The impact of nanoparticle shape on cellular internalisation and transport: what do the different analysis methods tell us?
Wenqian
Wang
abc,
Katharina
Gaus
bde,
Richard D.
Tilley
 abf and
J. Justin
Gooding
abf and
J. Justin
Gooding
 *abc
*abc
aSchool of Chemistry, The University of New South Wales, Sydney, NSW 2052, Australia. E-mail: justin.gooding@unsw.edu.au
bAustralian Centre for NanoMedicine, The University of New South Wales, Sydney, NSW 2052, Australia
cARC Centre of Excellence in Convergent Bio-Nano Science and Technology, The University of New South Wales, Sydney, NSW 2052, Australia
dEMBL Australia Node in Single Molecule Science, The University of New South Wales, Sydney, NSW 2052, Australia
eARC Centre of Excellence in Advanced Molecular Imaging, The University of New South Wales, Sydney, NSW 2052, Australia
fElectron Microscope Unit, Mark Wainwright Analytical Centre, The University of New South Wales, Sydney, NSW 2052, Australia
First published on 6th July 2019
Abstract
This article focuses on how nanoparticle shape affects the cellular internalisation and transport of nanoparticles inside cells and what the different analytical methods for determining nanoparticle internalisation by cells tell us. Rod-shaped nanoparticles typically show greater cellular internalisation relative to spherical nanoparticles, although there are studies with contradictory conclusions. The contradiction may be a result of differences in the materials being used in the comparison and/or a result of the analytical method employed. Finally, future opportunities in studying cellular internalisation with 3D cell-culture models and light-sheet microscopy are discussed.
Introduction
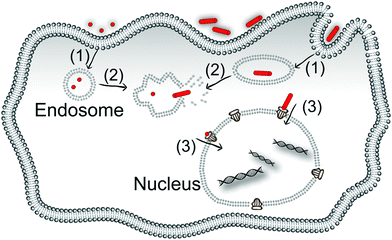
Targeting nanoparticles to specific subcellular structures inside the cell has the potential to significantly improve the treatment efficiency of nanomedicine because many drugs are effective in specific subcellular locations.1–3 The nucleus is one of the most important locations with regards to the viability and function of the cells. Preparing nanoparticles which can move into the nucleus and then release their cargo in this organelle is important for gene modification and anti-tumour therapy; two of the most intensely studied areas of research in nanomedicine.There are several cellular barriers that nanoparticles must cross before they can enter the nucleus. In the simplest case, these barriers include crossing the plasma membrane, escaping the endosomes or lysosomes, and crossing the nuclear envelope. Different nanoparticles can have different mechanisms and efficacies at surmounting these barriers, which in turn determines the efficiency of the delivery of the payload of the nanoparticles to the desired location in the cell (Scheme 1).
The plasma membrane is the first barrier for nanoparticle internalisation, and different kinds of nanoparticles cross the plasma membrane via different internalisation pathways. Very small nanoparticles (a few nanometres, like gold nanoparticles4 and carbon nanotubes5) can penetrate the plasma membrane directly or passively diffuse into the cell. Nanoparticles with sizes in the range of sub-micrometres to micrometres will be internalised mostly through pinocytosis and phagocytosis.6–10 Then, nanoparticles, which are already inside the cells through pinocytosis and phagocytosis, are typically encapsulated within endosomes, which they must escape, or they will finally be transferred to the lysosomes and excreted out of the cells. Thirdly, after escaping from the endosomes, nanoparticles can freely diffuse in the cytoplasm and have a chance to enter the nucleus. The efficiency of crossing the nuclear envelope is limited by the size of the nanoparticles, which must be small enough to pass through the nuclear pore complex.
