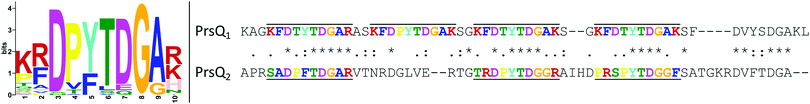Open Access Article
Open Access ArticleSpontaneous mutation in the AgrRS two-component regulatory system of Cupriavidus metallidurans results in enhanced silver resistance†
Kristel
Mijnendonckx
 *a,
Md Muntasir
Ali
ab,
Ann
Provoost
a,
Paul
Janssen
*a,
Md Muntasir
Ali
ab,
Ann
Provoost
a,
Paul
Janssen
 a,
Max
Mergeay
a,
Natalie
Leys
a,
Daniël
Charlier
b,
Pieter
Monsieurs‡
a and
Rob
Van Houdt
a,
Max
Mergeay
a,
Natalie
Leys
a,
Daniël
Charlier
b,
Pieter
Monsieurs‡
a and
Rob
Van Houdt
 *a
*a
aUnit of Microbiology, Belgian Nuclear Research Centre SCK·CEN, 2400 Mol, Belgium. E-mail: Kristel.mijnendonckx@sckcen.be; rvhoudto@sckcen.be
bResearch Group of Microbiology, Department of Bioengineering Sciences, Vrije Universiteit Brussel, Pleinlaan 2, B-1050 Brussel, Belgium
First published on 14th October 2019
Abstract
The uncontrolled and widespread use of (nano)silver compounds has led to the increased release of these compounds into the environment, raising concerns about their negative impact on ecosystems. Concomitantly, silver resistance determinants are widely spread among environmental and clinically relevant bacteria although the underlying mechanisms are not yet fully understood. We show that Cupriavidus metallidurans is able to adapt to toxic silver concentrations. However, none of the known silver resistance determinants present in C. metallidurans are involved in the adaptative response. Instead, increased silver resistance is achieved by the concerted action of a two-component system AgrR–AgrS, previously not associated with metal resistance, and two periplasmic proteins PrsQ1 and PrsQ2. Both proteins belong to an unique group of small, uncharacterized, secreted proteins restricted to the genera Cupriavidus and Ralstonia. This system gives C. metallidurans the ability to withstand much higher silver concentrations. The latter could be facilitated by the accumulation of silver ions and the formation of silver nanoparticles.
Significance to metallomicsIn this study, the molecular adaptation to silver exposure was studied for the model metal-resistant bacterium Cupriavidus metallidurans. Although it carries many known metal and silver resistance determinants, none were involved in the adaptation response. Our results showed that a single mutation resulted in the adaptation to high silver concentrations and previously uncharacterized periplasmic proteins played a crucial role in this adaptation process. The latter are part of a larger family within this and a closely related genus, suggesting a specific adaptation potential to metals in these genera that are able to live and thrive in harsh and oligotrophic environments. |
Introduction
In addition to many commercial and industrial uses, silver is widely used in health and hygiene applications such as water disinfection, topical creams for the treatment of burn wounds, dental amalgams and silver-impregnated polymers for medical devices.1–3 The rapid development of nanotechnology further expanded its use and applications and, today, silver nanoparticles (AgNPs) are the most commonly engineered nanomaterial. AgNPs are routinely incorporated in a wide range of domestic and personal care products such as laptops, refrigerators, dietary supplements, clothing, children's toys and many more.4 The toxicity of AgNPs can largely be attributed to the release of Ag+ ions.5–7 Silver ions interact with the cytoplasmic membrane compromising electron transfer and the proton motive force, ultimately resulting in cell death. In addition, once inside the cell, Ag+ ions can interact with enzymes, inhibit DNA replication and is expected to promote the production of reactive oxygen species.7–10 Due to the excessive use of silver compounds, an increased release of silver in the environment through point source direct discharge or through wastewater effluents is expected.11 In wastewater treatment plants, AgNPs are believed to be retained in sewage sludge that is often used as fertilizer for agricultural soils or send to landfills. In this manner, AgNPs can be transferred via surface runoff or leaching into the aquatic environment.12,13 Although the fate and transformations of silver compounds in the environment are largely unknown, the possible impact on the ecosystem is an emerging cause of concern.12,13 Moreover, concerns have been raised about the potential proliferation of silver-resistant bacteria in a manner similar to antibiotic resistant bacteria, as well as the co-selection of antibiotic resistance.14,15 In fact, silver-resistant bacteria have been isolated from both clinical and non-clinical environments.16–19 So far, the mechanisms of silver resistance have not been fully elucidated and different preventive strategies have been suggested. Next to chemical detoxification,20 bacterial silver resistance results from active efflux systems. These efflux clusters often additionally code for periplasmic binding proteins that act as chaperones or ‘metal sponges’ by specifically binding silver.21,22In the present study, we assessed the adaptation potential of Cupriavidus metallidurans to toxic silver concentrations. C. metallidurans is an environmental β-proteobacterium repeatedly isolated from metal-contaminated soils23–25 as well as other anthropogenic environments not typified by metal contamination,26–28 including medically relevant sources.29–32 It contains a large diversity of mobile genetic elements,33,34 facilitating horizontal gene transfer, and a number of systems involved in silver detoxification, including the silDCBA and cusDCBAF operons coding for HME-RND-driven efflux systems located on respectively pMOL30 and the chromid,35–38 and cupRAC coding for a PIB1-type ATPase located on the chromosome.37
Results
Spontaneous silver-resistant mutants
Spontaneous silver-resistant mutants were obtained from three different C. metallidurans strains (CH34, AE104 and NA4) by exposing each strain to a toxic concentration based on the minimal inhibitory concentration of silver for each strain. In a first series of experiments, spontaneous mutants of type strain CH34 resistant to silver were selected on LB agar supplemented with respectively 0.5 and 2 mM AgNO3 and designated as CH34S1 and CH34S2. To reduce metal complexation, a Tris-buffered mineral medium (MM284) was used in a second series of experiments with strain AE104 and NA4. Strain AE104 is a derivative of type strain CH34 that does not contain the two megaplasmids pMOL28 and pMOL30,39 which carry most of the metal detoxification functions.40 A spontaneous mutant resistant to silver was selected after incubation in MM284 supplemented with 3 μM AgNO3 and designated as AE104S. Strain NA4 is isolated from potable water from the Russian condensate recycle system SRV-K aboard the ISS.41,42 In contrast to CH34 where both megaplasmids have a key role in metal detoxification, most of the metal resistance determinants are located on the chromid.43 For NA4, a spontaneous mutant resistant to silver was selected after incubation in MM284 supplemented with 8 μM AgNO3 and designated as NA4S. The minimal inhibitory concentration (MIC) of AgNO3 for CH34S1, CH34S2, AE104S and NA4S was 4, 4, ±30 and ±10 times higher than for their parental strain, respectively (Fig. 1).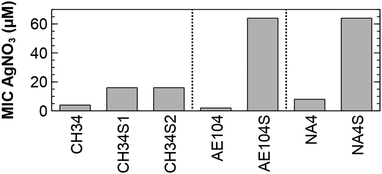 | ||
| Fig. 1 Minimal inhibitory concentration of AgNO3 for the silver-resistant mutants and respective parental strains. | ||
All silver-resistant mutants harbour a common subset of differentially expressed genes
Genome-wide expression profiling of CH34S1, CH34S2, AE104S and NA4S under non-selective growth conditions (i.e. in the absence of silver) showed a substantial number of differentially expressed genes in each silver-resistant mutant compared to their respective parental strain. In CH34S1, 301 genes were differentially expressed (133 up- and 168 down-regulated), 293 genes (158 up- and 135 down-regulated) were differentially expressed in CH34S2, 179 genes (75 up- and 104 down-regulated) in AE104S and 308 genes (204 up- and 104 down-regulated) in NA4S. The C. metallidurans systems known to be involved in silver detoxification (silDCBA and cusDCBAF operons coding for HME-RND-driven efflux systems, and cupRAC coding for a PIB1-type ATPase) were not differentially expressed in all mutants (ESI,† Table S1). In addition, exposure to 1 μM AgNO3 elicited a response in CH34S1 and CH34S2 similar to that in CH34 with upregulation of silDCBA, cusDCBAF and cupRAC (ESI,† Table S1). However, induction of these silver detoxification systems in the parental CH34 strain does not result in a silver resistant phenotype like that of the evolved strains, which indicated that other mechanisms are at play.On the other hand, only seven genes were commonly upregulated in all mutants under non-selective conditions (Table 1). In addition, their expression was not induced by AgNO3 (ESI,† Table S1). No commonly down-regulated genes were identified. The identified genes code for the response regulator AgrR, an outer membrane protein and five conserved hypothetical proteins. The response regulator AgrR, which is part of the two-component regulatory system AgrR–AgrS, has already been shown to be constitutively expressed in CH34S1.35 Its cognate target, located upstream of the agrRS locus and divergently transcribed, codes for the uncharacterized RND-driven efflux system AgrABC with characteristics of heavy metal efflux (HME) as well as hydrophilic/amphiphilic compounds (HAE) RND-driven efflux systems.35 Three of the conserved hypothetical proteins belong to an uncharacterized protein family of relatively small proteins, ranging in size from 69 to 165 amino acid residues. Strains CH34 and NA4 carry 22 and 26 genes encoding such proteins, respectively. Although their function remains to be elucidated, ten members of this family were found to be transcriptionally induced in CH34 in response to different heavy metals,37 including the pMOL30-encoded copQ, part of a complex 33-gene region involved in copper resistance, and czcJ, part of a 25-gene cluster involved in cadmium, zinc and cobalt resistance. Based on the first observations for the cop cluster, these homologs were previously designated as CopQ-like protein family members.44
