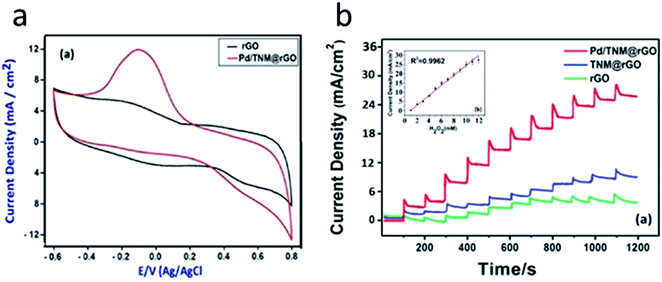 | ||
| Fig. 3 Operation of the amperometric sensor for hydrogen peroxide detection. (a) Cyclic voltammograms obtained with rGO electrode and PdNP/TNM/rGO in 0.1 M PBS, with 5 mM H2O2. (b) Amperometric responses of rGO, TNM/rGO, and Pd/TNM/rGO sensor to 1–12 mM H2O2 in 0.1 M PBS at pH 7.4 at applied potential of −0.1 V. Inset: corresponding calibration plot of Pd/TNM/rGO sensor. Reprinted from Analytica Chimica Acta, 989, S. Bozkurt, B. Tosun, B. Sen, S. Akocak, A. Savk, M. F. Ebeoğlugil and F. Sen, A hydrogen peroxide sensor based on TNM functionalized reduced graphene oxide grafted with highly monodisperse Pd nanoparticles, 88–94, Copyright (2017), with permission from Elsevier.62 | ||
3.5. Iron oxide nanoparticles
The iron oxide (Fe3O4) NPs catalyze the oxidation of hydrogen peroxide and thus can improve the response of oxidase-based biosensors.63 Although the catalytic activity of the iron oxide nanoparticles is lower than that of PtNPs and PdNPs, reagents for the Fe3O4 NPs synthesis are much cheaper. Furthermore, these NPs have magnetic properties and can be attached to the surface of the electrode using external magnetic field. Recently Fe3O4 NPs were used for creation of biosensors based on GOx64–66 and acetylcholine esterase/choline oxidase bienzyme system.67 Enhancement of electron transfer between the enzyme and electrode and improvement of the biosensor sensitivity were observed. In another work, urease was trapped in chitosan–Fe3O4 nanocomposite for the creation of urea biosensor.68 Authors stated that the composite material increased active surface area of the biosensor. Ultra-wide linear range (0.1 mM to 80 mM) of the urea biosensor based on urease/Fe3O4/chitosan was reported.69 Glucose biosensor based on GOx immobilized on Fe3O4 NPs/chitosan/graphene electrode also had wide linear range (up to 16 mM) with LOD of 16 μM.703.6. Nanostructured titanium dioxide
Titanium nanotubes have large active surface due to huge number of nanopores and internal cavity. This, along with chemical stability, non-toxicity and charge transfer capability, makes TiO2 nanotubes a promising material for the enzyme immobilization in electrochemical biosensors.38 TiO2 films are also perspective sensor components.71For example, a composite material based on titanium nanotubes and polyaniline was used to immobilize GOx.72 The biosensor was characterized by LOD of 0.5 μM and a dynamic range of glucose determination 10–2500 μM. However, the characteristics do not notably differ from those of similar glucose biosensors constructed without NMs.
The photoelectrochemical biosensor for the determination of hydrogen peroxide contained horseradish peroxidase and an array of TiO2 nanotubes covered with a polydopamine layer.39 The biosensor was highly sensitive and selective, LOD was 0.7 nM, the detection range – 1 nM–50 μM.
3.7. Zeolites and other nanosized aluminosilicates
Several types of nanosized aluminosilicates are known, in particular, zeolites, mesoporous silicon spheres, etc.73,74 Zeolites, in turn, are divided into Beta, A, Y, LTA, silicalites, and other types (Fig. 4). The basic property of these nanoparticles is a highly ordered structure with a complex pore and canal system, based on a crystalline lattice of aluminum, silicon, and oxygen atoms (sometimes of silicon and oxygen only). This significantly increases the crystal surface area for the enzyme absorption.75 Although aluminosilicates are inherently good adsorbents, they can be modified to further improve the enzyme immobilization.76 Commonly, surface modification consists in the attachment of functional molecules containing carboxyl groups, amino groups, etc. The modification enhances the electrostatic bonds between the nanoparticle and the enzyme and allows for the formation of strong covalent bonds, for example, between carboxyl groups on the nanoparticle surface and amino groups of the enzyme. Additionally, different variants of zeolites with integrated metal ions can be used in electrochemical biosensors as the charge carriers.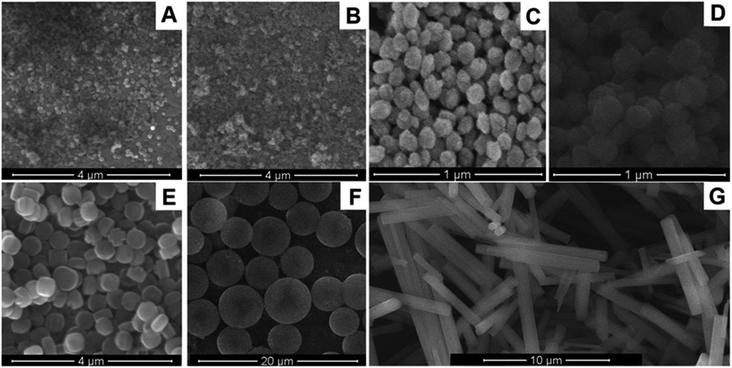 | ||
| Fig. 4 Morphology of nanosized aluminosilicates: nanozeolite beta (A), nanozeolite L (B), 80 nm silicalite-1 (C), 160 nm silicalite-1 (D), 450 nm silicalite-1 (E), mesoporous silica spheres (F), zeolite L (G). Reproduced from ref. 77 Copyright 2015 Springer. | ||
At present, various types of zeolites were integrated in amperometric, potentiometric, and conductometric biosensors together with the enzymes GOx,78–80 urease,81–84 acetylcholinesterase,77 butyrylcholinesterase,85 creatinine deiminase,86 glutamate oxidase,87etc. Zeolites in biosensors simplify the immobilization procedure and allow avoiding usage of toxic reagents, e.g., glutaraldehyde. Additionally, better reproducibility of the biosensor preparation was shown. In some works, a 20–30% increase in the sensitivity of zeolite-based biosensors was observed.
3.8. Inorganic quantum dots
Quantum dots are semiconductor nanoscale crystals with unique optical properties.88 They can consist of inorganic materials as well as of carbon or graphene. In the first place, quantum dots are used as fluorophores for generating light, so their application in electrochemical enzyme biosensors is rather unreasonable. Nevertheless, in some cases they were used.A photoelectrochemical biosensor was proposed, which was based on glucose dehydrogenase immobilized on the electrode covered with quantum dots (ZnS and CdS) and multi-walled carbon nanotubes.89 The biosensor was suitable for glucose determination in ampero- and photometric (at light irradiation) modes; the NMs, interacting with NADH formed in enzymatic reaction, generated the biosensor response. LOD was lower in case of the photoamperometric measurements than in usual amperometry (4 μM and 6 μM, respectively).
An amperometric biosensor for the determination of phenolic compounds contained tyrosinase, immobilized in a composite matrix of quantum dots (CdS) and chitosan.43 The biosensor had a wide linear range of catechol detection (from 1 nM to 100 μM) and low LOD (0.3 nM). According to the authors, it was due to the use of this highly porous, hydrophilic and biocompatible nanocomposite for the enzyme immobilization.
An amperometric biosensor proposed for the determination of cholesterol and its esters was based on the bi-enzyme system cholesterol esterase/cholesterol oxidase immobilized into a composite matrix of quantum dots (CdS) and chitosan.34 The authors believe that quantum dots mediated the electron transfer between the enzyme active center and the electrode. They also assumed an oriented enzyme immobilization near the quantum dot surface, which accelerated the charge transfer between the quantum dots and the enzymes as well as the participation of quantum dots in the electron transfer.90
The photoelectrochemical biosensor based on antibodies and horseradish peroxidase was proposed for the determination of carcinoembryonic antigen.91 In the presence of the antigen, peroxidase catalyzed a multistage reaction near the electrode surface and the formation of CdS quantum dots on the surface of graphene oxide. When exposed to light, the dots generated a photocurrent, which was detected as the biosensor signal. This biosensor had a wide linear detection range from 2.5 ng mL−1 to 50 μg mL−1 with LOD 0.72 ng mL−1.