The physicochemical properties of nanoparticles, such as size, surface charge, and shape can influence the internalisation efficiency of the nanoparticles and how they respond to the challenge of crossing the different cellular barriers. For spherical nanoparticles, 50 nm diameter is often regarded as the optimal size for cellular internalisation by non-phagocytic cells and the uptake efficiency will decrease for larger particles. However, there is less consensus on the optimal size for phagocytic cells.11,12 Apart from the effect of size, greater positive or negative surface charge can lead to an increase in uptake of the nanoparticles compared with less charged or uncharged nanoparticles.11,13 Positively charged particles are typically internalised more extensively than negatively charged nanoparticles, for a similar absolute value of the zeta potentials.14 In addition, the shape of nanoparticles has an effect on the cellular uptake efficiency. Most commonly, high aspect ratio nanoparticles such as rods and tubes show a higher cellular uptake efficiency and drug delivery efficiency than spheres.15–17 We use the word ‘most’ here as there are some contradictory studies that suggest the opposite.11,12 These contradictory results may be a result of the difference in size range, the material composition of the nanoparticles, different cell types or different measurement tools used to evaluate the uptake. In this focus paper, how the shape of nanoparticles influences the internalisation efficiency will be discussed along with the size of the nanoparticles in every dimension, different measurement tools employed and how the information provided by these methods can influence the conclusions arrived at.
The shape influences the internalisation of nanoparticles
Are rods better than spheres?
Incubating nanoparticles with cells seeded on a surface [in other words, two-dimension (2D) cell culture] is the most common experimental construct to study the cellular internalisation of nanoparticles. This experimental construct faces challenges with particle sedimentation, giving artificially high particle delivery efficiencies, but its commonality makes it highly useful for comparing studies on the impact of nanoparticle shape.18,19 The dominant conclusion of studies on nanoparticle shape and delivery to cells is that rod-shape nanoparticles have greater cellular internalisation than the equivalent spherical nanoparticles for a range of materials.19–26 For example, Zhang et al. reported that the number of internalised gold nanorods [12 nm (diameter) × 50 nm (length)] was almost 1.5 times higher than the gold nanospheres [22 nm] in HepG2 cells (human liver cancer cell line).23 Similarly, Meng et al. demonstrated that for three kinds of silica nanorods [70 nm (diameter) × 120 nm (length); 75 nm (diameter) × 175 nm (length); 60 nm (diameter) × 280 nm (length)], the amount of nanorods internalised was significantly greater than for silica nanospheres [110 nm]. This was most notable for the silica nanorods with the aspect ratio of 2.1–2.5 which exhibited the greatest amount of internalisation in HeLa cells (human cervical cancer cell line) and A549 cells (human lung cancer cell line) (Fig. 1).21 Agarwal et al. demonstrated that long polyethylene glycol diacrylate (PEGDA)-based hydrogel nanorods [100 nm × 100 nm × 800 nm] could be endocytosed to a greater extent than their shorter counterparts [100 nm × 100 nm × 400 nm]. In the same study, they also showed that large size nanodisks [325 nm (diameter) × 100 nm (length)] were internalised more efficiently than their small counterparts [220 nm (diameter) × 100 nm (length)].19 Importantly from these three examples, we noticed that this conclusion has been reached with both hard inorganic and soft polymer materials and for three of the most common materials, gold, silica and polyethylene glycol-based polymers, used in drug delivery.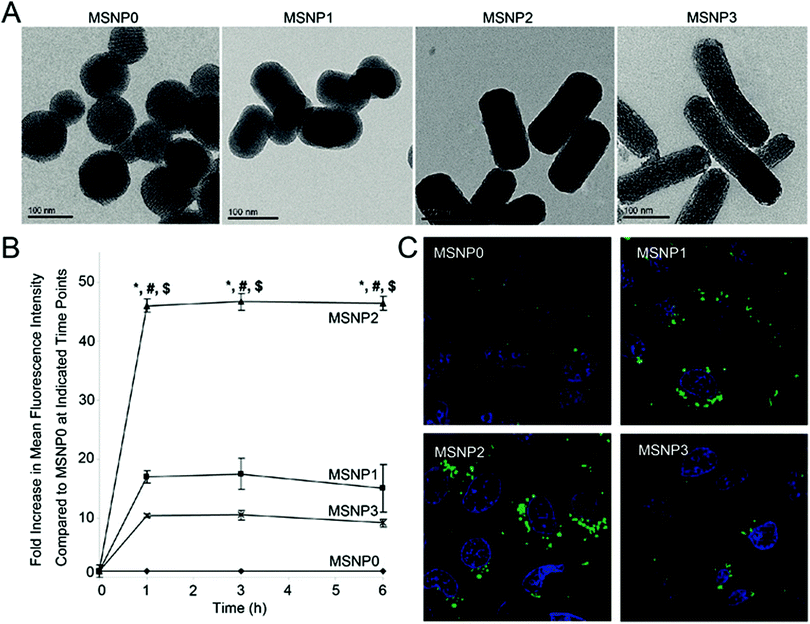 | ||