| CH34a | NA4a | Function | Ident%b | Transcription (fold change)c | AgrRd | |||
|---|---|---|---|---|---|---|---|---|
| CH34S1 | CH34S2 | AE104S | NA4S | |||||
| a CH34 (Rmet_) and NA4 (CmetNA4v1_) locus tag based on MaGe45 annotation v1 (ESI, Table S3). b % protein identity between CH34 and NA4 copy. c Fold change of gene expression measured by microarray expression profiling for each mutant compared to its parental strain in non-selective growth conditions. d AgrR binding site prediction (Only values above 0.8 are shown). | ||||||||
| 0477 | 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 157 157 |
Conserved hypothetical, CopQ-like | 100 | 9.0 | 9.1 | 3.9 | 15.8 | |
| 1751 | 170![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 096 096 |
DNA-binding response regulator, AgrR | 100 | 78.5 | 65.4 | 81.0 | 54.2 | 0.99 |
| 3571 | 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 101 101 |
Conserved hypothetical, CopQ-like, PsrQ1 | 100 | 23.8 | 22.0 | 21.2 | 2.9 | |
| 4461 | 570![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 171 171 |
Conserved hypothetical, CopQ-like, PsrQ2 | 100 | 81 | 88.6 | 106.6 | 26.2 | 0.83 |
| 4464 | 570![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 173 173 |
Conserved hypothetical, CzcL | 98 | 5.1 | 4.9 | 2.1 | 2.8 | 0.87 |
| 4595 | 570![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 182 182 |
Conserved hypothetical, CzcI2 | 100 | 7.7 | 8.2 | 3.1 | 2.7 | 0.84 |
| 5118 | 400![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 150 150 |
Outer membrane protein (porin), OmpC family | 100 | 8.6 | 9.0 | 5.8 | 3.6 | |
Whole genome sequencing of mutants corroborates the involvement of the two-component regulatory system AgrR–AgrS
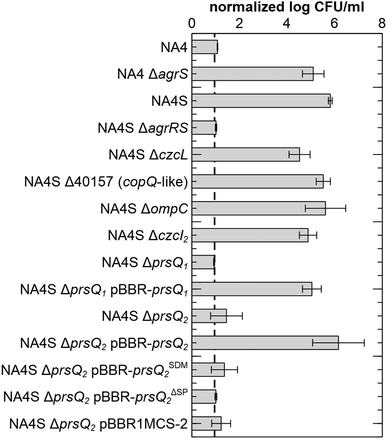
Sequencing of the CH34S1, CH34S2 and NA4S genomes showed one commonly affected gene, agrS, coding for the histidine kinase of the two-component regulatory system AgrR–AgrS (full list in ESI,† Table S2). In CH34S1 and CH34S2, agrS was inactivated by insertion of Insertion Sequence (IS) element ISRme3 and IS1086, respectively.34 Similarly, PCR and sequence analysis of AE104S showed an insertion of ISRme3 in agrS (data not shown). In NA4S, a C–T substitution occurred in agrS, which resulted in an arginine to cysteine substitution at residue 414 (of 469). Notably, agrA is inactivated by an IS insertion in the parental NA4 strain. In addition, CH34S2 carries a frameshift mutation in agrA (4 bp duplication) and expression of agrCBA in CH34S1 under non-selective conditions was comparable to that in the parental strain (putatively elicited by a 7 bp duplication in the promoter region of agrCBA) (ESI,† Tables S1 and S2). These results refuted a putative role of the AgrCBA RND-driven efflux system in the acquired silver resistance. However, all mutants showed an increased expression of agrR as well as a mutation in agrS, supporting an important role for this two-component regulatory system in the observed increased silver resistance. This role was further confirmed by deleting agrRS in NA4S, which decreased silver resistance to the parental level as well as by deleting agrS in NA4, which increased silver resistance to the level of NA4S (Fig. 2). Noteworthy, when grown in LB, the stationary phase cell density of NA4 ΔagrS was comparable to that of NA4S and slightly less than that of NA4 (Fig. S1, ESI†).Insertional inactivation and complementation elucidated an important role for prsQ1 and prsQ2
Next, the role of the commonly upregulated genes was identified. Because the observations were strikingly similar for the four mutants and since strain CH34 harbours almost three times more insertion sequence elements compared to NA4,43 affecting genome plasticity and adaptability,46 further work was carried out with NA4S. Insertional inactivation mutants were constructed for each gene and the absence of the corresponding mRNA confirmed (ESI,† Fig. S2). No significant differences in viable count on LB were observed (ESI,† Fig. S3). Next, silver resistance was assessed by total viable count on LB supplemented with 0.5 mM AgNO3. Negligible growth was observed for NA4, whereas 5.81 ± 0.08 log CFU ml−1 were counted for NA4S (Fig. 2). Noteworthy, CH34S1, CH34S2 and AE104S and their respective parental strains CH34 and AE104 showed comparable total viable counts on LB and LB supplemented with 0.5 mM AgNO3 as NA4S and NA4, respectively.The viable count on LB with 0.5 mM AgNO3 reduced to a level similar to that of NA4 for inactivation of agrRS and two of the copQ-like genes, consequently designated prsQ1 and prsQ2 (for periplasmic resistance to silver related to the CopQ-like family). Furthermore, plasmid-based complementation of NA4S prsQ1 and prsQ2 mutants restored transcription levels (ESI,† Fig. S2) and silver resistance (Fig. 2). As the function of the CopQ-like protein family members is unstudied, we confirmed that the actual protein is involved in the acquired silver resistance by introducing a −1 frame shift in the start codon of prsQ2 on pBBR-prsQ2, which abolished complementation (Fig. 2). In addition, we confirmed that translocation across the inner membrane is necessary by deleting the 26-aa signal peptide (based on PrediSi and SignalP signal peptide prediction) of prsQ2 on pBBR-prsQ2, which again abolished complementation (Fig. 2).
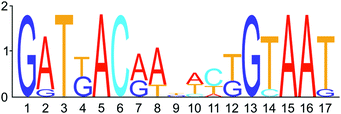
In a next step, the link between the commonly affected regulatory genes, agrS and agrR, and the other identified genes was evaluated. Phylogenetic footprinting was used to discover the DNA-binding motif for AgrR (Fig. 3). The promoter region of agrR from closely and more distantly related species was compared in silico by several motif detection algorithms (MotifSampler,47 AlignACE,48 MEME,49 Weeder50 and RSAT51), followed by a Markov clustering on the outcome of the models. In this way, a possible regulatory motif for agrR was found.
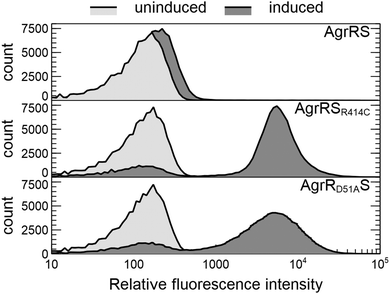
The latter was subsequently used in a genome-wide screening to identify possible AgrR target genes (Table 1). For the commonly upregulated genes, an AgrR binding motif was predicted (cut-off score 0.8) in the promoter region of agrR, prsQ2, czcL and czcI2. Only deletion of agrRS and prsQ2 reduced silver resistance of NA4S to the parental level. To identify AgrR regulation on prsQ2 expression further, a heterologous dual expression/reporter system was designed. In this Escherichia coli system, a gfp promoter probe vector to measure prsQ2 expression levels (PprsQ2-gfp) was combined with the regulatory genes placed under the control of the arabinose-inducible araBAD promoter. Arabinose induction of the NA4S-type agrRS (pBAD-agrRSR414C) strongly induced prsQ2 expression (>30-fold) in contrast to arabinose induction of native agrRS (Fig. 4). The agrRS operon in NA4S carried a C–T substitution in agrS, resulting in an arginine to cysteine substitution at residue 414. The Arg-414 residue corresponds for instance to Arg-434 of the histidine kinase PhoQ (part of the two-component regulatory system PhoP-PhoQ in E. coli), which is essential for catalysis.52 This indirectly indicated that AgrR most probably binds to the promoter region of prsQ2 in its unphosphorylated state. To support this, a variant of pBAD-agrRS was constructed in which the Asp-51 codon of agrR was replaced with an alanine codon. The Asp51 residue of AgrR corresponds for instance to Asp-55 of Spo0A (part of the two-component regulatory system Spo0A-KinD in Geobacillus stearothermophilus) and Asp-57 of CheY (part of the two-component regulatory system CheY-CheA in E. coli), which are both phosphorylation sites.53,54 Arabinose induction of agrR carrying a dephosphomimetic mutation (pBAD-agrRD51AS) strongly induced prsQ2 expression to levels similar to that of the arabinose-induced NA4S-type agrRS (pBAD-agrRSR414C) (Fig. 4).
Next to prsQ2, also deletion of prsQ1 reduced silver resistance of NA4S to the parental level. Although genome-wide expression profiling indicated increased expression in NA4S compared to NA4 (2.9-fold; Table 1), this could not be confirmed by a plasmid-based promoter probe reporter. No significant difference in fluorescence was measured for pSCK108-PprsQ1-gfp in NA4 and NA4S, respectively. In contrast, and in agreement with the high differential expression observed via genome-wide expression profiling, significant difference in fluorescence was measured for pSCK108-PprsQ2-mcherry in NA4 and NA4S, respectively (data not shown).