3.9. Doped inorganic NMs
Properties of NMs can be tuned by doping.92 Doping is a flexible way of NMs modification without significant change of their morphology, which leads to changes in electrical, optical, catalytic, and other properties.93 Doped inorganic NMs can be used as sensor components separately or in combination with biomaterial. For example, recently described thiourea sensor is based on cobalt oxide codoped manganese oxide nanoparticles.94 Ethanol95 and acetone96 sensors based on ternary-doped metal oxides are characterized by the extremely low LOD of 0.127 nM and 0.05 nM, respectively. Melamine sensor based on cadmium doped antimony oxide nanostructures had ultralow LOD (14 pM) as well.97 Ultra-sensitive photoelectrochemical glucose biosensor has been constructed from the nitrogen-doped carbon sheets and titanium dioxide nanoparticles.98 LOD was 13 nM, and linear range was shifted to low concentrations of glucose (0.05–10 μM). Nitrogen-doped zinc oxide thin films were used for construction of uric acid biosensor with a wide linear range (0.05–1.0 mM).993.10. Nanowires
Advances in nanotechnology make it possible to synthesize single nanowires and utilize them as sensitive elements of biosensors. In particular, nanowire field-effect transistors (FETs) attract significant attention.100 As common FETs, they contain a gate, channel, source, drain, and the body, but the channel consists of a nanowire instead of a macrostructure. Silicon and silica nanowires are most widely used due to their high compatibility with the standard fabrication technology.100 The main advantage of the nanowire-based FETs is high sensitivity – properties of nanowire (i.e., conductance) are easily changed by processes that take place on its surface, thus biosensors can generate signal even when very small amounts of target molecules are present in the sample. Another advantages are low cost and possibility of multiplexed sensing using several FET on a single platform.101 On the other hand, problem of high background noise arises.102 Thus, special signal processing algorithms should be often used to split noise and useful signal.103 Antibodies are often used as bioreceptors in such nanowire-based biosensors,104 but several enzymes were also tested. For example, GOx was immobilized on the surface of FET based on In2O3 nanoribbons.105 Low LOD of 10 nM glucose was achieved, but storage stability was rather poor – more than 50% of sensitivity was lost after 2 weeks.3.11. Nanorods
Nanorods are objects with length about 3–5 times larger than width. They are typically synthesized by direct chemical synthesis, which is not so convenient for biosensor applications as direct electrochemical synthesis on the transducer surface. Properties of nanorods are close to those of nanoparticles, and thus nanorods are applied in electrochemical biosensors mostly to improve electron transport and electrode surface area, but it should be noted that ZnO nanorods have semiconductor properties. Most commonly used nanorods are based on ZnO and gold.106 Few electrochemical biosensors based on enzymes and nanorods were recently described. For example, galactose oxidase was immobilized on ZnO nanorods for the creation of galactose biosensor.107 An interesting feature of the biosensor was linear range shifted towards high concentration of the target molecule (10–200 mM). The biosensor was selective against electroactive substances and did not change sensitivity during 4 weeks. In another work, ZnO nanorod arrays were used for GOx immobilization.108 However, analytical characteristics of the biosensor (sensitivity, selectivity, linear range) were not significantly different from similar amperometric biosensors without nanorods.4. Use of organic nanomaterials in enzyme-based biosensors
Organic NMs represented by CNTs,109 graphene,110 fullerenes,111 calixarenes,112 organic quantum dots, etc., are widely used in the development of electrochemical biosensors due to their properties, enhancing electrochemical signal (Table 3). Additionally, organic NMs often have high biocompatibility, although it is generally worse than that of inorganic NMs.6 The molecules with aromatic groups can be non-covalently bonded to the surface of graphene and CNTs via strong π–π interactions. The hydrophobic molecules can also interact with the hydrophobic surface of organic NMs. However, this also may cause fouling, which is a significant problem for biological applications of graphene- or CNT-based biosensors.| Sensitive element | Analyte | Method of detection | Limit of detection | Sample | Ref. |
|---|---|---|---|---|---|
| a SWV – square wave voltammetry; CA – chronoamperometry. | |||||
| GOx/CNT/mucin | Glucose | CA | 3 μM | Human blood plasma | 113 |
| Horseradish peroxidase/CNT | Hydrogen peroxide | CV | 22 nM | — | 114 |
| Urease/MWCNT | Urea | CA | 30 μM | Blood plasma | 115 |
| Lysine oxidase/MWCNT/SnO2/graphene/chitosan | Lysine | CA | 0.15 μM | Dietary supplements | 116 |
| Laccase/graphene quantum dots | Epinephrine | CV | 83 nM | Medicinal drug | 117 |
| Tyrosinase/graphene oxide | Phenolic compounds | CA | 30 nM | Tap water | 118 |
| Soybean peroxidase/reduced graphene oxide | Hydrogen peroxide | CA | 50 nM | Solution for contact lenses | 119 |
| GOx/reduced graphene oxide/Fe3O4 | Glucose | SWV, CA | — | — | 120 |
| Urease/fullerene C60/poly(n-butyl acrylate) | Urea | Potentiometry | 42 μM | Urea | 121 |
4.1. Carbon nanotubes
CNT is a graphene sheet rolled up into a tube, which is characterized by high electrical conductivity, mechanical strength, and large surface area. The CNTs compatibility with other NMs and electrode materials is also important for electrochemical biosensors.13 In the first place, conductive properties of CNTs are significant, because they enhance the enzyme-electrode charge transfer and, accordingly, sensitivity and response time of the biosensor. CNTs are classified as single- and multi-walled (SWCNT and MWCNT, respectively), depending on the number of graphene sheets rolled up into a tube. In biosensors, multi-walled CNTs are mostly used.However, freshly synthesized CNTs have poor solubility in water, thus, it is difficult to bring them together with the aqueous solutions of enzymes. To improve solubility and biocompatibility, CNTs are functionalized with carboxylic or amino groups, or used in combination with other materials such as polymers or nanoparticles of metals.2 The functionalized CNTs do not lose their initial high electrical conductivity and mechanical stability but can form suspensions in aqueous solutions and interact with enzymes via functional groups. This enables the development of CNT-based highly sensitive electrochemical biosensors.
CNTs are one of the most common NMs used in electrochemical enzyme-based biosensors. Numerous amperometric biosensors based on GOx and CNTs were described. For example, there is a biosensor, in which GOx was immobilized by cross-linking with glutaraldehyde, mucin and CNTs. It was shown that without CNTs, the biosensor was less sensitive and the analysis time was longer.113 In,122 MWCNTs modified with amino groups were applied when immobilizing GOx by cross-linking with glutaraldehyde; it resulted in a wider linear range of glucose determination and a higher biosensor sensitivity. In,123 the biosensor based on GOx and polyaniline-modified MWCNTs was proposed; in ref. 124 GOx, SWCNTs and thermally expanded graphite were used.
A biosensor based on horseradish peroxidase and CNTs was characterized by the linear range of determination of hydrogen peroxide in low concentrations – from 0.1 μM to 120 μM.114 The enzyme was modified with 4-aminothiophenol and immobilized by electropolymerization in CNT/polyaniline matrix (Fig. 5). LOD was 22 nM. Such biosensor can be adapted for highly sensitive detection of other target molecules by incorporation of the H2O2-producing enzymes.
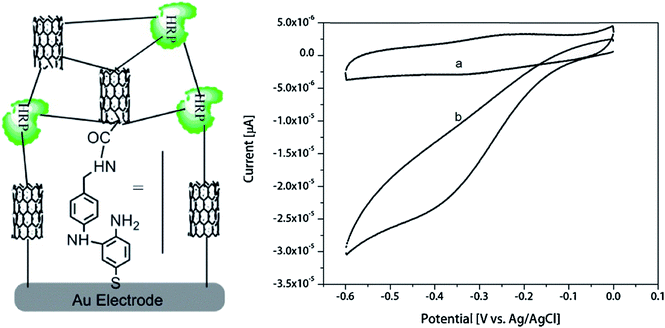 | ||
| Fig. 5 Schematics of the hydrogen peroxide biosensor based on oligoaniline-cross-linked HRP/CNT composite and cyclic voltammograms of the biosensor obtained in the absence of H2O2 (a) and in the presence of 5 μM H2O2 (b). Reprinted from Enzyme and Microbial Technology, 113, K. M. Kafi, M. Naqshabandi, M. M. Yusoff and M. J. Crossley, Improved peroxide biosensor based on Horseradish Peroxidase/Carbon Nanotube on a thiol-modified gold electrode, 67–74, Copyright (2017), with permission from Elsevier.114 | ||
Other enzymes were also used together with CNTs in biosensors. The urea biosensor contained MWCNTs, conducting polymer poly(o-toluidine) and urease.115 It had a wide range of urea determination – from 0.1 to 11 mM, which almost completely covered the possible urea concentrations in blood samples, but LOD was rather high (30 μM). CNTs were also used to create the biosensors based on laccase,125 peroxidase from zucchini (Cucurbita pepo),126 lysine oxidase,116 and others.
4.2. Carbon quantum dots
Carbon quantum dots, or simply carbon dots, are nanosized carbon structures.127 They are usually a few nanometers in size, chemically inert, suitable for various surface modifications, biocompatible, and cheap. Their greatest advantage is photoluminescent properties (not important for electrochemical sensing, though). Numerous optical (primarily fluorescent) biosensors with carbon dots have been described, however they are beyond the scope of this review.128–130 We found only one electrochemical enzyme-based biosensor with carbon quantum dots. It was a horseradish peroxidase-based amperometric biosensor for hydrogen peroxide determination, which used carbon dots (2.8 ± 0.5 nm) deposited on the surface of glassy carbon electrode.131 The dots had amino acids on their surface and served as a matrix for the enzyme immobilization. The biosensor was highly sensitive, with LOD of 1.8 nM and a linear range of 5–590 nM.Furthermore, the sensor based on carbon dots modified by magnetite particles was able to detect NADH in the range of 0.2–5 μM.132 Thus, these nanoparticles may be useful in electrochemical biosensors based on enzymes, which produce or use NADH.