Fig. 1 (A) Transmission electron microscopy images of silica nanoparticles with different aspect ratio values. Aspect ratio values of MSNP0, MSNP1, MSNP2, and MSNP3 are 1–1.2, 1.5–1.7, 2.1–2.5, 4.0–4.5 respectively. (B) Fold increase in mean fluorescence intensity of the FITC-labelled silica rods compared to spheres at 0 to 6 h. HeLa cells were exposed to different FITC-labelled silica nanoparticles at 20 μg mL−1, and flow cytometry was conducted at the indicated time points. *![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) p < 0.05 compared with the spherical FITC-labelled particle (MSNP0); # p < 0.05 compared with the spherical FITC-labelled particle (MSNP0); #![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) p < 0.05 compared with FITC-labelled MSNP1; $ p < 0.05 compared with FITC-labelled MSNP1; $![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) p < 0.05 compared with FITC-labelled MSNP3. (C) Fluorescent image of the FITC-labelled silica nanoparticles (green) with different aspect ratio values inside HeLa cells. The nuclei (blue) were stained with Hoechst 33342. Reprinted with permission from ref. 21. Copyright 2011 American Chemical Society. p < 0.05 compared with FITC-labelled MSNP3. (C) Fluorescent image of the FITC-labelled silica nanoparticles (green) with different aspect ratio values inside HeLa cells. The nuclei (blue) were stained with Hoechst 33342. Reprinted with permission from ref. 21. Copyright 2011 American Chemical Society. | ||
Two other conclusions that suggest that rod-shaped nanoparticles may be superior for drug delivery relative to spherical nanoparticles are first, that rod-shaped nanoparticles have longer retention time inside the cells than the spherical ones.25,27 This longer residence time inside cells can be attributed to non-spherical particles having higher efficiency of escaping organelles than spherical ones.28–30 Second, several studies suggest that rod-shaped nanoparticles that are less than 40 nm in width can pass across the nuclear envelope through the nuclear pore complex and carry more cargo into the nucleus where it is needed, than spheres which have similar diameter.31–33 This conclusion is consistent with electron microscopy studies on the central opening size of the nuclear pore complex.34–36
Three-dimensional (3D) cell cultures, including balls of cancer cells such as spheroids and organoids, have been proposed as superior in vitro models for the study of the internalisation of nanoparticles in vivo than conventional 2D cultures.37 Unlike 2D cell culture, a 3D cell culture allows cells to grow in all directions which provides an opportunity to study nanoparticles transport in solid tissues in a more controlled microenvironment than in real tissue. The conclusions on the effect of nanoparticle shape on cellular internalisation does not change greatly when going from 2D to 3D cultures. Again, in 3D, rod-shaped nanoparticles exhibit greater internalisation efficiency. Rod-shaped nanoparticles also exhibit better 3D penetration and much more uniform distribution inside the spheroid than their spherical counterparts,38–40 which does significantly influence the therapeutic and diagnostic efficiency of nanomedicine. Agarwal et al. demonstrated that high aspect ratio rods [325 nm (diameter) × 100 nm (length)] showed maximal delivery and more uniform penetration compared with low aspect ratio ones [220 nm (diameter) × 100 nm (length)].38 The penetration ability of high aspect ratio nanoparticles has also been demonstrated within living tissue. Fernandes et al. reported that rod-shaped nanoparticles [20 nm (diameter) × 60 nm (length)] were found in the skin tissue in higher numbers than their spherical counterparts [15 nm].39 Black et al. also reported that gold nanorods [6–9 nm (diameter) × 30–50 nm (length)] had higher tumour tissue penetration than their spherical counterparts [50–60 nm].40
Are spheres better than rods?