Sequence analysis and structural prediction of PrsQ1, PrsQ2 and other CopQ-like family members
After confirmation that the actual PrsQ1 and PrsQ2 proteins are involved in the acquired resistance (see above), we further examined their protein sequence and structure to get possible information on their function. De novo prediction of sequence motifs was performed at the MEME-server49 for the CopQ-like family member protein sets of C. metallidurans strains CH34 and NA4 (22 and 26 sequences, respectively). This indicated that they invariably contain a signal peptide and one or more typical Dx[YF]xxG domain motifs (Fig. 5). Searching for CopQ-like family members across the available protein sequences of 7927 complete genomes based on an extended 10-aa search pattern, the presence of a signal peptide and size, retrieved 787 protein hits. All hits belonged to genomes of the Cupriavidus and Ralstonia genera. Finally, all hits were used to predict a general sequence motif in CopQ-like proteins (Fig. 5).A preliminary prediction towards a structural model of the PrsQ1 and PrsQ2 proteins by using the PHYRE2 (http://www.sbg.bio.ic.ac.uk/phyre2) and RaptorX (http://raptorx.uchicago.edu/) prediction software55,56 indicated that these proteins do not fold. Instead, they form intrinsically disordered polypeptides, which could be explained by the abundance of chain-disturbing glycines (allow tight turns) and prolines (reduce structural flexibility) that are seemingly evenly spaced. The latter may cause the aromatic rings of phenylalanines and tyrosines to be more exposed than in normally folded proteins.
Formation of silver nanoparticles
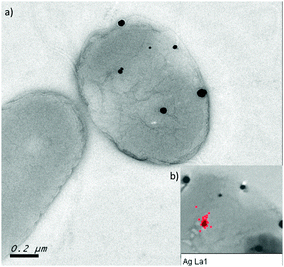
Finally, transmission electron microscopy was performed to study the location and distribution of silver. The silver-resistant mutant NA4S was grown in the presence of 0.5 mM AgNO3 and analysis showed the formation of nanoparticles, which mostly contained Ag as indicated by EDX (Fig. 6). Neither growth nor nanoparticle formation was observed in these conditions for the parental strain NA4 (data not shown).Discussion
The broad-spectrum antimicrobial activities of silver (nanoparticles) resulted in the extensive use in a wide range of medical applications as well as consumer and personal care products.1,4 Furthermore, the increase in occurrence and number of antibiotic-resistant strains contributed to the resurgence in the interest and use, including combating multi-drug resistant bacteria.57,58 The potential of microorganisms to develop resistance to silver and AgNPs59,60 and the incidence of silver-resistant determinants in many environments, including those where the use of silver compounds was restricted, raises the concern for their widespread use and co-selection for antibiotic resistance. Together with the health and environmental risks associated with the increased use of nanotechnology, the European Commission advised that more data is needed to better understand the bacterial response to ionic silver and silver nanoparticles.61 Since efflux-pump related silver resistance is well studied, but less is known for other resistance and adaptation mechanisms, we addressed in this study the bacterial adaptation to toxic silver concentrations. We showed that C. metallidurans was able to adapt to toxic silver concentrations. C. metallidurans was selected because it carries many distinct and well-studied heavy metal resistance determinants, including several involved in silver resistance. However, none of these systems were involved in the adaptive response to silver described here. Specific for silver resistance determinants, the HME-RND-driven efflux system encoded by the silDCBA operon and the cop cluster (for which some genes are shown to be involved in Ag+ resistance) are located on plasmid pMOL3038,40 and therefore neither present in AE104 nor in its silver-resistant mutant AE104S. In addition, the HME-RND-driven efflux system encoded by the cusDCBAF operon36 and cupRAC coding for a PIB1-type ATPase37 were not commonly differentially expressed (ESI,† Table S1). Furthermore, cupA in CH34S2 was inactivated by ISRme5 (ESI,† Table S2). Instead, sequencing, whole genome expression profiling and insertional mutation indicated the involvement of the AgrRS two-component system and two uncharacterized periplasmic proteins, designated PrsQ1 and PrsQ2 for periplasmic resistance to silver related to the uncharacterized CopQ-like protein family. Contrary to observations involving the Cus or Sil HME-RND-driven efflux systems,59,60 for which mutations within the associated sensor kinase resulted in an increased silver resistance via increased expression of the RND-driven efflux pump, this is not the case for the AgrCBA efflux system and associated AgrRS. Mutations in a sensor kinase that result in the constitutive and stress-independent transcriptional activation of its cognate RND-driven efflux system are often at the base of an increased resistance phenotype and have also been shown for other metals and antibiotics.62,63 However, in our study, the AgrRS two-component regulatory system increased the expression of non-cognate targets, including prsQ2, resulting in increased silver resistance.The pivotal mechanistic determinants mediating the increased silver resistance are the small periplasmic proteins PrsQ1 and PrsQ2. Both belong to a previously uncharacterized protein family whose members can only be found in the related genera Cupriavidus and Ralstonia.44 A recent sequence analysis of more than 28 million protein sequences showed that all 787 predicted CopQ-like proteins belong to the Cupriavidus and Ralstonia genera. Structure prediction of PrsQ1 and PrsQ2 indicated that they could be intrinsically disordered proteins. Such disordered proteins are common in nature and include SilE and PcoE, which have been shown to be involved in silver resistance.22,64 Both SilE and PcoE act as a ‘metal sponge’, fold upon binding to Ag+ and transport Ag+ to the associated HME-RND efflux pump.21,22 Although the function of PsrQ1 and PsrQ2 still needs to be determined, they do not contain conserved histidine and methionine residues within specific sequence motifs that are involved in Ag+ binding like SilE and PcoE.22,64
As the increased silver resistance is not mediated by efflux, we hypothesize that the formation of silver nanoparticles prevents ionic silver to exert its action in the cytoplasm. The latter provides C. metallidurans the ability to withstand much higher silver concentrations than efflux-mediated resistance. Nanoparticle formation has been studied in silver-resistant bacteria as a way to mitigate silver stress.20,65,66 However, the underlying mechanisms have not yet been studied. Our results are similar to those of a synthetic silver-binding peptide that increases silver tolerance in E. coli when expressed in the periplasm but not when expressed in the cytoplasm.67 Similarly, the synthetic peptide does not contain conserved histidine and methionine residues and transmission electron microscopy showed the presence of electron-dense silver nanoparticles.67
A more detailed study of this protein family is required as many of the encoding genes are highly induced in response to different metals.37 As such, these studies will not only gain insights in the different effects and potential adverse outcomes of ionic silver and silver nanoparticles but also in metal (silver) resistance mechanisms as well as novel routes for metal nanoparticle formation and metal processing in biotechnical and biomedical applications.
Experimental
Strains and growth conditions
Bacterial strains and plasmids used in this study are summarized in Tables 2 and 3, respectively. C. metallidurans strains were routinely cultured at 30 °C in Lysogeny Broth (LB; Thermofisher Scientific) or Tris-buffered mineral medium supplemented with 0.2% (wt/vol) gluconate (MM284).39E. coli strains were routinely cultured at 37 °C in LB. For culturing on agar plates, 2% agar (Oxoid, Belgium) was added. Liquid cultures were grown in the dark on a rotary shaker at 150 rpm. When appropriate, the following chemicals were added to the growth medium at the indicated final concentrations: kanamycin sulfate (50 μg ml−1 for E. coli or 1500 μg ml−1 for C. metallidurans) (Km50; Km1500), tetracycline hydrochloride (20 μg ml−1) (Tc20) and ampicillin sodium salt (100 μg ml−1) (Ap100) (PanReac AppliChem, Germany), chloramphenicol (30 μg ml−1) (Cm30), L-arabinose (0.02%) (Sigma-Aldrich, Belgium), 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-Gal; 40 μg ml−1) (Thermofisher Scientific, Belgium) and isopropyl β-D-1-thiogalactopyranoside (IPTG; 0.1 mM) (Thermofisher Scientific, Belgium).| Strain | Genotype/relevant characteristics | Ref. |
|---|---|---|
| AgR (silver resistant); KmR and km (kanamycin resistance and resistance gene); SmR (streptomycin resistant); TcR and tet (tetracycline resistance and resistance gene). | ||
| Escherichia coli | ||
| DG1 | mcrA Δmrr-hsdRMS-mcrBC (rB−mB−) Φ80lacZΔM15 ΔlacX74 recA1 araD139 Δ(ara-leu)7697 galU galK rpsL endA1 nupG | Eurogentec, B |
| MFDpir | MG1655 RP4-2-Tc::[ΔMu1::aac(3)IV-ΔaphA-Δnic35-ΔMu2::zeo] ΔdapA::(erm-pir) ΔrecA | 68 |
| HB101 | F−mcrB mrr hsdS20(rB−mB−) recA13 leuB6 ara-14 proA2 lacY1 galK2 xyl-5 mtl-1 rpsL20 (SmR) glnV44 λ− | Lab collection |