4.3. Graphene quantum dots
Graphene quantum dots are nanosized graphene sheets similar to other quantum dots by their properties (biocompatibility, low toxicity, photostability, excellent fluorescence, high surface area). They are rarely used in conventional electrochemical biosensors, however an amperometric biosensor was developed for the determination of epinephrine in pharmaceuticals based on laccase immobilized with graphene quantum dots on the surface of a glassy carbon electrode.117 The biosensor was characterized by a linear range of 1–120 μM and a LOD of 83 nM. High selectivity was demonstrated, no impact of interfering substances (ascorbic and uric acids, cysteine, glutathione, and tryptophan) on the biosensor response was revealed. In another work laccase was immobilized on graphene quantum dots and molybdenum disulphide (MoS2) nanoflakes for the creation of the amperometric biosensor for polyphenol detection.133 Synergetic effect between the two NMs was found, and nanocomposite matrix was favorable for the enzyme immobilization. Graphene quantum dots were used for GOx adsorption in glucose biosensors.134,135 Direct electron transfer was observed, and the biosensors had better analytical characteristics than similar biosensors without quantum dots. Porosity of the NM and presence of hydrophilic and hydrophobic sited favored the GOx adsorption.4.4. Graphene and its derivatives
Graphene is a one-atom thick sheet of carbon atoms arranged in a honeycomb-like structure. It is characterized by large surface area, excellent electrical and thermal conductivity, high mechanical stiffness.136 Most of these parameters significantly exceed those of CNTs.137Due to the large surface area of graphene, it is an effective substrate for the enzyme immobilization. The direct electron transfer between the enzyme and electrode is also enhanced because of high conductivity of graphene.138 However, like CNTs, graphene is practically insoluble in water and many other solvents. To improve the hydrophilicity, additional nitrogen atoms are introduced into graphene structure, or graphene oxide (GO) is used, which relatively easily forms stable dispersions in an aqueous medium.139 On the other hand, additional groups notably weaken conducting properties of graphene; therefore, GO is further reduced to rGO to decrease the number of oxygen-containing groups and achieve a balance between electric conductivity and solubility.
Graphene electrodes can be created either by direct printing of graphene ink or by printing a sacrificial polymer pattern followed by the graphene deposition.140,141 On the other hand, graphene electrodes can be produced from carbon-based polymers (i.e., polyimide) by CO2 laser irradiation.142 In both cases the electrodes of relatively complex pattern (e.g., interdigitated) can be easily obtained in large quantities suitable for the creation of amperometric, impedimetric or other types of biosensors. Although the specialized materials printer as well as CO2 laser cost tens of thousands of USD, they assure the manufacture of hundreds of uniform electrodes per hour. Furthermore, raw materials for the process are cheap, and the electrode design can be modified quickly and conveniently in computer aided design software. Thus, graphene electrodes are promising for mass production and have clear advantages over the electrodes produced by traditional methods and subsequently modified with graphene. However, such electrodes are rarely utilized in enzyme-based biosensors; recent examples include diamine oxidase-based biosensor for the detection of biogenic amines (e.g., histamine),143 GOx-based glucose biosensor,144 and phosphotriesterase-based biosensor for the detection of organophosphate pesticides.25
Still, although the graphene electrode itself is porous, its surface area can be further increased by creation of the conductive porous graphene/polymer composites as well as other porous graphene structures.145
An amperometric biosensor based on tyrosinase and GO for the determination of phenolic compounds is described in.118 First, the glassy carbon electrode was modified with GO; afterwards glutaraldehyde was linked to free hydroxyl groups. Another end of glutaraldehyde formed covalent bonds with tyrosinase. A linear range of catechol determination was 50 nM to 50 μM, LOD – 30 nM.
A biosensor based on acetylcholinesterase and a field-effect transistor modified with reduced GO was proposed.146 It was used to determine the acetylcholinesterase activity and the concentration of the enzyme inhibitors donepezil and rivastigmine (drugs).
A biosensor based on soybean peroxidase and rGO was proposed for determination of hydrogen peroxide.119 Usage of the NM made it possible to decrease the working potential to −0.09 V vs. Ag/AgCl reference electrode and to obtain LOD of 50 nM. The presence of interfering substances (ascorbic and uric acids, dopamine) did not affect the results of hydrogen peroxide determination.
In most works, single-enzyme biosensors are described. However, co-immobilization of several enzymes is also possible. For example, a bienzyme biosensor for the triglyceride and glycerol determination was proposed.147 Lipase and glycerol dehydrogenase were co-immobilized on the surface of rGO modified with a mediator toluidine blue (Fig. 6). Lipase split triglycerides to glycerol and fatty acids, and then glycerol dehydrogenase oxidized glycerol with subsequent reduction of the mediator. The mediator was oxidized on the electrode surface. The biosensor demonstrated high selectivity, rapid response (12 s) and retained 90% of the initial sensitivity after 11 week. The triglyceride content in human blood serum was successfully analyzed.
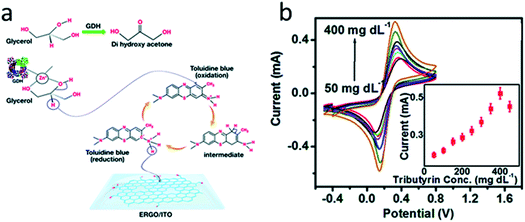 | ||
| Fig. 6 (a) Chemical reactions that are the basis for the glycerol biosensor operation. Glycerol dehydrogenase (GDH), toluidine blue and electrochemically reduced graphene oxide (ERGO) are deposited on indium-tin oxide (ITO) electrode. Second enzyme (lipase) is not shown. (b) Response studies of the biosensor to varying concentrations of triglyceride (tributyrin). Inset: corresponding calibration curve. Reproduced from ref. 147 with permission from The Royal Society of Chemistry. | ||
A nanocomposite consisting of rGO and 50 nm Fe3O4 nanoparticles was used for GOx adsorption in glucose biosensor.120 The authors showed that the application of the nanocomposite allowed 4-fold improvement of conductivity of the electrode surface. The linear range of glucose determination was 0.5–10 mM. At a working potential −0.4 V vs. Ag/AgCl reference electrode the biosensor was not sensitive to dopamine, ascorbic and uric acids.
4.5. Fullerenes
Fullerenes are closed spherical molecules with a pentagonal honeycomb structure made of carbon atoms. Due to high conductivity and large area of the hydrophobic surface on which the organic molecules could be adsorbed, fullerenes are promising for use in electrochemical biosensors as adsorbents and mediators to enhance the transport of electrons between an enzyme and an electrode.148,149 Fullerenes are designated by the number of carbon atoms in the molecule, for example, C60 – fullerene composed of 60 carbon atoms. The most commonly used fullerenes are C60 and C70.Fullerenes are not as common in electrochemical enzyme biosensors as CNTs and graphene derivatives, despite their high similarity. Fullerenes C60 and C70 were used as carriers for ascorbate oxidase in the biosensor system for the determination of ascorbic acid and phenols.150 It turned out that much more enzyme molecules are immobilized on the surface of fullerenes than on SWCNTs and MWCNTs. Furthermore, fullerene C60 was used as a urease carrier in the potentiometric biosensor for the urea determination.121 The fullerene-urease conjugate was captured in a polymeric membrane, which provided excellent stability of immobilized urease, so the biosensor responses decreased by 5% after 140 days of dry storage at +4 °C.
The examples of other fullerene biosensors are the laccase-based biosensor for the determination of polyphenols in wine,149 the glutathione reductase-based biosensor for the glutathione determination,151 and a number of GOx-based biosensors for glucose determination in real samples.152,153
Furthermore, a glucose biosensor based on glucose dehydrogenase and AuNPs covered with C70 fullerene was described.154 This biosensor utilized multistep electron transfer: first the enzyme oxidized glucose and reduced NAD+, then NADH was reduced on the surface of the fullerene, the latter transferred electrons to AuNPs and then to the electrode (Fig. 7). The biosensor operated at relatively high working potential (+0.7 V), thus the negatively charged Nafion® membrane was placed over the enzyme membrane to repel interfering anions (i.e., ascorbic and uric acids). The biosensor appeared to be highly selective.
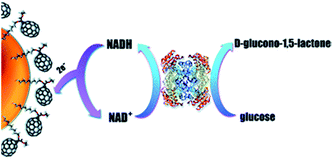 | ||
| Fig. 7 Working principle of the glucose biosensor based on glucose dehydrogenase, fullerene C70, and AuNPs. Electrons from glucose are transferred to NAD+, fullerene, AuNPs, and finally to the working electrode. Reproduced from ref. 154 – published by The Royal Society of Chemistry. | ||
Catalytic properties of fullerenes are poorly investigated. Nevertheless, it was shown that fullerene C60 catalyzes oxidation of nandrolone (steroid hormone),155 bisphenol A,156 dopamine and ascorbic acid.157
4.6. Calixarenes
Calixarenes are the cup-shaped chemical compounds consisting of several cyclic phenolic oligomers. Different variants of calixarene molecules can be synthesized and usually used as selective binding agents in sensors and chromatography columns. However, sometimes calixarenes and their derivatives are used as matrices for the enzyme immobilization.158,159Effective GOx immobilization on a calixarene/AuNPs modified electrode was achieved.112 Conductive polymer poly(2-(2-octyldodecyl)-4,7-di(selenoph-2-yl)-2H-benzo[d][1,2,3]triazole) was deposited on the electrode to improve conductance, then calixarene/AuNP suspension was drop-casted and calixarene formed thiol bonds with AuNPs (Fig. 8). Finally, GOx was covalently attached to the calixarene by N-(3-dimethylaminopropyl)-N-ethylcarbodiimide hydrochloride (EDC)/N-hydroxysuccinimide (NHS) crosslinking agents. The biosensor operated at the working potential of −0.7 V vs. Ag wire and was successfully used to determine glucose in beverages.