There are however contradictory studies by several other groups that report spherical particles show advantages over their rod counterparts during cellular uptake, and increasing the aspect ratio of the nanoparticles decreases the total cell internalisation.27,41–47 For example, Zhang et al. demonstrate that small spherical poly(acrylic acid)-b-polystyrene nanoparticles [11 nm] have a higher cellular uptake efficiency than large rod-shaped nanoparticles [20 nm (diameter) × 180 nm (length); 30 nm (diameter) × 1000 nm (length)] when they were incubated with CHO cells (Chinese hamster ovary cells).27 Zhao et al. also reported that increasing the aspect ratio decreased the amount of cellular uptake with rod-shaped fructose-based polymeric nanoparticles that have aspect ratios over 13.46,47 Chithrani et al. demonstrated gold nanospheres [14 nm] were taken up more than gold nanorods [14 (diameter) × 40 nm (length); 14 nm (diameter) × 74 nm (length)] by HeLa cells.44 Shi et al. demonstrated that short rod-shaped polymeric nanoparticles [25 (diameter) × 74 nm (length)] could accumulate more in HeLa cells than the long rod-shaped nanoparticles [18 (diameter) × 102 nm (length)] even though both showed a similar percentage of cellular association.41 Similar results also exist when using 3D cell culture as a study model; Dias et al. suggested that spherical gold core–mesoporous silica shell nanoparticles [110 nm] displayed a more homogeneous distribution and penetration in 3D tumour spheroids than the rod-shaped ones [47 nm (diameter) × 70 nm (length)].48All these results are consistent only in the conclusion that the shape of the nanoparticles can influence cellular uptake. The results really reflect the complexity of cellular uptake mechanisms and intracellular transport routes and processes. Differences in the cell types could influence the conclusions, for example, it is believed that macrophages always uptake much more spherical nanoparticles than their rod counterparts.45,49,50 Reasons for differing results also can be attributed to differences in the surface charge density and surface chemistry of the carriers (and how these influence the fouling of the nanoparticle in the biological milieu), the rigidity of the particles, the distribution of the nanoparticle size in a sample, and difference in the material compositions.
Comments about different results possibly being due to cell types, differences in particle flexibility, simply differences in size, surface chemistry, the stability of the surface chemistry, how the nanoparticle-protein corona forms during nanoparticle fouling in biological solutions, and the actual size distributions are all valid but somehow do not illuminate as to actually why we see the differences for specific situations.
For example, for methacrylate nanoparticles, two studies by Shimoni et al. and Zhao et al. compare spheres to nanorods and show that the spheres have greater uptake. In both these studies, the nanorods are very long, 900 nm in one case, 1300 nm in the other.42,46,47 At these lengths it is likely that the rods are quite flexible and hence perhaps the spheres are compared with ill-defined shaped nanoparticles. When in our own work we compared the uptake of rods versus much longer flexible worm-like nanoparticles, we saw greater uptake of nanorods than the worms.31 Similarly, divergent views of rods versus spheres for the polystyrene particles could be due to the differences in size. In one example where more spheres are internalised than nanorods, the spheres were 100 nm in diameter while the nanorods were 84 nanometers in their shortest dimensions.43 In a contradictory study, nanorods of 80 nm in their shortest dimensions were more efficiently internalised over spheres 220 nm in diameter.24 Further ambiguity in these two studies arises from the fact that the surface coating for the former is sulphonate terminated43 whilst the latter is carboxylate terminated.24 Differences in the surface chemistry [CTAB (Fig. 2(A–E)) versus a polyethyleneimine–bovine serum albumin coat (Fig. 2(F–H))] can also be surmised as the reason for divergent results on the uptake of nanorods versus spheres in two studies when the nanoparticles are made from gold.23,51 However, in this latter case the conclusions are drawn from very different methods to determine the uptake and these differences in the measurement method could also be the reason (mass spectrometry versus flow cytometry).