| Cupriavidus metallidurans | ||
| CH34T | Isolated from a decantation tank at a Belgian zinc factory | 39 |
| AE104 | Plasmidless derivative of CH34 | 39 |
| NA4 | Isolated from ISS SRV-K filter reactor effluent returned on Soyuz 10S (2004) (original code: 0502478-1) | 69 |
| CH34S1 | Spontaneous silver resistant mutant of CH34, AgR | A. Bossus |
| CH34S2 | Spontaneous silver resistant mutant of CH34, AgR | A. Bossus |
| AE104S | Spontaneous silver resistant mutant of AE104, AgR | This study |
| NA4ΔagrS | agrR::3xFLAG agrS::km, KmR | This study |
| NA4S | Spontaneous silver resistant mutant of NA4, AgR | This study |
| NA4SΔ40157 | 40157::tet, TcR | This study |
| NA4SΔagrRS | agrRS::tet, TcR | This study |
| NA4SΔczcI2 | czcI 2::tet, TcR | This study |
| NA4SΔczcL | czcL::tet, TcR | This study |
| NA4SΔompC | ompC::tet, TcR | This study |
| NA4SΔprsQ1 | prsQ 1::tet, TcR | This study |
| NA4SΔprsQ2 | prsQ 2::tet, TcR | This study |
| Plasmid | Genotype/relevant characteristics | Ref. |
|---|---|---|
| ApR (ampicillin resistant); CmR (chloramphenicol resistant); KmR and km (kanamycin resistant and resistance gene); TcR and tet (tetracycline resistant and resistance gene); PBAD (arabinose inducible promoter); GenBio (General Biosystems, Inc., Morrisville, USA). | ||
| pACYC184 | p15A ori, CmR, TcR | Lab collection |
| pBAD33 | p15A ori, PBAD, araC, CmR | 73 |
| pBAD33-agrRS | agrRS of NA4 in pBAD33, KmR | This study |
| pBAD33-agrRD51AS | pBAD33-agrRS with 152A > C 153T > C in agrR, KmR | This study |
| pBAD33-agrRSR414C | agrRS of NA4S in pBAD33, KmR | This study |
| pBBR1MCS-2 | pBBR1 ori, Mob+, lacZ, KmR | 74 |
| pBBR-prsQ1 | prsQ 1 of NA4 in pBBR1MCS-2, KmR | This study |
| pBBR-prsQ2 | prsQ 2 of NA4 in pBBR1MCS-2, KmR | This study |
| pBBR-prsQSDM2 | pBBR-prsQ2 harbouring a frameshift in the start codon of prsQ2, KmR | This study |
| pBBR-prsQΔSP2 | pBBR-prsQ2 with 26-bp deletion of the prsQ2 signal sequence, KmR | This study |
| pK18mob | pMB1 ori, Mob+, lacZ, KmR | 75 |
| pK18mob-40157::tet | tet gene flanked by 1 kb DNA region up- and downstream of CmetNA4v1_40157, KmR, TcR | This study |
| pK18mob-agrRS | agrRS gene region of NA4 in pK18mob, KmR | This study |
| pK18mob-agrRS::tet | pK18mob-agrRS derivative, agrRS::tet, KmR, TcR | This study |
| pK18mob-czcI2::tet | tet gene flanked by 1 kb DNA region up- and downstream of czcI2 NA4, KmR, TcR | This study |
| pK18mob-czcL::tet | pK18mob-prsQ2 derivative, czcL::tet, KmR, TcR | This study |
| pK18mob-ompC::tet | tet gene flanked by 1 kb DNA region up- and downstream of ompC NA4, KmR, TcR | This study |
| pK18mob-prsQ1::tet | tet gene flanked by 1 kb DNA region up- and downstream of prsQ1 NA4, KmR, TcR | This study |
| pK18mob-prsQ2 | prsQ 2–czcL gene region of NA4 in pK18mob, KmR | This study |
| pK18mob-prsQ2::tet | pK18mob-prsQ2 derivative, prsQ2::tet, KmR, TcR | This study |
| pKT230 | IncQ, Mob+, KmR | 76 |
| pRK600 | ColE1 oriV; RP4 Tra+ RP4 oriT, CmR; helper in triparental matings | Lab collection |
| pPROBE-TT | pBBR1 ori, Mob+, promoterless gfp, TcR | 77 |
| pPTT-prsQ2 | Promoter region of prsQ2 NA4 in pPROBE-TT, TcR | This study |
| pT18mob | pMB1 ori, Mob+, lacZ, TcR | This study |
| pT18mob-agrS::km | km gene flanked by 1 kb DNA region up- and downstream of agrS, agrR::3xFLAG, KmR | This study |
| pSCK108 | IncQ, Mob+, lacZ, KmR | This study |
| pSCK108-PprsQ1-gfp | SacI/BamHI fragment of pUC57-PprsQ1-gfp in pSCK108, KmR | This study |
| pSCK108-PprsQ2-mcherry | BamHI/HindIII fragment of pUC57-PprsQ2-mcherry in pSCK108, KmR | This study |
| pUC57-PprsQ1-gfp | 350 bp 5′ region of prsQ1 fused to gfp (synthetic construct), ApR | GenBio |
| pUC57-PprsQ2-mcherry | 350 bp 5′ region of prsQ1 fused to mcherry (synthetic construct), ApR | GenBio |
Generation of spontaneous silver resistant mutants
Spontaneous silver resistant mutants of C. metallidurans CH34, its plasmidless derivative AE104 and C. metallidurans NA4 were generated by exposure to a toxic concentration of AgNO3 (Sigma-Aldrich, Belgium). To this end, CH34S1 and CH34S2 were selected on LB agar supplemented with respectively 0.5 and 2 mM AgNO3, AE104S and NA4S were generated in MM284 supplemented with respectively 3 μM and 8 μM AgNO3.Whole-genome sequencing
Total DNA (10 μg) of C. metallidurans CH34S1, CH34S2 and NA4S was extracted using the QIAamp DNA Mini Kit (Qiagen). The quantity and quality of the DNA was analysed using a NanoDropTM 1000 spectrophotometer (Thermofisher Scientific). A 50 bp paired-end sequencing approach with an insert size of 300 bp was performed by Baseclear on the Illumina GAIIx platform. The paired-end sequencing reads were aligned to the reference genomes (i.e. the genome sequence of C. metallidurans strain CH34 and strain NA4, respectively) using the BWA-MEM algorithm version 0.7.13.70 Genomic rearrangements were identified using an in-house developed Perl script specifically looking for wrongly mapped paired-end reads. An increase in the insert size combined with a peak in incorrectly aligned paired reads at the breakpoints indicates a deletion, where the size of the deletion can be derived from the predict insert size based on the mapping of both mapped reads. Two neighbouring peaks (<100 bp) – respectively on the forward and reverse strand – in incorrectly aligned paired-end reads indicate an insertion of a new DNA fragment at that position. SNPs and small indels were identified using the Genome Analysis Tool Kit version 3.6 using the Haplotyper command, after sorting and indexing the BWA alignment using the samtools software (version 1.3.1).71 Sequencing data are available within the Sequencing Read Archive (SRA) of NCBI using the accession numbers SRR6340090, SRR6340091 and SRR6340092 for the strains NA4S, CH34S1 and CH34S2 respectively.Whole genome expression analysis
Gene expression in the different spontaneous silver resistant mutants was compared with the parental strain under non-selective growth conditions. To this end, three independent stationary phase cultures of C. metallidurans CH34, AE104, NA4 and the spontaneous silver resistant mutants CH34S1, CH34S2, AE104S and NA4S were diluted 1/500 in 25 ml MM284. These subcultures were allowed to grow until a cell density of 5 × 108 cells per ml (OD600 = 0.5) was reached. Each culture was subdivided in 2 ml portions and cells were harvested by centrifugation for 2 min at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g. Bacterial pellets were flash frozen by immersion into liquid nitrogen and were kept frozen at −80 °C at all times. RNA extraction, labelling and hybridization, microarray spotting, scanning and data analysis was performed according to the work of Monsieurs et al.37 The full description of the microarray data have been deposited at the Gene Expression Omnibus website (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE107669. Annotation of C. metallidurans NA4 was based on GenBank entry JFZE00000000.1 and MaGe72 annotation v1 (ESI,† Table S3).
000g. Bacterial pellets were flash frozen by immersion into liquid nitrogen and were kept frozen at −80 °C at all times. RNA extraction, labelling and hybridization, microarray spotting, scanning and data analysis was performed according to the work of Monsieurs et al.37 The full description of the microarray data have been deposited at the Gene Expression Omnibus website (http://www.ncbi.nlm.nih.gov/geo/) under accession number GSE107669. Annotation of C. metallidurans NA4 was based on GenBank entry JFZE00000000.1 and MaGe72 annotation v1 (ESI,† Table S3).
Construction of insertional inactivation mutants
All PCR reactions were performed with Phusion High-Fidelity Polymerase (Thermofisher Scientific, Belgium) unless stated otherwise. Primers used are presented in ESI,† Table S4. The C. metallidurans NA4 agrRS and prsQ2-czcL gene regions, including 0.5 kb up- and downstream, were amplified by PCR (DreamTaq Polymerase, Thermofisher Scientific, Belgium) using the primer pairs agrR_FW-agrS_RV and prsQ2-czcL_FW-RV providing amplicons with a size of 3.3 kb and 3 kb, and EcoRI/HindIII and BamHI/HindIII recognition sites, respectively. The amplicons were subsequently cloned in pK18mob digested with EcoRI/HindIII and BamHI/HindIII, respectively. The gene region of C. metallidurans NA4 CmetNA4v1_570182 (czcI2) including 1 kb up- and downstream was amplified with the primers czcI_FW-RV providing an amplicon of 2.4 kb. This amplicon was subsequently cloned in pK18mob, which was linearized with EcoRI and amplified with the primers pK18mob_FW-RV (3.6 kb), using the GeneArt™ seamless cloning and assembly enzyme mix (Thermofisher Scientific, Belgium). The resulting constructs pK18mob-agrRS, pK18mob-prsQ2 and pK18mob-czcI2 were transformed to E. coli DG1 chemocompetent cells (Eurogentec, Belgium).Next, pK18mob_agrRS, pK18mob-czcI2, pK18mob-czcL and pK18mob-prsQ2 were used as PCR (DreamTaq Polymerase, Thermofisher Scientific, Belgium) template using the outward-oriented primers agrRS_OUT_FW-RV, czcI_OUT_FW-RV, czcL_OUT_FW-RV and prsQ2_OUT_FW-RV, respectively. A tetracycline resistance cassette, amplified from pACYC184 with primers tet_FW-RV, was cloned as a BspTI/BcuI fragment in each BspTI/BcuI digested amplicon. The resulting constructs pK18mob-agrRS::tet, pK18mob-czcI2::tet, pK18mob-czcL::tet and pK18mob-prsQ2::tet were transformed to E. coli DG1.