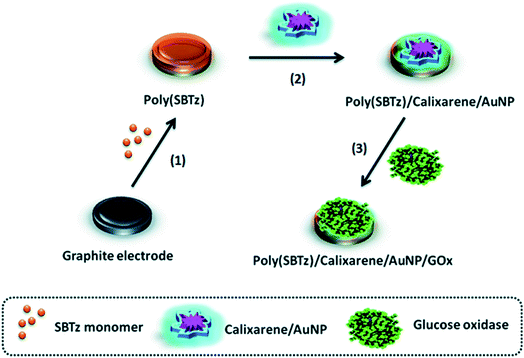 | ||
| Fig. 8 Preparation of the glucose biosensor based on conductive polymer poly(2-(2-octyldodecyl)-4,7-di(selenoph-2-yl)-2H-benzo[d][1,2,3]triazole) (poly[SBTz]), calixarene, AuNPs and GOx. Graphite electrode is modified with conductive polymer, calixarene, and AuNPs followed by GOx immobilization. Reproduced from ref. 112 with permission from The Royal Society of Chemistry. | ||
A series of biosensors using calixarenes were developed for glucose determination in beverages;160 calixarenes can be also promising in the conductometric enzyme biosensors for ammonia detection.161
4.7. Conducting polymers
Conducting (or conductive) polymers are large organic molecules that are electrically conductive. Examples of conducting polymers include poly(acetylene), poly(thiophene), poly(pyrrole), and poly(aniline).162 They are widely used in electrochemical biosensors to enhance electron transfer between the enzyme and the electrode and to improve conditions of the enzyme immobilization.163 Thus, conducting polymers are usually used in amperometric biosensors based on oxidoreductase enzymes, where charge transfer plays especially important role. Generally, biosensors based on conducting polymers are characterized by higher sensitivity than similar biosensors without polymers due to greatly increased electron transfer between the enzyme active center and the electrode surface.164 Another reason for popularity of the conducting polymers in biosensors is the ease of their synthesis, which is usually done electrochemically directly on the electrode surface. Entrapment of enzymes can occur during synthesis, which simplifies the biosensor preparation. Examples of biosensors based on conducting polymers include glucose biosensor based on GOx,165–167 H2O2 biosensor based on horseradish peroxidase,165 urea biosensor based on urease,168 and cholesterol biosensor based on cholesterol oxidase.1695. Application of composite nanomaterials in electrochemical biosensors
The combination of several types of NMs in a single biosensor is gaining popularity. Composite nanoparticles containing several metal oxides can be prepared.170,171 Furthermore, metal-based NMs can be combined with CNTs,172,173 graphene, rGO,174 polymers,175etc., and used to immobilize enzymes. This can lead to a synergistic effect of NMs on the characteristics of biosensors. In more detail, an application of hybrid composite materials in biosensors was reviewed in.176Most often GOx is used in combination with several NMs. For example, a glucose biosensor contained GOx immobilized on a gold electrode modified with rGO and platinum–gold nanoparticles.177 A LOD (1 μM) was not outstanding, but the linear range was very wide (0.01–8 mM). The biosensor showed good storage stability – after one month its sensitivity decreased by only 18%. The working potential was relatively high (+0.6 V) despite the application of NMs.
In another work GOx was immobilized along with CNTs and PtNPs; the hydrogen peroxide reduction was observed at the working potential −0.1 V.35 The biosensor was characterized by a very fast response (2 s), a LOD of 6 μM, and a linear range of 50 μM to 5 mM. In the similar work, glutamate oxidase was immobilized together with carbon nanotubes and AuNPs; a low potential (+0.135 V) was used for hydrogen peroxide oxidation.37 The biosensor had a fast response (2 s), a LOD of 1.6 μM, and a linear range of 5–500 μM.
A glucose biosensor was proposed based on GOx and nanocomposite consisting of nanosheets of rGO and AgNPs.178 The nanocomposite had high biocompatibility and conductivity, and the biosensor was characterized by excellent analytical parameters – a LOD of 25 nM, a high sensitivity of 15.32 mA M−1 cm−2 and a linear range from 150 nM to 10 mM.
Another glucose biosensor was based on field-effect transistor (FET) and GOx, immobilized with graphene, modified with flower-shaped Pd nanostructures (Fig. 9).179 The biosensor had relatively complex structure: graphene was located in the bottom layer and functioned as a working electrode, then Pd nanoflowers were electrodeposited and patterned with gold, next Nafion®/rGO membrane was deposited by spin coating, and finally GOx/GO dispersion was drop-casted. Thus, GOx was entrapped in a matrix formed by GO and rGO. The biosensor had a low LOD (1 nM) and was insensitive to common interferents (uric and ascorbic acids).
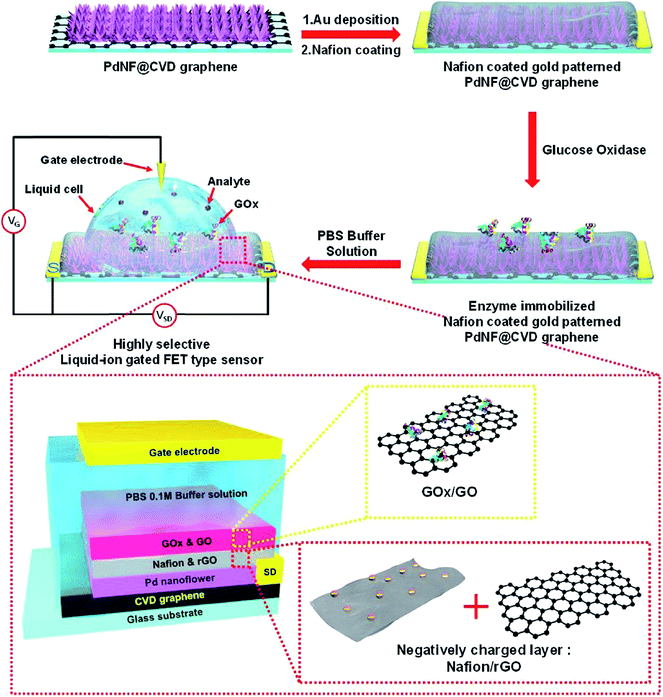 | ||
| Fig. 9 Schematic diagram of the fabrication of FET glucose biosensor (top) and structure of the biosensor sensitive element (bottom). For the biosensor fabrication, Pd nanoflowers were electrodeposited and patterned with gold, then Nafion®/rGO membrane was deposited by spin coating, and finally GOx/GO dispersion was drop-casted. Reprinted from Sensors and Actuators B: Chemical, 264, D. H. Shin, W. Kim, J. Jun, J. S. Lee, J. H. Kim and J. Jang, Highly selective FET-type glucose sensor based on shape-controlled palladium nanoflower-decorated graphene, 216–223, Copyright (2018), with permission from Elsevier.179 | ||
A biosensor for catechol determination has been recently described which was based on tyrosinase immobilized together with MWCNTs and gold nanowires.180 Due to the synergetic effect of the two nanostructures, it was possible to achieve a low LOD (0.027 μM).
An ultrasensitive biosensor for tyrosine determination was based on tyrosine hydroxylase immobilized in a composite material consisting of chitosan, nanoparticles of platinum/palladium alloy, graphene and MWCNTs.181 The biosensor was highly selective and had a very low LOD (9 pM).
In another work,182 a printed carbon electrode was modified by a composite membrane containing CNTs, nanosized zirconium oxide, Prussian blue, and a polymer Nafion® membrane. Additionally, acetylcholinesterase was immobilized on the surface of magnetic nanoparticles (Fe3O4/Au) through covalent bonds Au–S. Under the action of an external magnetic field, the nanoparticles with acetylcholinesterase came into a contact with the modified electrode to form a biosensor. After analysis, the magnetic field was switched off, the nanoparticles with the enzyme were washed out, and the modified electrode was reused to create new biosensors. In such a way, a high sensitivity to the organophosphorus pesticide dimethoate was achieved. LOD was 5.6 × 10−4 ng mL−1, the linear range 1.0 × 10−3 to 10 ng mL−1.
6. Problems associated with the application of nanomaterials in enzyme-based biosensors
From the above data it can be concluded that the application of NMs in enzyme-based biosensors is very attractive for the researchers and this field is rapidly expanding without any problems. However, a few negative tendencies in the NMs application should be noted.One issue is that instead of a comprehensive evaluation of benefits of one certain NM the researchers often make a quick study and move to another kind of NMs and their combinations. This is partially explained by the fact that it is much more difficult to publish a second article describing the biosensor with the same NM, since the work will be considered less novel. As a result of such rush for novelty, a large number of papers about enzyme/NM-based biosensors are published each month, but it is very difficult to analyze them since each research group tries to use a unique NM only to distinguish their work from others. Indeed, almost any newly described biosensor contains one or several NMs, and a combination of NMs is one of the easiest ways to stand out from dozens of similar publications. Such approach often allows publication of the manuscript even if no clear advantage over biosensors without NMs is presented.