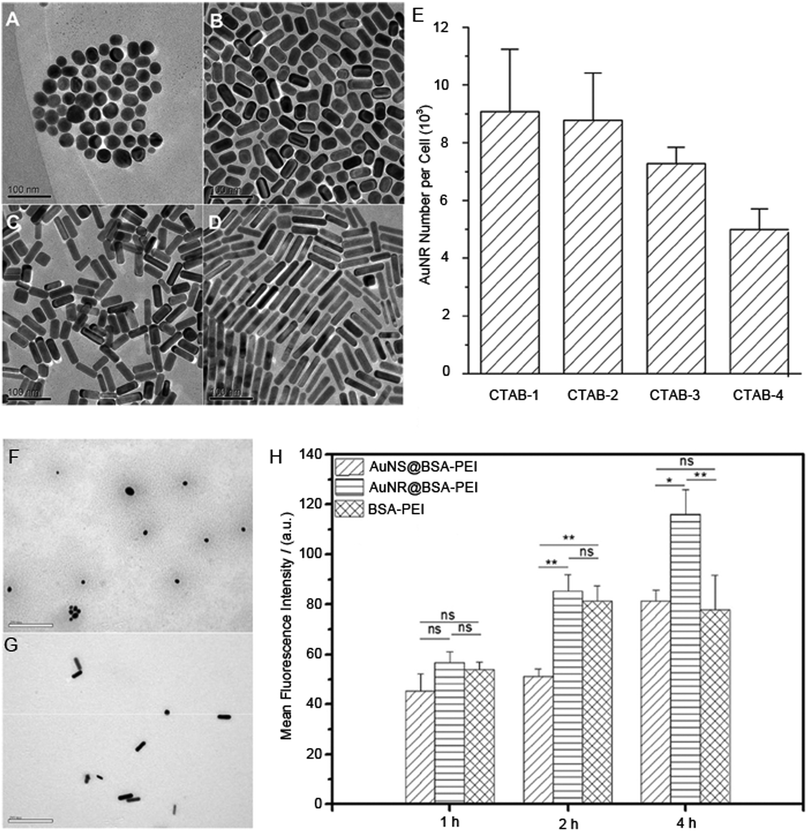 | ||
| Fig. 2 Transmission electron microscopy images of the CTAB-coated gold nanoparticles with different aspect ratios (A) CTAB-1, (B) CTAB-2, (C) CTAB-3, and (D) CTAB-4. (E) The internalised number of gold nanoparticles with different aspect ratios coated by CTAB, which was measured by a mass spectrometry-based method. Transmission electron microscopy images (F), polyethyleneimine-bovine serum albumin-coated gold nanospheres and (G) polyethyleneimine-bovine serum albumin-coated gold nanorods. (H) Cellular internalisation efficiency of polyethyleneimine-bovine serum albumin-coated gold nanospheres (AuNS@BSA-PEI) and nanorods (AuNR@BSA-PEI) in HepG2 cells with the incubation time of 1 h, 2 h and 4 h, which was measured by the fluorescence-based quantitative technique. Reprinted with permission from ref. 23 and 51. Copyright 2010 and 2017 Elsevier Ltd. | ||
What should be evident, even from these basic comparisons, is that the experimental conditions reported are often insufficient to allow a complete comparison between studies or the conditions are too different to be able to ascertain which of the differences are responsible. In relation to the former, insufficient information, it is for this reason, we and many others advocate studies to employ minimum information reporting standards as suggested in the recent MIRIBEL initiatives, which includes three categories: material characterization, biological characterization and details of experimental protocols.52 Such initiatives will at least mean that certain basic particle characterization information will be available to give comparisons where there is reduced ambiguity and hopefully allow us to start to deconvolute important variables in drug delivery of nanoparticles.
Furthermore, as indicated above for the different conclusions between studies on the shape of gold nanoparticles, different measurement methods can give different conclusions. In relation to understanding the impact of shape, we feel the impact of the measurement technique has not been well discussed. As such, next, we will discuss common measurement tools used for studying the cellular uptake to give an overview of the types of information each method is ideal for, what the challenges are, and what other tools might be needed. The measurement tools can be subdivided into methods that give the amount of internalised nanoparticles and those that enable imaging of the nanoparticles within the cell.