GeneArt™ was applied to obtain different pK18mob constructs in which the 1 kb up- and downstream DNA region of prsQ1, CmetNA4v1_40157 and ompC flanked a tetracycline cassette. To this end, the 1 kb up- and downstream region of prsQ1, CmetNA4v1_40157 and ompC were amplified with the primers prsQ1_5_FW-RV and prsQ1_3_FW-RV, 40157_5_FW-RV and 40157_3_FW-RV and ompC_5_FW-RV and ompC_3_FW-RV, respectively. A tetracycline resistance cassette was amplified from pACYC184 with primers tet-5 and tet-3 and the EcoRI-linearized pK18mob was amplified with the primers pK18mob_FW-RV (3.6 kb). Assembly of the products resulted in pK18mob-prsQ1::tet, pK18mob-40157::tet and pK18mob-ompC::tet, which were transformed to E. coli DG1. All plasmids were confirmed by sequencing with tet-5_OUT and tet-3_OUT primers.
For constructing pT18mob-agrS::km, the agrRS gene region of C. metallidurans NA4 was amplified with primer pair (agrS_5_FW/agrS_3_RV) and cloned via GeneArt™ in pT18mob (linear PCR fragment with primer pair pK18mob_FW-RV). Next, pTmob-agrS was linearized with primer pair (agrS_3FL_FW/agrS_5FL_RV) and fused via GeneArt™ to a PCR fragment of pSUB11 (primer pair 3xFLAGKM_FW-RV) creating a C-terminal 3xFLAG tagged AgrR while replacing agrS by a kanamycin resistance cassette. The resulting pT18mob-agrS::km construct was transformed to E. coli DG1.
For insertion mutagenesis, all constructs (pK18mob-40157::tet, pK18mob-agrRS::tet, pK18mob-czcI2::tet, pK18mob-czcL::tet, pK18mob-ompC::tet, pK18mob-prsQ1::tet, pK18mob-prsQ2::tet, and pT18mob-agrS::km) were conjugated into C. metallidurans NA4S (or NA4 for pT18mob-agrS::tet) either with E. coli HB101 pRK600 as helper (triparental) or via E. coli MFDpir as donor (biparental). For each mutant, a tetracycline-resistant and kanamycin-sensitive exconjugant was selected (kanamycin-resistant and tetracycline-sensitive in the case of agrS::km), and confirmed being a genuine mutant by PCR and sequencing, yielding NA4SΔ40157, NA4SΔagrRS, NA4SΔczcI2, NA4SΔczcL, NA4SΔompC, NA4SΔprsQ1, NA4SΔprsQ2 and NA4ΔagrS, respectively.
Construction of plasmids
PCR amplification of prsQ1 and prsQ2 (including their promoter region) was performed on genomic DNA from C. metallidurans NA4 with the respective primer pairs prsQ1_FW-RV (0.6 kb) and prsQ2_FW-RV (0.6 kb). These PCR products were cloned as BamHI/HindIII fragments into pBBR1MCS-2. The resulting pBBR-prsQ1 and pBBR-prsQ2 plasmids from transformants selected on LB Km50 were further confirmed by sequencing with the primers pBBR_FW and pBBR_RV. Afterwards, pBBR-prsQ1 and pBBR-prsQ2 were transformed by electroporation to NA4SΔprsQ1 and NA4SΔprsQ2, respectively.The agrRS genes from C. metallidurans NA4 and NA4S as well as agrRD51AS (using pBBR-agrRD51AS as template, see below) were amplified by PCR with the primer pair agrRS_BAD_FW-agrS_RV (2.1 kb), providing BamHI/HindIII recognition sites. These amplicons were subsequently cloned in pBAD3378 and transformed to E. coli DG1, resulting in the plasmids pBAD-agrRS, pBAD-agrRSR414C and pBAD-agrRD51AS.
For the transcriptional prsQ2 promoter-gfp fusion, a 0.3 kb DNA fragment upstream the prsQ2 start codon was amplified by PCR using the primer pairs PprsQ2_FW-RV, providing BamHI/XbaI recognition sites. This amplicon was subsequently cloned into pPROBE-TT77 containing a promoterless gfp and transformed into E. coli DG1 competent cells, resulting in pPTT-prsQ2.
Plasmid-based promoter probe reporters in C. metallidurans were constructed in the broad-host-range vector pSCK108. The latter was constructed by ligating a BsiWI-digested PCR fragment of pK18mob (primer pair pK18mob_inv_FW-RV) to an Acc65I-digested PCR fragment of pKT230 (primer pair KT261_FW-RV). pSCK108-PprsQ1-gfp was constructed by cloning a SacI/BamHI fragment of pUC57-PprsQ1-gfp, carrying a 350-bp 5′ region of prsQ1 fused to gfp, in SacI/BamHI-digested pSCK108. pSCK108-PprsQ2-mcherry was constructed by cloning a BamHI/HindIII fragment of pUC57-PprsQ2-mcherry, carrying a 350 bp 5′ region of prsQ2 fused to mcherry, in BamHI/HindIII-digested pSCK108. pUC57-PprsQ1-gfp and pUC57-PprsQ2-mcherry were custom synthesized by General Biosystems, Inc. (Morrisville, USA).
Plasmid pT18mob was constructed by replacing the kanamycin cassette of pK18mob with the tetracycline cassette of pACYC184 via GeneArt™ cloning (PCR fragment of pK18mob (primer pair pK18_inv_FW-RV) and PCR fragment of pACYC184 (primer pair pACYC184_tet_FW-RV)).
Site-directed mutagenesis
Site-directed mutagenesis was performed with the “Phusion Site-Directed Mutagenesis Kit” (Thermo Scientific, Belgium). Plasmid pBBR-prsQ2 was used as a template for PCR amplification. Phosphorylated primer pairs prsQSDM2_FW-RV and prsQΔSP2_FW-RV were used for constructing a frame shift mutation in the prsQ2 start codon and a 26-bp deletion of the prsQ2 signal sequence, respectively. Plasmid pBBR_agrRS was used as template for PCR amplification with the phosphorylated primer pair agrR_SDM_FW-RV, which introduced 152A > C 153T > C, resulting to D51A in AgrR. The resulting linear fragments were purified using the “Wizard® SV Gel and PCR Clean-Up System” (Promega, the Netherlands), subsequently self-ligated, and finally transformed to E. coli DG1. The resulting pBBR-prsQSDM2 and pBBR-prsQΔSP2 plasmids from transformants selected on LB Km50 were further confirmed by sequencing prior to transformation to C. metallidurans NA4SΔprsQ2.cDNA amplification and RT-PCR
Total RNA was extracted from the silver-resistant mutant NA4S, all insertional inactivation mutants and complementation strains using the SV Total RNA Isolation System (Promega, the Netherlands). To this end, cultures were grown in MM284 media until an OD of 0.6–0.8 was reached. Each culture was subdivided in 2 ml portions and cells were harvested by centrifugation for 2 min at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g. Bacterial pellets were flash frozen by immersion into liquid nitrogen and were kept frozen at −80 °C at all times. From each strain, single-stranded complementary DNA (cDNA) was synthesised from 1 μg total RNA by using random hexamers as primers and MultiScribe® Reverse Transcriptase (ThermoFisher Scientific, Belgium) and the quantity of synthesised cDNA was measured by a NanoDrop® 1000 spectrophotometer (ThermoFisher Scientific, Belgium). This cDNA (50 ng) was used to check the absence or presence of mRNA for inactivated or reintroduced genes by PCR amplification (ESI,† Table S4; primers with _cDNA tag).
000g. Bacterial pellets were flash frozen by immersion into liquid nitrogen and were kept frozen at −80 °C at all times. From each strain, single-stranded complementary DNA (cDNA) was synthesised from 1 μg total RNA by using random hexamers as primers and MultiScribe® Reverse Transcriptase (ThermoFisher Scientific, Belgium) and the quantity of synthesised cDNA was measured by a NanoDrop® 1000 spectrophotometer (ThermoFisher Scientific, Belgium). This cDNA (50 ng) was used to check the absence or presence of mRNA for inactivated or reintroduced genes by PCR amplification (ESI,† Table S4; primers with _cDNA tag).
The expression of prsQ2 was analysed by RT-PCR using the MESA Blue qPCR MasterMix Plus for SYBR® Assay Low ROX (Eurogentec, Belgium) according to the manufacturer's protocol with primers prsQ1_cDNA_FW-RV (ESI,† Table S4). Expression was compared with that of uvrD, which was amplified with primers uvrD_cDNA_FW-RV (ESI,† Table S4). RT-PCRs were performed with a 7500 Real-Time PCR System (Applied Biosystems, UK).