Furthermore, some authors report unbelievably good results (i.e., extremely low LOD of a biosensor) attributing them to the influence of NMs. This allows researchers to claim that they have created the most sensitive or otherwise great biosensor and thus get published in the top-rated journals. In our opinion, it would be quite difficult, if even possible, to reproduce such results. Although we cannot estimate the exact amount of papers with partially or completely fake results, we consider that the amount is sufficiently high to raise a concern about the repeatability of experiments, especially in case of reporting LOD below 10 nM, or extremely good stability over several months. Meanwhile, the researchers remain relatively safe, since nobody will repeat their work because replication studies are usually not funded and cannot be published due to the lack of novelty.183 Furthermore, even if somebody replicates a work (unsuccessfully), it is easy to explain the different results by improper synthesis of NMs, variance in methodology, materials, equipment, etc. Unfortunately, such action makes it difficult to other researchers to publish real, but worse results. The problem of replication is common for different research areas, and hopefully some solutions will be found in future.184
7. Conclusions
The development of enzyme-based electrochemical biosensors based on nanomaterials is an important direction of biosensor research and it will remain an important field in the nearest future. Today, the modified biosensors are proposed for many fields, including the food industry, clinical diagnosis, biomedicine, environmental monitoring. The main effects of NMs application in electrochemical enzyme-based biosensors can be divided into three groups: (1) enhancement of the charge transfer between the enzyme and the electrode (up to direct electron transfer); (2) improvement of conditions of enzyme immobilization and stability; (3) catalysis of electrochemical reactions. Although there is a large variety of NMs that differ in chemical structure, properties, and morphology, positive effects of NMs in biosensors are caused mostly by the three factors: large surface area-to-volume ratio, high conductance, and excellent biocompatibility. In few cases (PtNPs and PdNPs), catalytic properties of NMs are also important. Application of NMs is also favored by their simple synthesis, which is often performed electrochemically directly on the electrode surface.Wide perspectives are opening for biosensors based on nanocomposites and newly discovered nanostructures. Utilization of nanomaterials with specifically tailored properties (i.e., designed receptor cites) is also growing. Application of NMs is expected to improve selectivity, sensitivity, storage stability, and other analytical characteristics of electrochemical biosensors. Such improvements will make biosensors more robust and increase their practical application.
8. Glossary
| AgNP | Silver nanoparticle |
| AuE | Gold electrode |
| AuNP | Gold nanoparticle |
| CA | Chronoamperometry |
| CNT | Carbon nanotube |
| CV | Cyclic voltammetry |
| FET | Field-effect transistor |
| GCE | Glassy carbon electrode |
| GO | Graphene oxide |
| GOx | Glucose oxidase |
| ITO | Indium tin oxide |
| LOD | Iimit of detection |
| MWCNT | Multi-walled carbon nanotube |
| NAD+ | Oxidized nicotinamide adenine dinucleotide |
| NADH | Reduced nicotinamide adenine dinucleotide |
| NM | Nanomaterial |
| NP | Nanoparticle |
| NT | Nanotube |
| PdNP | Palladium nanoparticle |
| PtNP | Platinum nanoparticle |
| rGO | Reduced graphene oxide |
| SPE | Screen-printed electrode |
| SWV | Square wave voltammetry |
| SWCNT | Single-walled carbon nanotube |
| TNM | tert-Nonyl mercaptan |
| QDs | Quantum dots |
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the State Budget Program “Support for the Development of Priority Areas of Scientific Research” (Code: 6541230) through a program of National Academy of Sciences of Ukraine “Smart sensor devices of a new generation based on modern materials and technologies”.References
- D. R. Thevenot, K. Tóth, R. A. Durst and G. S. Wilson, Pure Appl. Chem., 1999, 71, 2333–2348 CAS.
- Y. Zeng, Z. Zhu, D. Du and Y. Lin, J. Electroanal. Chem., 2016, 781, 147–154 CrossRef CAS.
- W. Jin and G. Maduraiveeran, J. Anal. Sci. Technol., 2018, 9, 18 CrossRef.
- M. N. Arshad, T. A. Sheikh, M. M. Rahman, A. M. Asiri, H. M. Marwani and M. R. Awual, J. Organomet. Chem., 2017, 827, 49–55 CrossRef CAS.
- M. M. Hussain, M. M. Rahman, M. N. Arshad and A. M. Asiri, ACS Omega, 2017, 2, 420–431 CrossRef PubMed.
- S. Kurbanoglu, S. A. Ozkan and A. Merkoçi, Biosens. Bioelectron., 2017, 89, 886–898 CrossRef CAS PubMed.
- M. M. Rahman and A. M. Asiria, RSC Adv., 2015, 5, 63252–63263 RSC.
- M. R. Awual, M. Khraisheh, N. H. Alharthi, M. Luqman, A. Islam, M. Rezaul Karim, M. M. Rahman and M. A. Khaleque, Chem. Eng. J., 2018, 343, 118–127 CrossRef CAS.
- M. R. Awual, N. H. Alharthi, Y. Okamoto, M. R. Karim, M. E. Halim, M. M. Hasan, M. M. Rahman, M. M. Islam, M. A. Khaleque and M. C. Sheikh, Chem. Eng. J., 2017, 320, 427–435 CrossRef CAS.
- M. R. Awual, M. M. Hasan, G. E. Eldesoky, M. A. Khaleque, M. M. Rahman and M. Naushad, Chem. Eng. J., 2016, 290, 243–251 CrossRef CAS.
- M. R. Awual, N. H. Alharthi, M. M. Hasan, M. R. Karim, A. Islam, H. Znad, M. A. Hossain, M. E. Halim, M. M. Rahman and M. A. Khaleque, Chem. Eng. J., 2017, 324, 130–139 CrossRef CAS.
- M. R. Awual, A. M. Asiri, M. M. Rahman and N. H. Alharthi, Chem. Eng. J., 2019, 363, 64–72 CrossRef CAS.
- G. Maduraiveeran and W. Jin, Trends Environ. Anal. Chem., 2017, 13, 10–23 CrossRef CAS.
- M. M. Alam, M. A. Rashed, M. M. Rahman, M. M. Rahman, Y. Nagao and M. A. Hasnat, RSC Adv., 2018, 8, 8071–8079 RSC.
- M. M. Hussain, A. M. Asiri, M. N. Arshad and M. M. Rahman, New J. Chem., 2018, 42, 1169–1180 RSC.
- S. Nawaz, S. Khan, U. Farooq, M. S. Haider, N. M. Ranjha, A. Rasul, A. Nawaz, N. Arshad and R. Hameed, Des. Monomers Polym., 2018, 21, 18–32 CrossRef CAS.
- H. Li and D. Xu, TrAC, Trends Anal. Chem., 2014, 61, 67–73 CrossRef CAS.
- Y. Wang, K. Qu, L. Tang, Z. Li, E. Moore, X. Zeng, Y. Liu and J. Li, TrAC, Trends Anal. Chem., 2014, 58, 54–70 CrossRef CAS.
- V. V. Pokropivny and V. V. Skorokhod, Mater. Sci. Eng., C, 2007, 27, 990–993 CrossRef CAS.
- M. M. Rahman, Sens. Actuators, B, 2018, 264, 84–91 CrossRef CAS.
- A. M. Asiri, M. M. Hussain, M. N. Arshad and M. M. Rahman, New J. Chem., 2018, 42, 4465–4473 RSC.
- M. M. Rahman, M. M. Alam, A. M. Asiri and M. R. Awual, New J. Chem., 2017, 41, 9159–9169 RSC.
- A. Umar, M. M. Rahman, S. H. Kim and Y.-B. Hahn, Chem. Commun., 2008, 166–168 RSC.
- A. A. P. Khan, A. Khan, M. M. Rahman, A. M. Asiri and M. Oves, Int. J. Biol. Macromol., 2017, 98, 256–267 CrossRef CAS PubMed.
- J. A. Hondred, J. C. Breger, N. J. Alves, S. A. Trammell, S. A. Walper, I. L. Medintz and J. C. Claussen, ACS Appl. Mater. Interfaces, 2018, 10, 11125–11134 CrossRef CAS PubMed.
- I. Rassas, M. Braiek, A. Bonhomme, F. Bessueille, G. Rafin, H. Majdoub and N. Jaffrezic-Renault, Mater. Sci. Eng., C, 2019, 95, 152–159 CrossRef CAS PubMed.
- M. Badea, A. Curulli and G. Palleschi, Biosens. Bioelectron., 2003, 18, 689–698 CrossRef CAS PubMed.
- K. Kerman, M. Saito, E. Tamiya, S. Yamamura and Y. Takamura, TrAC, Trends Anal. Chem., 2008, 27, 585–592 CrossRef CAS.
- N. Li, P. Zhao and D. Astruc, Angew. Chem., Int. Ed., 2014, 53, 1756–1789 CrossRef CAS PubMed.
- R. G. Weiner and S. E. Skrabalak, Angew. Chem., Int. Ed., 2015, 54, 1181–1184 CrossRef CAS PubMed.
- K. D. Gilroy, A. Ruditskiy, H.-C. Peng, D. Qin and Y. Xia, Chem. Rev., 2016, 116, 10414–10472 CrossRef CAS PubMed.
- R. Sinha, M. Ganesana, S. Andreescu and L. Stanciu, Anal. Chim. Acta, 2010, 661, 195–199 CrossRef CAS PubMed.
- N. Chauhan and C. S. Pundir, Electrochim. Acta, 2012, 67, 79–86 CrossRef CAS.