Quantifying the internalisation of nanoparticles
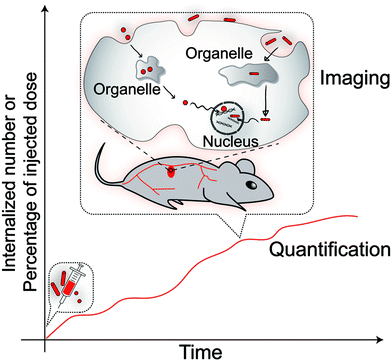
To quantify the internalised nanoparticles, “internalised number of nanoparticles” and “percentage of injected dose (%ID)” in the cells of interest are two important parameters (Scheme 2). There are many analytical methods which have been used to obtain these two parameters. These methods are summarized in Table 1 with a list of their strengths and weaknesses and conclusions that can be drawn from each family of techniques. We also provide references where each technique has been used to derive information about the impact of the nanoparticle shape on cellular uptake.| Methods | Strengths | Weaknesses | What conclusions it can give | Ref. |
|---|---|---|---|---|
| Mass spectrometry-based methods | The amount of associated particles would be quantified on its original state, and there is no need for modifying specific reporter molecules. | 1. The cells need to be lysed before nanoparticles quantification. | Mass of specific elements from nanoparticles associated with cells. | 45 |
| 2. They could not distinguish internalised particles from all associated particles. | 44, 50 and 51 | |||
| Fluorescence-based quantitative techniques | 1. The amount of internalised nanoparticles can be quantified by fluorescent-based techniques in living cells. | 1. The detachment of fluorescent dyes will influence the quantification of the internalised nanoparticles. | Fluorescent intensity of the nanoparticles associated with cells. | 19–26 and 28 |
| 2. Fluorescent-based techniques have a high sensitivity and they can reach a very low detection limit with a high fluorophores labelling ratio. | 2. There are several uncertainties about the quantification based on fluorescence techniques, which includes the auto-fluorescence induced noise, possible bleaching or quenching of fluorescence signal at high concentration of the fluorophores. | 42, 43, 46, 47 and 49 | ||
| 3. This technique can distinguish internalised particles from associated particles. | ||||
| Radioactivity quantification | 1. The amount of internalised nanoparticles can be quantified based on radiation intensity in living cells. | 1. The detachment of labelled radioisotopes will influence the quantification of the internalised nanoparticles. | The radiation intensity of the nanoparticles associated with cells. | 41 and 56 |
| 2. This technique has good tissue penetration ability. | 2. The radiation intensity of the attached radioisotopes will decrease along with time. | |||
| Electron Microscope | 1. This microscope has the highest spatial resolution among all kinds of microscopes. | 1. Cells must be fixed before imaging. | Location of the nanoparticles within/out of cells. | 21, 22, 28 and 58 |
| 2. There is no need to modify any reporter molecules on the nanoparticles for observation. | 2. The sample preparation process can introduce artefacts when accidentally precipitating on the sample. | 41, 43, 44 and 51 | ||
| Fluorescent microscope | 1. Living cells can be observed under a fluorescent microscope. | 1. The resolution of the fluorescence microscope is limited by the wavelength range of visible light. | 1. Location of the nanoparticles within/out of cells. | 19–22, 24–26, 28 and 58 |
| 2. Fluorescent microscope can realise imaging and quantification for the internalised nanoparticles at the same time. | 2. Fluorescent dye detachment will influence the observation of the nanoparticles; Fluorescent labelling is not feasible for all nanoparticles. | 2. Fluorescent intensity of the nanoparticles associated with cells. | 27, 42, 43, 46, 47, 51 and 56 | |
| 3. The fluorescent dye-tagged nanoparticles may move differently from the non-tagged ones. | ||||
| Single/multiple-particle tracking | 1. The trajectory of the nanoparticles entering cells and moving within cells can be mapped with this technique. | 1. The concentration of studied nanoparticles within cells needs to be low. | 1. The trajectory of the nanoparticles entering cells and moving within cells. | 28 and 41 |
| 2. The movement of the nanoparticles in the cells can be quantified by this technique based on the trajectory of nanoparticles. | 2. Lots of tracking experiments need to be done to find major moving behaviour of the internalised nanoparticles. | 2. Diffusion coefficient of the nanoparticles during they enter cells or move within the cells. | ||