Total viable count
Three independent colonies of each strain were cultivated up to stationary phase in LB medium at 30 °C. Afterwards, 109 CFU ml−1 were pelleted, washed with MgSO4 (10 mM) and resuspended in MgSO4 (10 mM). Cell suspensions of 100 μl of a serial ten-fold dilution in MgSO4 (10 mM) were spread on LB agar plates and on LB agar plates supplemented with 0.5 mM AgNO3. The plates were incubated at 30 °C and colony forming units were counted after 3 days. To calculate the survival, the viable count on LB agar supplemented with 0.5 mM AgNO3 was normalized to the viable count on LB agar.Phylogenetic footprinting
A phylogenetic footprinting approach with the promoter region of agrR was performed in silico by combining the output of different motif detection algorithms (MotifSampler,47 AlignACE,48 MEME,49 Weeder50 and RSAT51) followed by a Markov clustering of the output motif models. The regulatory motif obtained in this manner was used for a genome wide screening of all upstream regions in type strain C. metallidurans CH34 to identify putative binding sites for AgrR.In vivo cross-regulation in E. coli
Plasmid pPTT-prsQ2 was transformed by electroporation in E. coli DG1 carrying pBAD-agrRS, pBAD-agrRSR414C, pBAD-agrRD51AS and pBAD-agrR, respectively. Possible cross-regulation was tested by comparing Gfp production in each construct, either un-induced or induced with 0.2% arabinose. To this end, overnight LB cultures of three independent colonies (grown in the presence of Tc (20 μg ml−1) and Cm (30 μg ml−1)), were diluted 10 times in 2 ml LB with the same antibiotics and 0.2% arabinose on the one hand and water on the other hand. These cultures were incubated for 6 hours at 30 °C, diluted 100-fold in 10 mM MgSO4 (filtered with 0.2 μm filter) and analysed with a C6 Accuri™ flow cytometer (BD Biosciences, Belgium). The flow cytometer is equipped with a 20 mW 488 nm laser, two scatter detectors and four fluorescence detectors (530/30 nm, 585/40 nm, >670 nm and 675/25 nm). The flow cytometer is operated with Milli-Q water (Merck Millipore, Belgium) as sheath fluid. Gfp production was measured with filter FL1 (excitation 488 nm/emission 533/30 nm) and 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells were recorded. The respective cell populations were delimited to eliminate background signals originating from cell debris. All data analysis was performed with the CFlow and FlowJo V10 Software.
000 cells were recorded. The respective cell populations were delimited to eliminate background signals originating from cell debris. All data analysis was performed with the CFlow and FlowJo V10 Software.
Motif discovery CopQ-like proteins
The available protein sequences of 7927 complete genomes were retrieved from the NCBI ftp server (http://ftp://ftp.ncbi.nlm.nih.gov/genomes/genbank/bacteria/ on October 4, 2017) and concatenated in one FastA file. This FastA file, containing 28![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 248
248![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 917 protein sequences, was queried by the multi-threaded pattern-matching tool Biogrep 1.079 using an extended 10-aa search pattern [GSRDEQ][GSAKRTPN]xD[PVT][YF][TS][DEQ]G[ASG] covering the Dx[YF]xxG domain motif characteristic for CopQ-like family proteins. This extended search pattern is based on a multiple alignment and additional visual inspection of the protein sequences of CopQ-like family members identified in several Cupriavidus and Ralstonia proteomes by BLASTp analysis of the non-redundant protein sequence database with PrsQ1 as a query. When applied in Biogrep analysis, this pattern detected exactly and exclusively all 22 CopQ-like family members in strain CH34.
917 protein sequences, was queried by the multi-threaded pattern-matching tool Biogrep 1.079 using an extended 10-aa search pattern [GSRDEQ][GSAKRTPN]xD[PVT][YF][TS][DEQ]G[ASG] covering the Dx[YF]xxG domain motif characteristic for CopQ-like family proteins. This extended search pattern is based on a multiple alignment and additional visual inspection of the protein sequences of CopQ-like family members identified in several Cupriavidus and Ralstonia proteomes by BLASTp analysis of the non-redundant protein sequence database with PrsQ1 as a query. When applied in Biogrep analysis, this pattern detected exactly and exclusively all 22 CopQ-like family members in strain CH34.
Transmission electron microscopy
Bacterial cells were adhered to a copper EM-grid (300 mesh) by depositing 3 μl of cell suspension on the grid and drying in an evacuated desiccator. Bacteria were observed in a JEOL JEM2100 LaB6 TEM operated at 200 kV, equipped with an Oxford Instruments (UK) EDX-system and STEM detectors. Images were recorded with a Gatan multiscan 1k × 1k pxls camera. For X-ray spectra, areas of the sample were scanned using the bright field STEM detector.Conclusions
In summary, we showed that C. metallidurans is able to adapt to toxic silver concentrations by activating latent mechanisms that are not related to the known metal or silver resistance determinants. A single mutation was sufficient for activation and provided resistance to much higher silver concentrations. Previously uncharacterized small periplasmic proteins carrying distinctive sequence motifs played a central role in this adaptation. Moreover, they appear to be exclusively found in members of the Cupriavidus and Ralstonia genera, suggesting a specific adaptation potential to metal and putative other stressors in these genera that are able to live and thrive in harsh and oligotrophic environments. Finally, the increased resistance is most probably facilitated by the accumulation of silver ions and the formation of silver nanoparticles.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the European Space Agency (ESA-PRODEX) and the Belgian Science Policy (Belspo) through the COMICS project (C90356). The authors want to thank Dr Pieter Baatsen from the VIB BIO Imaging Core electron microscopy facility (KULeuven, Belgium) for conducting the TEM and EDX analysis of the silver resistant mutant.Notes and references
- S. Silver, T. Phung le and G. Silver, Silver as biocides in burn and wound dressings and bacterial resistance to silver compounds, J. Ind. Microbiol. Biotechnol., 2006, 33, 627–634 CrossRef CAS PubMed.
- V. Edwards-Jones, The benefits of silver in hygiene, personal care and healthcare, Lett. Appl. Microbiol., 2009, 49, 147–152 CrossRef CAS PubMed.
- K. Chaloupka, Y. Malam and A. M. Seifalian, Nanosilver as a new generation of nanoproduct in biomedical applications, Trends Biotechnol., 2010, 28, 580–588 CrossRef CAS PubMed.
- M. E. Vance, T. Kuiken, E. P. Vejerano, S. P. McGinnis, M. F. Hochella, Jr., D. Rejeski and M. S. Hull, Nanotechnology in the real world: Redeveloping the nanomaterial consumer products inventory, Beilstein J. Nanotechnol., 2015, 6, 1769–1780 CrossRef CAS PubMed.
- N. Duran, M. Duran, M. B. de Jesus, A. B. Seabra, W. J. Favaro and G. Nakazato, Silver nanoparticles: A new view on mechanistic aspects on antimicrobial activity, Nanomedicine, 2016, 12, 789–799 CrossRef CAS PubMed.
- V. De Matteis, M. A. Malvindi, A. Galeone, V. Brunetti, E. De Luca, S. Kote, P. Kshirsagar, S. Sabella, G. Bardi and P. P. Pompa, Negligible particle-specific toxicity mechanism of silver nanoparticles: The role of Ag+ ion release in the cytosol, Nanomedicine, 2015, 11, 731–739 CrossRef CAS PubMed.
- K. Mijnendonckx, N. Leys, J. Mahillon, S. Silver and R. V. Houdt, Antimicrobial silver: uses, toxicity and potential for resistance, Biometals, 2013, 26, 609–621 CrossRef CAS PubMed.
- O. Gordon, T. Vig Slenters, P. S. Brunetto, A. E. Villaruz, D. E. Sturdevant, M. Otto, R. Landmann and K. M. Fromm, Silver coordination polymers for prevention of implant infection: thiol interaction, impact on respiratory chain enzymes, and hydroxyl radical induction, Antimicrob. Agents Chemother., 2010, 54, 4208–4218 CrossRef CAS PubMed.
- A. D. Russell and W. B. Hugo, Antimicrobial activity and action of silver, Prog. Med. Chem., 1994, 31, 351–370 CrossRef CAS PubMed.
- H. J. Park, J. Y. Kim, J. Kim, J. H. Lee, J. S. Hahn, M. B. Gu and J. Yoon, Silver-ion-mediated reactive oxygen species generation affecting bactericidal activity, Water Res., 2009, 43, 1027–1032 CrossRef CAS PubMed.
- E. McGillicuddy, I. Murray, S. Kavanagh, L. Morrison, A. Fogarty, M. Cormican, P. Dockery, M. Prendergast, N. Rowan and D. Morris, Silver nanoparticles in the environment: Sources, detection and ecotoxicology, Sci. Total Environ, 2017, 575, 231–246 CrossRef CAS PubMed.
- K. Schlich, T. Klawonn, K. Terytze and K. Hund-Rinke, Hazard assessment of a silver nanoparticle in soil applied via sewage sludge, Environ. Sci. Eur., 2013, 25, 17 CrossRef.
- G. E. Batley, J. K. Kirby and M. J. McLaughlin, Fate and risks of nanomaterials in aquatic and terrestrial environments, Acc. Chem. Res., 2013, 46, 854–862 CrossRef CAS PubMed.
- SCENIHR, Assessment of the antibiotic resistance effects of biocides, European Commission, Brussels, 2009.
- N. Seltenrich, Nanosilver: weighing the risks and benefits, Environ. Health Perspect., 2013, 121, A220–A225 Search PubMed.
- P. J. Finley, R. Norton, C. Austin, A. Mitchell, S. Zank and P. Durham, Unprecedented silver resistance in clinically isolated Enterobacteriaceae: Major implications for burn and wound management, Antimicrob. Agents Chemother., 2015, 59, 4734–4741 CrossRef CAS PubMed.