- H. Dhyani, M. A. Ali, S. P. Pal, S. Srivastava, P. R. Solanki, B. D. Malhotra and P. Sen, RSC Adv., 2015, 5, 45928–45934 RSC.
- H. N. Choi, J. H. Han, J. A. Park, J. M. Lee and W.-Y. Lee, Electroanalysis, 2007, 19, 1757–1763 CrossRef CAS.
- S. H. Lim, J. Wei, J. Lin, Q. Li and J. KuaYou, Biosens. Bioelectron., 2005, 20, 2341–2346 CrossRef CAS PubMed.
- B. Batra and C. S. Pundir, Biosens. Bioelectron., 2013, 47, 496–501 CrossRef CAS PubMed.
- F. Wu, J. Xu, Y. Tian, Z. Hu, L. Wang, Y. Xian and L. Jin, Biosens. Bioelectron., 2008, 24, 198–203 CrossRef CAS PubMed.
- J. Li, X. Li, Q. Zhao, Z. Jiang, M. Tadé, S. Wang and S. Liu, Sens. Actuators, B, 2018, 255, 133–139 CrossRef CAS.
- F. W. P. Ribeiro, M. F. Barroso, S. Morais, S. Viswanathan, P. de Lima-Neto, A. N. Correia, M. B. P. P. Oliveira and C. Delerue-Matos, Bioelectrochemistry, 2014, 95, 7–14 CrossRef CAS PubMed.
- M. A. Abrar, Y. Dong, P. K. Lee and W. S. Kim, Sci. Rep., 2016, 6, 30565 CrossRef PubMed.
- T. Liu, M. Xu, H. Yin, S. Ai, X. Qu and S. Zong, Microchim. Acta, 2011, 175, 129–135 CrossRef CAS.
- E. Han, Y. Yang, Z. He, J. Cai, X. Zhang and X. Dong, Anal. Biochem., 2015, 486, 102–106 CrossRef CAS PubMed.
- J. A. Hondred, J. C. Breger, N. T. Garland, E. Oh, K. Susumu, S. A. Walper, I. L. Medintz and J. C. Claussen, Analyst, 2017, 142, 3261–3271 RSC.
- G. A. Valencia, L. C. de Oliveira Vercik and A. Vercik, J. Polym. Eng., 2014, 34, 633–638 CAS.
- C. Vaghela, M. Kulkarni, S. Haram, R. Aiyer and M. Karve, Int. J. Biol. Macromol., 2018, 108, 32–40 CrossRef CAS PubMed.
- Y. Zhou, M. Wang, H. Yin and S. Ai, Microchim. Acta, 2017, 184, 3301–3308 CrossRef CAS.
- V. Pavlov, Y. Xiao and I. Willner, Nano Lett., 2005, 5, 649–653 CrossRef CAS PubMed.
- M. Taguchi, N. Schwalb, Y. Rong, D. C. Vanegas, N. Garland, M. Tan, H. Yamaguchi, J. C. Claussen and E. S. McLamore, Analyst, 2016, 141, 3367–3378 RSC.
- E. Lebègue, C. M. Anderson, J. E. Dick, L. J. Webb and A. J. Bard, Langmuir, 2015, 31, 11734–11739 CrossRef PubMed.
- Y. Li, C. Sella, F. Lemaître, M. Guille Collignon, L. Thouin and C. Amatore, Electroanalysis, 2013, 25, 895–902 CrossRef CAS.
- B. Rismetov, T. A. Ivandini, E. Saepudin and Y. Einaga, Diamond Relat. Mater., 2014, 48, 88–95 CrossRef CAS.
- H. Olivia, B. V. Sarada, K. Honda and A. Fujishima, Electrochim. Acta, 2004, 49, 2069–2076 CrossRef CAS.
- L. Wang, J. Li, Y. Pan, L. Min, Y. Zhang, X. Hu and Z. Yang, Microchim. Acta, 2017, 184, 2357–2363 CrossRef CAS.
- S. B. Adeloju and S. Hussain, Microchim. Acta, 2016, 183, 1341–1350 CrossRef CAS.
- F. N. Crespilho, R. M. Iost, S. A. Travain, O. N. Oliveira and V. Zucolotto, Biosens. Bioelectron., 2009, 24, 3073–3077 CrossRef CAS PubMed.
- T. Kavetskyy, O. Smutok, M. Gonchar, O. Demkiv, H. Klepach, Y. Kukhazh, O. Šauša, T. Petkova, V. Boev, V. Ilcheva, P. Petkov and A. L. Stepanov, J. Appl. Polym. Sci., 2017, 134, 45278 CrossRef.
- J. Wang, X. Chen, K. Liao, G. Wang and M. Han, Nanoscale Res. Lett., 2015, 10, 311 CrossRef PubMed.
- M. Barquero-Quirós and M. Arcos-Martínez, Sensors, 2016, 16, 1588 CrossRef PubMed.
- V. Guzsvány, J. Anojčić, E. Radulović, O. Vajdle, I. Stanković, D. Madarász, Z. Kónya and K. Kalcher, Microchim. Acta, 2017, 184, 1987–1996 CrossRef.
- Q. Zeng, J.-S. Cheng, X.-F. Liu, H.-T. Bai and J.-H. Jiang, Biosens. Bioelectron., 2011, 26, 3456–3463 CrossRef CAS PubMed.
- S. Bozkurt, B. Tosun, B. Sen, S. Akocak, A. Savk, M. F. Ebeoğlugil and F. Sen, Anal. Chim. Acta, 2017, 989, 88–94 CrossRef CAS PubMed.
- L. Gao, K. Fan and X. Yan, Theranostics, 2017, 7, 3207–3227 CrossRef CAS PubMed.
- N. Sanaeifar, M. Rabiee, M. Abdolrahim, M. Tahriri, D. Vashaee and L. Tayebi, Anal. Biochem., 2017, 519, 19–26 CrossRef CAS PubMed.
- C. He, M. Xie, F. Hong, X. Chai, H. Mi, X. Zhou, L. Fan, Q. Zhang, T. Ngai and J. Liu, Electrochim. Acta, 2016, 222, 1709–1715 CrossRef CAS.
- N. Mohamad Nor, K. Abdul Razak and Z. Lockman, Electrochim. Acta, 2017, 248, 160–168 CrossRef CAS.
- E. Ö. Bolat, G. A. Tığ and Ş. Pekyardımcı, J. Electroanal. Chem., 2017, 785, 241–248 CrossRef CAS.
- M. W. Akram, M. F. Alam, H. N. Ji, A. Mahmood, T. Munir, M. Z. Iqbal, M. R. Saleem, N. Amin and A. G. Wu, IOP Conf. Ser.: Mater. Sci. Eng., 2019, 474, 012060 Search PubMed.
- A. Ali, M. S. AlSalhi, M. Atif, A. A. Ansari, M. Q. Israr, J. R. Sadaf, E. Ahmed, O. Nur and M. Willander, J. Phys.: Conf. Ser., 2013, 414, 012024 CrossRef.
- W. Zhang, X. Li, R. Zou, H. Wu, H. Shi, S. Yu and Y. Liu, Sci. Rep., 2015, 5, 11129 CrossRef PubMed.
- M. M. Rahman, V. G. Alfonso, F. Fabregat-Santiago, J. Bisquert, A. M. Asiri, A. A. Alshehri and H. A. Albar, Microchim. Acta, 2017, 184, 2123–2129 CrossRef CAS.
- J. Zhu, X. Liu, X. Wang, X. Huo and R. Yan, Sens. Actuators, B, 2015, 221, 450–457 CrossRef CAS.
- W.-H. Chen, S.-J. Huang, H.-H. Ko, A.-Y. Lo, H.-K. Lee, L.-L. Wu, C.-F. Cheng and S.-B. Liu, Stud. Surf. Sci. Cat., 2005, 156, 657–662 CrossRef CAS.
- S. Mintova, J.-P. Gilson and V. Valtchev, Nanoscale, 2013, 5, 6693 RSC.
- M. G. Valdés, A. I. Pérez-Cordoves and M. E. Díaz-García, TrAC, Trends Anal. Chem., 2006, 25, 24–30 CrossRef.
- C. Ispas, I. Sokolov and S. Andreescu, Anal. Bioanal. Chem., 2009, 393, 543–554 CrossRef CAS PubMed.
- I. Kucherenko, O. Soldatkin, B. O. Kasap, S. K. Kirdeciler, B. A. Kurc, N. Jaffrezic-Renault, A. Soldatkin, F. Lagarde and S. Dzyadevych, Nanoscale Res. Lett., 2015, 10, 209 CrossRef PubMed.
- R. Nenkova, J. Wu, Y. Zhang and T. Godjevargova, Anal. Biochem., 2013, 439, 65–72 CrossRef CAS PubMed.
- O. O. Soldatkin, B. Ozansoy Kasap, B. Akata Kurc, A. P. Soldatkin, S. V. Dzyadevych and A. V. El'skaya, Biopolym. Cell, 2014, 30, 291–298 Search PubMed.
- V. N. Pyeshkova, O. Y. Dudchenko, O. O. Soldatkin, I. S. Kucherenko, B. Ozansoy Kasap, B. Akata Kurc and S. V. Dzyadevych, Biopolym. Cell, 2014, 30, 462–468 Search PubMed.
- M. Hamlaoui, K. Reybier, M. Marrakchi, N. Jaffrezic-Renault, C. Martelet, R. Kherrat and A. Walcarius, Anal. Chim. Acta, 2002, 466, 39–45 CrossRef CAS.