| Auto/pair correlation microscopy | 1. By doing a line scan with a confocal microscope, the route of the nanoparticles entering cells and moving within cells can be known, and transit time of the nanoparticles moving along with the scanning line can be quantified as well. | It can only analyse the nanoparticle movement from one cell at each time. | 1. A number of mobile nanoparticles at each position on the scanning line. | 31 |
| 2. This technique can realise studying the nanoparticle movement on a population level. | 2. Transit time of the nanoparticles moving between two positions on the scanning line. | |||
Mass spectrometry-based methods, such as inductively coupled plasma mass spectrometry, laser desorption/ionization mass spectrometry, and high-resolution secondary ion mass spectrometry, have been widely used for the study nanoparticles that are composed of elements which are not abundant in the cells such as Au and Si.39,44,45,51 One of the greatest advantages of this technique is no modification of the nanoparticles is required and hence the influence from the reporter molecules on internalisation efficiency need not be considered. Furthermore, mass spectrometry is inherently quantitative such that the number of nanoparticles can be determined by converting the number of atoms to number of nanoparticles based on the size of the studied nanoparticles.44 The percentage of injected dose delivered (%ID) in target tissue/cultured cells can be acquired by directly comparing the amount of specific element at different time points with that at the starting time point. There are challenges with this method that limit its popularity for quantifying internalisation efficiency: (1) the cells need to be lysed before quantification to extract the nanoparticles, therefore, the recovery ratio of intact cell associated nanoparticles from the cells will determine the quantification accuracy; (2) the method will also count any nanoparticles which are just attached to the exterior surface of the cells (associated nanoparticles), although there are approaches to identify the surface-bound nanoparticles.53 This uncertainty impacts on the studies of nanoparticle shape because the surface contact area between the different shaped nanoparticles and the plasma membrane can be significantly different, which can even influence how the plasma membrane transforms to encapsulate the nanoparticles for endocytosis.54,55
Both fluorescence spectroscopy and radioisotope quantification can also be used to quantify internalisation of the nanoparticles provided that they can be conjugated with the appropriate label.19–26,28,41–43,46,47,49,56 With such labelling methods, they can be non-destructive to the cells such that the amount of internalised nanoparticles could be quantified within living cells and over time. Furthermore, these methods are highly sensitive and able to distinguish minor differences in internalisation between two different shaped nanoparticles. Such methods also have the potential to distinguish between the nanoparticles attached to the plasma membrane and those internalised. There are some challenges facing the labelling methods. First, one needs the labelling efficiency with different shaped nanoparticles before a quantitative comparison can be made. Second, uncertainty can exist in relation to the reporter molecules due to whether detachment of fluorescent dyes/radioisotopes from the nanoparticles has occurred, from photo-bleaching or quenching of fluorescence dyes, and radiation decay of radioisotopes. Third, some contradictory results come from various evaluation methods used in different papers as well. For example, (1) the percentage of cells with internalised nanoparticles was used to compare the internalisation efficiency between different nanoparticles;26,42 (2) other papers evaluated the internalisation efficiency by using the ratio of internalised nanoparticles relative to the number of cell-associated nanoparticles (number of all the cell-associated nanoparticles is equal to the sum of the number of nanoparticles inside cell and attached at the cellular surface).22,42,56 Overall, different evaluation systems describe the internalisation of different shaped nanoparticles from different viewpoints.