- G. L. McHugh, R. C. Moellering, C. C. Hopkins and M. N. Swartz, Salmonella typhimurium resistant to silver nitrate, chloramphenicol, and ampicillin, Lancet, 1975, 1, 235–240 CrossRef CAS.
- C. Haefeli, C. Franklin and K. Hardy, Plasmid-determined silver resistance in Pseudomonas stutzeri isolated from a silver mine, J. Bacteriol., 1984, 158, 389–392 CAS.
- P. Choudhury and R. Kumar, Multidrug- and metal-resistant strains of Klebsiella pneumoniae isolated from Penaeus monodon of the coastal waters of deltaic Sundarban, Can. J. Microbiol., 1998, 44, 186–189 CrossRef CAS PubMed.
- T. Klaus, R. Joerger, E. Olsson and C. G. Granqvist, Silver-based crystalline nanoparticles, microbially fabricated, Proc. Natl. Acad. Sci. U. S. A., 1999, 96, 13611–13614 CrossRef CAS PubMed.
- S. Silver, A. Gupta, K. Matsui and J. F. Lo, Resistance to Ag+ cations in bacteria: environments, genes and proteins, Met. – Based Drugs, 1999, 6, 315–320 CrossRef CAS PubMed.
- M. Zimmermann, S. R. Udagedara, C. M. Sze, T. M. Ryan, G. J. Howlett, Z. Xiao and A. G. Wedd, PcoE - A metal sponge expressed to the periplasm of copper resistance Escherichia coli. Implication of its function role in copper resistance, J. Inorg. Biochem., 2012, 115, 186–197 CrossRef CAS PubMed.
- J. Goris, P. D. Vos, T. Coenye, B. Hoste, D. Janssens, H. Brim, L. Diels, M. Mergeay, K. Kersters and P. Vandamme, Classification of metal-resistant bacteria from industrial biotopes as Ralstonia campinensis sp. nov., Ralstonia metallidurans sp. nov. and Ralstonia basilensis, Int. J. Syst. Evol. Microbiol., 2001, 51, 1773–1782 CrossRef CAS PubMed.
- H. Brim, M. Heyndrickx, P. de Vos, A. Wilmotte, D. Springael, H. G. Schlegel and M. Mergeay, Amplified rDNA restriction analysis and further genotypic characterization of metal-resistant soil bacteria and related facultative hydrogenotrophs, Syst. Appl. Microbiol., 1999, 22, 258–268 CrossRef CAS PubMed.
- L. Diels and M. Mergeay, DNA probe-mediated detection of resistant bacteria from soils highly polluted by heavy metals, Appl. Environ. Microbiol., 1990, 56, 1485–1491 CAS.
- M. T. La Duc, W. Nicholson, R. Kern and K. Venkateswaran, Microbial characterization of the Mars Odyssey spacecraft and its encapsulation facility, Environ. Microbiol., 2003, 5, 977–985 CrossRef CAS PubMed.
- C. M. Ott, R. J. Bruce and D. L. Pierson, Microbial characterization of free floating condensate aboard the Mir space station, Microb. Ecol., 2004, 47, 133–136 CrossRef CAS PubMed.
- P. Monsieurs, K. Mijnendonckx, A. Provoost, K. Venkateswaran, C. M. Ott, N. Leys and R. Van Houdt, Genome sequences of Cupriavidus metallidurans Strains NA1, NA4, and NE12, isolated from space equipment, Genome Announcements, 2014, 2(4), e00719–14 CrossRef PubMed.
- T. Coenye, T. Spilker, R. Reik, P. Vandamme and J. J. Lipuma, Use of PCR analyses to define the distribution of Ralstonia species recovered from patients with cystic fibrosis, J. Clin. Microbiol., 2005, 43, 3463–3466 CrossRef CAS PubMed.
- R. Van Houdt, P. Monsieurs, K. Mijnendonckx, A. Provoost, A. Janssen, M. Mergeay and N. Leys, Variation in genomic islands contribute to genome plasticity in Cupriavidus metallidurans, BMC Genomics, 2012, 13, 111 CrossRef CAS PubMed.
- S. Langevin, J. Vincelette, S. Bekal and C. Gaudreau, First case of invasive human infection caused by Cupriavidus metallidurans, J. Clin. Microbiol., 2011, 49, 744–745 CrossRef PubMed.
- P. Monsieurs, A. Provoost, K. Mijnendonckx, N. Leys, C. Gaudreau and R. Van Houdt, Genome sequence of Cupriavidus metallidurans Strain H1130, isolated from an invasive human infection, Genome Announcements, 2013, 1(6), e01051–13 CrossRef PubMed.
- R. Van Houdt, S. Monchy, N. Leys and M. Mergeay, New mobile genetic elements in Cupriavidus metallidurans CH34, their possible roles and occurrence in other bacteria, Antonie van Leeuwenhoek, 2009, 96, 205–226 CrossRef CAS PubMed.
- K. Mijnendonckx, A. Provoost, P. Monsieurs, N. Leys, M. Mergeay, J. Mahillon and R. V. Houdt, Insertion sequence elements in Cupriavidus metallidurans CH34: Distribution and role in adaptation, Plasmid, 2011, 65, 193–203 CrossRef CAS PubMed.
- V. Auquier, Docteur en Sciences Agronomiques et Ingénierie Biologique, Université Libre de Bruxelles, 2006 Search PubMed.
- M. Mergeay, S. Monchy, T. Vallaeys, V. Auquier, A. Benotmane, P. Bertin, S. Taghavi, J. Dunn, D. van der Lelie and R. Wattiez, Ralstonia metallidurans, a bacterium specifically adapted to toxic metals: towards a catalogue of metal-responsive genes, FEMS Microbiol. Rev., 2003, 27, 385–410 CrossRef CAS PubMed.
- P. Monsieurs, H. Moors, R. Van Houdt, P. J. Janssen, A. Janssen, I. Coninx, M. Mergeay and N. Leys, Heavy metal resistance in Cupriavidus metallidurans CH34 is governed by an intricate transcriptional network, Biometals, 2011, 24, 1133–1151 CrossRef CAS PubMed.
- B. Bersch, K. M. Derfoufi, F. De Angelis, V. Auquier, E. Ngonlong Ekende, M. Mergeay, J. M. Ruysschaert and G. Vandenbussche, Structural and metal binding characterization of the C-terminal metallochaperone domain of membrane fusion Protein SilB from Cupriavidus metallidurans CH34, Biochemistry, 2011, 50, 2194–2204 CrossRef CAS PubMed.
- M. Mergeay, D. Nies, H. G. Schlegel, J. Gerits, P. Charles and F. V. Gijsegem, Alcaligenes eutrophus CH34 is a facultative chemolithotroph with plasmid-bound resistance to heavy metals, J. Bacteriol., 1985, 162, 328–334 CAS.
- S. Monchy, M. A. Benotmane, P. Janssen, T. Vallaeys, S. Taghavi, D. van der Lelie and M. Mergeay, Plasmids pMOL28 and pMOL30 of Cupriavidus metallidurans are specialized in the maximal viable response to heavy metals, J. Bacteriol., 2007, 189, 7417–7425 CrossRef CAS PubMed.
- L. Bobe, A. Kochetkov, V. Soloukhin, P. Andreichuk, N. Protasov and Y. Sinyak, International conference on environmental systems, SAE International, San Francisco, CA, USA, 2008.
- K. Mijnendonckx, A. Provoost, C. M. Ott, K. Venkateswaran, J. Mahillon, N. Leys and R. Van Houdt, Characterization of the survival ability of Cupriavidus metallidurans and Ralstonia pickettii from space-related environments, Microb. Ecol., 2013, 56, 347–360 CrossRef PubMed.
- M. M. Ali, A. Provoost, L. Maertens, N. Leys, P. Monsieurs, D. Charlier and R. Van Houdt, Genomic and transcriptomic changes that mediate increased platinum resistance in Cupriavidus metallidurans, Genes, 2019, 10, E63 CrossRef PubMed.
- P. J. Janssen, R. Van Houdt, H. Moors, P. Monsieurs, N. Morin, A. Michaux, M. A. Benotmane, N. Leys, T. Vallaeys, A. Lapidus, S. Monchy, C. Medigue, S. Taghavi, S. McCorkle, J. Dunn, D. van der Lelie and M. Mergeay, The complete genome sequence of Cupriavidus metallidurans strain CH34, a master survivalist in harsh and anthropogenic environments, PLoS One, 2010, 5, e10433 CrossRef PubMed.
- D. Vallenet, L. Labarre, Z. Rouy, V. Barbe, S. Bocs, S. Cruveiller, A. Lajus, G. Pascal, C. Scarpelli and C. Medigue, MaGe: a microbial genome annotation system supported by synteny results, Nucleic Acids Res., 2006, 34, 53–65 CrossRef CAS PubMed.
- J. Vandecraen, M. Chandler, A. Aertsen and R. Van Houdt, The impact of insertion sequences on bacterial genome plasticity and adaptability, Crit. Rev. Microbiol., 2017, 43, 709–730 CrossRef CAS PubMed.
- M. Claeys, V. Storms, H. Sun, T. Michoel and K. Marchal, MotifSuite: workflow for probabilistic motif detection and assessment, Bioinformatics, 2012, 28, 1931–1932 CrossRef CAS PubMed.
- F. P. Roth, J. D. Hughes, P. W. Estep and G. M. Church, Finding DNA regulatory motifs within unaligned noncoding sequences clustered by whole-genome mRNA quantitation, Nat. Biotechnol., 1998, 16, 939 CrossRef CAS PubMed.
- T. L. Bailey and C. Elkan, Fitting a mixture model by expectation maximization to discover motifs in biopolymers, Proc. Int. Conf. Intell. Syst. Mol. Biol., 1994, 2, 28–36 CAS.