- M. K. Shelyakina, O. O. Soldatkin, V. M. Arkhypova, B. O. Kasap, B. Akata and S. V. Dzyadevych, Nanoscale Res. Lett., 2014, 9, 124 CrossRef PubMed.
- O. O. Soldatkin, I. S. Kucherenko, S. V. Marchenko, B. Ozansoy Kasap, B. Akata, A. P. Soldatkin and S. V. Dzyadevych, Mater. Sci. Eng., C, 2014, 42, 155–160 CrossRef CAS PubMed.
- I. S. Kucherenko, O. O. Soldatkin, B. O. Kasap, S. Öztürk, B. Akata, A. P. Soldatkin and S. V. Dzyadevych, Electroanalysis, 2012, 24, 1380–1385 CrossRef CAS.
- E. Soy, V. Arkhypova, O. Soldatkin, M. Shelyakina, S. Dzyadevych, J. Warzywoda, A. Sacco and B. Akata, Mater. Sci. Eng., C, 2012, 32, 1835–1842 CrossRef CAS.
- B. Ozansoy Kasap, S. V. Marchenko, O. O. Soldatkin, S. V. Dzyadevych and B. Akata Kurc, Nanoscale Res. Lett., 2017, 12, 162 CrossRef PubMed.
- O. V. Soldatkina, O. O. Soldatkin, B. O. Kasap, D. Y. Kucherenko, I. S. Kucherenko, B. A. Kurc and S. V. Dzyadevych, Nanoscale Res. Lett., 2017, 12, 260 CrossRef CAS PubMed.
- L. Cui, C. Li, B. Tang and C. Zhang, Analyst, 2018, 143, 2469–2478 RSC.
- B. Ertek and Y. Dilgin, Bioelectrochemistry, 2016, 112, 138–144 CrossRef CAS PubMed.
- G. Zhiguo, Y. Shuping, L. Zaijun, S. Xiulan, W. Guangli, F. Yinjun and L. Junkang, Electrochim. Acta, 2011, 56, 9162–9167 CrossRef.
- X. Zeng, W. Tu, J. Li, J. Bao and Z. Dai, ACS Appl. Mater. Interfaces, 2014, 6, 16197–16203 CrossRef CAS PubMed.
- T. C. Bharat, Shubham, S. Mondal, H. S. Gupta, P. K. Singh and A. K. Das, Mater. Today: Proc., 2019, 11, 767–775 CAS.
- X. Chen, Y. Lou, S. Dayal, X. Qiu, R. Krolicki, C. Burda, C. Zhao and J. Becker, J. Nanosci. Nanotechnol., 2005, 5, 1408–1420 CrossRef CAS PubMed.
- M. M. Rahman, J. Ahmed and A. M. Asiri, Biosens. Bioelectron., 2018, 99, 586–592 CrossRef CAS PubMed.
- M. M. Rahman, M. M. Alam, A. M. Asiri and M. A. Islam, RSC Adv., 2017, 7, 22627–22639 RSC.
- M. M. Rahman, M. M. Alam, A. M. Asiri and M. A. Islam, Talanta, 2017, 170, 215–223 CrossRef CAS PubMed.
- M. M. Rahman and J. Ahmed, Biosens. Bioelectron., 2018, 102, 631–636 CrossRef CAS PubMed.
- R. Atchudan, N. Muthuchamy, T. N. J. I. Edison, S. Perumal, R. Vinodh, K. H. Park and Y. R. Lee, Biosens. Bioelectron., 2019, 126, 160–169 CrossRef CAS PubMed.
- K. Jindal, M. Tomar and V. Gupta, Analyst, 2013, 138, 4353 RSC.
- P. Ambhorkar, Z. Wang, H. Ko, S. Lee, K. Koo, K. Kim and D. Cho, Micromachines, 2018, 9, 679 CrossRef PubMed.
- A. Gao, X. Yang, J. Tong, L. Zhou, Y. Wang, J. Zhao, H. Mao and T. Li, Biosens. Bioelectron., 2017, 91, 482–488 CrossRef CAS PubMed.
- Y. Kutovyi, I. Zadorozhnyi, H. Hlukhova, V. Handziuk, M. Petrychuk, A. Ivanchuk and S. Vitusevich, Nanotechnology, 2018, 29, 175202 CrossRef CAS PubMed.
- Y. Kutovyi, I. Zadorozhnyi, V. Handziuk, H. Hlukhova, N. Boichuk, M. Petrychuk and S. Vitusevich, Phys. Status Solidi A, 2019, 256, 1800636 CrossRef.
- M.-A. Doucey and S. Carrara, Trends Biotechnol., 2019, 37, 86–99 CrossRef CAS PubMed.
- Q. Liu, Y. Liu, F. Wu, X. Cao, Z. Li, M. Alharbi, A. N. Abbas, M. R. Amer and C. Zhou, ACS Nano, 2018, 12, 1170–1178 CrossRef CAS PubMed.
- X. Huang, S. Neretina and M. A. El-Sayed, Adv. Mater., 2009, 21, 4880–4910 CrossRef CAS PubMed.
- K. Khun, Z. H. Ibupoto, O. Nur and M. Willander, J. Sens., 2012, 2012, 1–7 CrossRef.
- N. S. Ridhuan, K. Abdul Razak and Z. Lockman, Sci. Rep., 2018, 8, 13722 CrossRef PubMed.
- A. Battigelli, C. Ménard-Moyon, T. Da Ros, M. Prato and A. Bianco, Adv. Drug Delivery Rev., 2013, 65, 1899–1920 CrossRef CAS PubMed.
- Y. Shao, J. Wang, H. Wu, J. Liu, I. A. Aksay and Y. Lin, Electroanalysis, 2010, 22, 1027–1036 CrossRef CAS.
- S. Pilehvar and K. De Wael, Biosensors, 2015, 5, 712–735 CrossRef CAS PubMed.
- T. C. Gokoglan, S. Soylemez, M. Kesik, H. Unay, S. Sayin, H. B. Yildiz, A. Cirpan and L. Toppare, RSC Adv., 2015, 5, 35940–35947 RSC.
- F. N. Comba, M. R. Romero, F. S. Garay and A. M. Baruzzi, Anal. Biochem., 2018, 550, 34–40 CrossRef CAS PubMed.
- A. K. M. Kafi, M. Naqshabandi, M. M. Yusoff and M. J. Crossley, Enzyme Microb. Technol., 2018, 113, 67–74 CrossRef CAS PubMed.
- R. Y. A. Hassan, A. M. Kamel, M. S. Hashem, H. N. A. Hassan and M. A. Abd El-Ghaffar, J. Solid State Electrochem., 2018, 22, 1–7 CrossRef.
- C. Kaçar, P. E. Erden and E. Kılıç, Appl. Surf. Sci., 2017, 419, 916–923 CrossRef.
- S. Baluta, A. Lesiak and J. Cabaj, Electroanalysis, 2018, 30, 1773–1782 CrossRef CAS.
- Y. Wang, F. Zhai, Y. Hasebe, H. Jia and Z. Zhang, Bioelectrochemistry, 2018, 122, 174–182 CrossRef CAS PubMed.
- C. H. Díaz Nieto, A. M. Granero, J. C. Lopez, G. D. Pierini, G. J. Levin, H. Fernández and M. A. Zon, Sens. Actuators, B, 2018, 263, 377–386 CrossRef.
- Y. Wang, X. Liu, X. Xu, Y. Yang, L. Huang, Z. He, Y. Xu, J. Chen and Z. Feng, Mater. Res. Bull., 2018, 101, 340–346 CrossRef CAS.
- K. Saeedfar, L. Heng, T. Ling and M. Rezayi, Sensors, 2013, 13, 16851–16866 CrossRef PubMed.
- O. A. Biloivan, N. S. Rogaleva and Y. I. Korpan, Biopolym. Cell, 2010, 26, 56–61 CAS.
- S. Cogal, G. Celik Cogal and A. U. Oksuz, Int. J. Polym. Mater. Polym. Biomater., 2018, 67, 454–461 CrossRef CAS.
- V. A. Arlyapov, S. S. Kamanin, O. A. Kamanina and A. N. Reshetilov, Nanotechnol. Russ., 2017, 12, 658–666 CrossRef CAS.
- M. Romero-Arcos, M. G. Garnica-Romo and H. E. Martínez-Flores, Procedia Technology, 2017, 27, 279–281 CrossRef.
- S. R. Benjamin, R. S. Vilela, H. S. de Camargo, M. I. Guedes, F. Fernandes and F. Colmati, Int. J. Electrochem. Sci., 2018, 563–586 CrossRef CAS.
- Z. L. Wu, Z. X. Liu and Y. H. Yuan, J. Mater. Chem. B, 2017, 5, 3794–3809 RSC.
- X. Sun and Y. Lei, TrAC, Trends Anal. Chem., 2017, 89, 163–180 CrossRef CAS.
- Y. Choi, Y. Choi, O.-H. Kwon and B.-S. Kim, Chem.–Asian J., 2018, 13, 586–598 CrossRef CAS PubMed.
- H. Shi, J. Wei, L. Qiang, X. Chen and X. Meng, J. Biomed. Nanotechnol., 2014, 10, 2677–2699 CrossRef CAS PubMed.