- G. Pavesi, G. Mauri and G. Pesole, An algorithm for finding signals of unknown length in DNA sequences, Bioinformatics, 2001, 17(suppl. 1), S207–S214 CrossRef PubMed.
- A. Medina-Rivera, M. Defrance, O. Sand, C. Herrmann, J. A. Castro-Mondragon, J. Delerce, S. Jaeger, C. Blanchet, P. Vincens, C. Caron, D. M. Staines, B. Contreras-Moreira, M. Artufel, L. Charbonnier-Khamvongsa, C. Hernandez, D. Thieffry, M. Thomas-Chollier and J. van Helden, RSAT 2015: Regulatory Sequence Analysis Tools, Nucleic Acids Res., 2015, 43, W50–W56 CrossRef CAS PubMed.
- A. Marina, C. Mott, A. Auyzenberg, W. A. Hendrickson and C. D. Waldburger, Structural and mutational analysis of the PhoQ histidine kinase catalytic domain. Insight into the reaction mechanism, J. Biol. Chem., 2001, 276, 41182–41190 CrossRef CAS PubMed.
- R. J. Lewis, J. A. Brannigan, K. N. Muchová, I. Barák and A. J. Wilkinson, Phosphorylated aspartate in the structure of a response regulator protein, J. Mol. Biol., 1999, 294, 9–15 CrossRef CAS PubMed.
- G. S. Lukat, B. H. Lee, J. M. Mottonen, A. M. Stock and J. B. Stock, Roles of the highly conserved aspartate and lysine residues in the response regulator of bacterial chemotaxis, J. Biol. Chem., 1991, 266, 8348–8354 CAS.
- M. Kallberg, H. Wang, S. Wang, J. Peng, Z. Wang, H. Lu and J. Xu, Template-based protein structure modeling using the RaptorX web server, Nat. Protoc., 2012, 7, 1511–1522 CrossRef CAS PubMed.
- L. A. Kelley, S. Mezulis, C. M. Yates, M. N. Wass and M. J. Sternberg, The Phyre2 web portal for protein modeling, prediction and analysis, Nat. Protoc., 2015, 10, 845–858 CrossRef CAS PubMed.
- J. R. Morones-Ramirez, J. A. Winkler, C. S. Spina and J. J. Collins, Silver enhances antibiotic activity against Gram-negative bacteria, Sci. Transl. Med., 2013, 5, 190ra181 Search PubMed.
- J. A. Garza-Cervantes, A. Chavez-Reyes, E. C. Castillo, G. Garcia-Rivas, O. Antonio Ortega-Rivera, E. Salinas, M. Ortiz-Martinez, S. L. Gomez-Flores, J. A. Pena-Martinez, A. Pepi-Molina, M. T. Trevino-Gonzalez, X. Zarate, M. Elena Cantu-Cardenas, C. Enrique Escarcega-Gonzalez and J. R. Morones-Ramirez, Synergistic antimicrobial effects of silver/transition-metal combinatorial treatments, Sci. Rep., 2017, 7, 903 CrossRef PubMed.
- J. L. Graves, Jr., M. Tajkarimi, Q. Cunningham, A. Campbell, H. Nonga, S. H. Harrison and J. E. Barrick, Rapid evolution of silver nanoparticle resistance in Escherichia coli., Front. Genet., 2015, 6, 42 Search PubMed.
- C. P. Randall, A. Gupta, N. Jackson, D. Busse and A. J. O'Neill, Silver resistance in Gram-negative bacteria: a dissection of endogenous and exogenous mechanisms, J. Antimicrob. Chemother., 2015, 70, 1037–1046 CAS.
- SCENIHR, Opinion on the guidance on the determination of potential health effects of nanomaterials Used in Medical Devices, European Commission, Luxemburg, 2015.
- K. Perron, O. Caille, C. Rossier, C. Van Delden, J. L. Dumas and T. Kohler, CzcR-CzcS, a two-component system involved in heavy metal and carbapenem resistance in Pseudomonas aeruginosa, J. Biol. Chem., 2004, 279, 8761–8768 CrossRef CAS PubMed.
- I. Marchand, L. Damier-Piolle, P. Courvalin and T. Lambert, Expression of the RND-type efflux pump AdeABC in Acinetobacter baumannii is regulated by the AdeRS two-component system, Antimicrob. Agents Chemother., 2004, 48, 3298–3304 CrossRef CAS PubMed.
- K. R. Asiani, H. Williams, L. Bird, M. Jenner, M. S. Searle, J. L. Hobman, D. J. Scott and P. Soultanas, SilE is an intrinsically disordered periplasmic “molecular sponge” involved in bacterial silver resistance, Mol. Microbiol., 2016, 101, 731–742 CrossRef CAS PubMed.
- R. Y. Parikh, S. Singh, B. L. Prasad, M. S. Patole, M. Sastry and Y. S. Shouche, Extracellular synthesis of crystalline silver nanoparticles and molecular evidence of silver resistance from Morganella sp.: towards understanding biochemical synthesis mechanism, ChemBioChem, 2008, 9, 1415–1422 CrossRef CAS PubMed.
- P. J. Finley, R. Norton, C. Austin, A. Mitchell, S. Zank and P. Durham, Unprecedented Silver Resistance in Clinically Isolated Enterobacteriaceae: Major Implications for Burn and Wound Management, Antimicrob. Agents Chemother., 2015, 59, 4734–4741 CrossRef CAS PubMed.
- R. H. Sedlak, M. Hnilova, C. Grosh, H. Fong, F. Baneyx, D. Schwartz, M. Sarikaya, C. Tamerler and B. Traxler, Engineered Escherichia coli silver-binding periplasmic protein that promotes silver tolerance, Appl. Environ. Microbiol., 2012, 78, 2289–2296 CrossRef CAS PubMed.
- L. Ferrieres, G. Hemery, T. Nham, A. M. Guerout, D. Mazel, C. Beloin and J. M. Ghigo, Silent mischief: bacteriophage Mu insertions contaminate products of Escherichia coli random mutagenesis performed using suicidal transposon delivery plasmids mobilized by broad-host-range RP4 conjugative machinery, J. Bacteriol., 2010, 192, 6418–6427 CrossRef CAS PubMed.
- K. Mijnendonckx, A. Provoost, C. M. Ott, K. Venkateswaran, J. Mahillon, N. Leys and R. Van Houdt, Characterization of the Survival Ability of Cupriavidus metallidurans and Ralstonia pickettii from Space-Related Environments, Microb. Ecol., 2013, 65, 347–360 CrossRef CAS PubMed.
- H. Li and R. Durbin, Fast and accurate short read alignment with Burrows-Wheeler transform, Bioinformatics, 2009, 25, 1754–1760 CrossRef CAS PubMed.
- H. Li, B. Handsaker, A. Wysoker, T. Fennell, J. Ruan, N. Homer, G. Marth, G. Abecasis and R. Durbin, and S. Genome Project Data Processing, The Sequence Alignment/Map format and SAMtools, Bioinformatics, 2009, 25, 2078–2079 CrossRef CAS PubMed.
- D. Vallenet, L. Labarre, Z. Rouy, V. Barbe, S. Bocs, S. Cruveiller, A. Lajus, G. Pascal, C. Scarpelli and C. Medigue, MaGe: a microbial genome annotation system supported by synteny results, Nucleic Acids Res., 2006, 34, 53–65 CrossRef CAS PubMed.
- L. M. Guzman, D. Belin, M. J. Carson and J. Beckwith, Tight regulation, modulation, and high-level expression by vectors containing the arabinose pBAD promoter, J. Bacteriol., 1995, 177, 4121–4130 CrossRef CAS PubMed.
- M. E. Kovach, P. H. Elzer, D. S. Hill, G. T. Robertson, M. A. Farris, R. M. Roop, 2nd and K. M. Peterson, Four new derivatives of the broad-host-range cloning vector pBBR1MCS, carrying different antibiotic-resistance cassettes, Gene, 1995, 166, 175–176 CrossRef CAS PubMed.
- A. Schafer, A. Tauch, W. Jager, J. Kalinowski, G. Thierbach and A. Puhler, Small mobilizable multi-purpose cloning vectors derived from the Escherichia coli plasmids pK18 and pK19: selection of defined deletions in the chromosome of Corynebacterium glutamicum, Gene, 1994, 145, 69–73 CrossRef CAS PubMed.
- M. Bagdasarian, R. Lurz, B. Ruckert, F. C. H. Franklin, M. M. Bagdasarian, J. Frey and K. N. Timmis, Specific-Purpose Plasmid Cloning Vectors 2. Broad Host Range, High Copy Number, RSF1010-Derived Vectors, and a Host-Vector System for Gene Cloning in Pseudomonas, Gene, 1981, 16, 237–247 CrossRef CAS PubMed.
- W. G. Miller, J. H. Leveau and S. E. Lindow, Improved gfp and inaZ broad-host-range promoter-probe vectors, Mol. Plant-Microbe Interact., 2000, 13, 1243–1250 CrossRef CAS PubMed.
- S. T. Cardona and M. A. Valvano, An expression vector containing a rhamnose-inducible promoter provides tightly regulated gene expression in, Burkholderia cenocepacia Plasmid, 2005, 54, 219–228 CrossRef CAS PubMed.
- K. Jensen, PhD, Massachusetts Institute of Technology, 2006.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9mt00123a |
| ‡ Present address: Unit Health, Flemish Institute for Technological Research (VITO), Belgium. |
| This journal is © The Royal Society of Chemistry 2019 |