- Y. Su, X. Zhou, Y. Long and W. Li, Microchim. Acta, 2018, 185, 114 CrossRef PubMed.
- T. C. Canevari, F. H. Cincotto, D. Gomes, R. Landers and H. E. Toma, Electroanalysis, 2017, 29, 1968–1975 CrossRef CAS.
- I. Vasilescu, S. A. V. Eremia, M. Kusko, A. Radoi, E. Vasile and G.-L. Radu, Biosens. Bioelectron., 2016, 75, 232–237 CrossRef CAS PubMed.
- S. Gupta, T. Smith, A. Banaszak and J. Boeckl, Nanomaterials, 2017, 7, 301 CrossRef PubMed.
- H. Razmi and R. Mohammad-Rezaei, Biosens. Bioelectron., 2013, 41, 498–504 CrossRef CAS PubMed.
- L. Tang, Y. Wang, Y. Li, H. Feng, J. Lu and J. Li, Adv. Funct. Mater., 2009, 19, 2782–2789 CrossRef CAS.
- S. Kumar, W. Ahlawat, R. Kumar and N. Dilbaghi, Biosens. Bioelectron., 2015, 70, 498–503 CrossRef CAS PubMed.
- S. Palanisamy, S. K. Ramaraj, S.-M. Chen, T. C. K. Yang, P. Yi-Fan, T.-W. Chen, V. Velusamy and S. Selvam, Sci. Rep., 2017, 7, 41214 CrossRef CAS PubMed.
- V. V. Neklyudov, N. R. Khafizov, I. A. Sedov and A. M. Dimiev, Phys. Chem. Chem. Phys., 2017, 19, 17000–17008 RSC.
- J. A. Hondred, L. R. Stromberg, C. L. Mosher and J. C. Claussen, ACS Nano, 2017, 11, 9836–9845 CrossRef CAS PubMed.
- A. Moya, G. Gabriel, R. Villa and F. Javier del Campo, Curr. Opin. Electrochem., 2017, 3, 29–39 CrossRef CAS.
- R. Ye, D. K. James and J. M. Tour, Acc. Chem. Res., 2018, 51, 1609–1620 CrossRef CAS PubMed.
- D. Vanegas, L. Patiño, C. Mendez, D. Oliveira, A. Torres, C. Gomes and E. McLamore, Biosensors, 2018, 8, 42 CrossRef PubMed.
- Z. Zhang, M. Song, J. Hao, K. Wu, C. Li and C. Hu, Carbon, 2018, 127, 287–296 CrossRef CAS.
- M. Wang, X. Duan, Y. Xu and X. Duan, ACS Nano, 2016, 10, 7231–7247 CrossRef CAS PubMed.
- M.-S. Chae, Y. K. Yoo, J. Kim, T. G. Kim and K. S. Hwang, Sens. Actuators, B, 2018, 272, 448–458 CrossRef CAS.
- S. K. Bhardwaj, R. Chauhan, P. Yadav, S. Ghosh, A. K. Mahapatro, J. Singh and T. Basu, Biomater. Sci., 2019, 7, 1598–1606 RSC.
- V. Perumal and U. Hashim, J. Appl. Biomed., 2014, 12, 1–15 CrossRef.
- C. Lanzellotto, G. Favero, M. L. Antonelli, C. Tortolini, S. Cannistraro, E. Coppari and F. Mazzei, Biosens. Bioelectron., 2014, 55, 430–437 CrossRef CAS PubMed.
- A. Barberis, Y. Spissu, A. Fadda, E. Azara, G. Bazzu, S. Marceddu, A. Angioni, D. Sanna, M. Schirra and P. A. Serra, Biosens. Bioelectron., 2015, 67, 214–223 CrossRef CAS PubMed.
- M. Carano, S. Cosnier, K. Kordatos, M. Marcaccio, M. Margotti, F. Paolucci, M. Prato and S. Roffia, J. Mater. Chem., 2002, 12, 1996–2000 RSC.
- L.-H. Lin and J.-S. Shih, J. Chin. Chem. Soc., 2011, 58, 228–235 CrossRef CAS.
- W. Zhilei, L. Zaijun, S. Xiulan, F. Yinjun and L. Junkang, Biosens. Bioelectron., 2010, 25, 1434–1438 CrossRef PubMed.
- P. Piotrowski, K. Jakubow, B. Kowalewska and A. Kaim, RSC Adv., 2017, 7, 45634–45640 RSC.
- R. N. Goyal, V. K. Gupta and N. Bachheti, Anal. Chim. Acta, 2007, 597, 82–89 CrossRef CAS PubMed.
- J. A. Rather and K. De Wael, Sens. Actuators, B, 2013, 176, 110–117 CrossRef CAS.
- R. N. N. Goyal, V. K. K. Gupta, N. Bachheti and R. A. A. Sharma, Electroanalysis, 2008, 20, 757–764 CrossRef CAS.
- A. Ikeda and S. Shinkai, Chem. Rev., 1997, 97, 1713–1734 CrossRef CAS PubMed.
- N. Morohashi, F. Narumi, N. Iki, T. Hattori and S. Miyano, Chem. Rev., 2006, 106, 5291–5316 CrossRef CAS PubMed.
- D. O. Demirkol, H. B. Yildiz, S. Sayın and M. Yilmaz, RSC Adv., 2014, 4, 19900–19907 RSC.
- O. Y. Saiapina, S. G. Kharchenko, S. G. Vishnevskii, V. M. Pyeshkova, V. I. Kalchenko and S. V. Dzyadevych, Nanoscale Res. Lett., 2016, 11, 105 CrossRef CAS PubMed.
- M. R. Karim, M. M. Alam, M. O. Aijaz, A. M. Asiri, M. A. Dar and M. M. Rahman, Talanta, 2019, 193, 64–69 CrossRef CAS PubMed.
- M. Gerard, Biosens. Bioelectron., 2002, 17, 345–359 CrossRef CAS PubMed.
- N. Aydemir, J. Malmström and J. Travas-Sejdic, Phys. Chem. Chem. Phys., 2016, 18, 8264–8277 RSC.
- B. Weng, A. Morrin, R. Shepherd, K. Crowley, A. J. Killard, P. C. Innis and G. G. Wallace, J. Mater. Chem. B, 2014, 2, 793–799 RSC.
- J. G. Ayenimo and S. B. Adeloju, Food Chem., 2017, 229, 127–135 CrossRef CAS PubMed.
- P. Krzyczmonik, E. Socha and S. Skrzypek, Bioelectrochemistry, 2015, 101, 8–13 CrossRef CAS PubMed.
- S. Ivanova, Y. Ivanov and T. Godjevargova, Open J. Appl. Biosens., 2013, 02, 12–19 CrossRef.
- E. Cevik, A. Cerit, N. Gazel and H. B. Yildiz, Electroanalysis, 2018, 30, 2445–2453 CrossRef CAS.
- J. Ahmed, M. M. Rahman, I. A. Siddiquey, A. M. Asiri and M. A. Hasnat, Electrochim. Acta, 2017, 246, 597–605 CrossRef CAS.
- M. M. Rahman, N. A. Alenazi, M. A. Hussein, M. M. Alam, K. A. Alamry and A. M. Asiri, Mater. Res. Express, 2018, 5, 065019 CrossRef.
- M. M. Hussain, M. M. Rahman and A. M. Asiri, J. Environ. Sci., 2017, 53, 27–38 CrossRef PubMed.
- M. M. Rahman, J. Ahmed and A. M. Asiri, RSC Adv., 2017, 7, 14649–14659 RSC.
- M. K. Alam, M. M. Rahman, M. Abbas, S. R. Torati, A. M. Asiri, D. Kim and C. Kim, J. Electroanal. Chem., 2017, 788, 66–73 CrossRef CAS.
- M. A. Hussein, M. M. Alam, N. A. Alenazi, K. A. Alamry, A. M. Asiri and M. M. Rahman, J. Polym. Res., 2018, 25, 262 CrossRef.
- M. A. Daniele, M. Pedrero, S. Burrs, P. Chaturvedi, W. W. A. Wan Salim, F. Kuralay, S. Campuzano, E. McLamore, A. A. Cargill, S. Ding and J. C. Claussen, in Nanobiosensors and Nanobioanalyses, Springer, Japan, Tokyo, 2015, pp. 137–166 Search PubMed.
- M. F. Hossain and J. Y. Park, PLoS One, 2017, 12, e0173553 CrossRef PubMed.
- S. Morteza Naghib, Int. J. Electrochem. Sci., 2018, 1013–1026 CrossRef.
- D. H. Shin, W. Kim, J. Jun, J. S. Lee, J. H. Kim and J. Jang, Sens. Actuators, B, 2018, 264, 216–223 CrossRef CAS.
- S. Kurbanoglu and S. A. Ozkan, Electrocatalysis, 2018, 9, 252–257 CrossRef CAS.
- K. Varmira, G. Mohammadi, M. Mahmoudi, R. Khodarahmi, K. Rashidi, M. Hedayati, H. C. Goicoechea and A. R. Jalalvand, Talanta, 2018, 183, 1–10 CrossRef CAS PubMed.
- N. Gan, X. Yang, D. Xie, Y. Wu and W. Wen, Sensors, 2010, 10, 625–638 CrossRef CAS PubMed.
- M. C. Makel and J. A. Plucker, Educ. Res., 2014, 43, 304–316 CrossRef.
- M. R. Munafò and G. Davey Smith, Nature, 2018, 553, 399–401 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2019 |