Imaging the internalisation of nanoparticles
Imaging methods can show the distribution and movements of nanoparticles inside cells and also provide some evidence to support the corresponding quantification result. The common imaging methods are summarized in Table 1 as well. Microscopy methods are widely used to observe how the nanoparticles interacted with cells and where they are located within the cells. There are of course many forms of microscopy available which have their own strengths and weaknesses. Of all the microscopy methods, an electron microscope (EM) has the highest resolution (down to several nanometres), so that single nanoparticles can easily be resolved. Most of the inorganic nanoparticles can be visualized without specific labelling because of different contrast between the cell and inorganic nanoparticles in an electron microscope. Using an electron microscope, it has been observed that the shape of nanoparticles can influence cell membrane penetration.21,28–30,51,57,58 Han et al. reported that rod-shaped nanoparticles, after 1 h incubation, could induce transformation of the plasma membrane to encapsulate the rods but spherical nanoparticles could not.58 And Meng et al. reported that cells exposed to rod-shaped nanoparticles had more prominent membrane ruffles and filopodia formation than spherical ones.21 The key limitation of electron microscopy is the methods are vacuum-based methods and as such only fixed cells can be imaged.59 This limits electron microscopy when one seeks to follow dynamic processes.Fluorescent microscopy is an even more widely used imaging tool that can be used on living cells, with high temporal resolution. Fluorescence microscopy is inherently quantitative, is incredibly versatile and with advances in instruments and imaging processes is providing progressively richer and richer information.27,43,49,56,60–62 Fluorescent microscopy, of course, faces the same challenges as flow cytometry with regards to tagging the nanoparticles with fluorescent dyes. Issues such as detachment or possible bleaching must also be considered. Furthermore, because of the resolution of the conventional fluorescent microscope of >250 nm, depending on the wavelength of light, it can be challenging to determine at times whether a nanoparticle is inside a small-sized organelle or whether it is just on the surface of such features.63
Although a fluorescent microscope has lower resolution than an electron microscope, the ability of fluorescent microscopes to perform living cell imaging is vital for studying the dynamic process of the internalisation of nanoparticles. Understanding the dynamic movement of different shaped nanoparticles inside cells is necessary since the whole internalisation process is dynamic. Furthermore, as the nanoparticles that are targeted for the nucleus must travel through the entire cell to get to their destination, fluorescent intensity measurements can only inform about the number of nanoparticles in a given location but not whether they are moving. Knowledge on the nanoparticle movement is important as a high fluorescence intensity might be indicative of a barrier in which the nanoparticles become stuck. In order to perform dynamic measurements of nanoparticle movement, continuous imaging and a specifically designed algorithm are required to track the movement of the nanoparticles and record the time information of cellular uptake. Take as an example “single/multiple-particle tracking”. Such a method can detect the cellular uptake process of one or several nanoparticles and identify the moving behaviour of the nanoparticles based on their trajectory.28,64 Shi et al. demonstrated that the movement of long rod-shaped polymer nanoparticles was slightly greater than the short rod counterparts in the cytoplasm with multiple-particle tracking.41 However, to realize the single/multiple-particle tracking successfully, the nanoparticles should be isolated from each other to easily track the localization of individual particles. Therefore, dilution of the fluorescent labelled nanoparticles is necessary to allow the software/algorithm to track the positions of individual nanoparticles.65 As such a challenge with single particle tracking is to obtain a statistically relevant amount of data.
Auto/pair-correlation microscopy is another dynamic fluorescent microscopy analysis method to study the number and translocation time of moving nanoparticles internalised in a living cell.66–68 By performing a line scan across the cell with internalised nanoparticles, the fluorescent signal of each pixel on the line is collected, and the intensity is related to the local number of nanoparticles at the pixel. With repeated scans of the line, analysing the fluorescence intensity fluctuation of each pixel with an auto-correlation function, provides a determination of how many fluorescent nanoparticles are moving at each pixel of the line, which is called “number of moving particles”. The number of moving nanoparticles can be very different from the local number of nanoparticles since not every nanoparticle inside cells is moving. By comparing the fluorescence intensity fluctuation of two pixels that are spatially separated and applying a pair-correlation function, the average transit time that the nanoparticles take to move between those two pixels can also be acquired. Such an approach can be used to evaluate whether there is a barrier and how fast the nanoparticles cross the barrier. Recently, we measured the cellular uptake of four different shaped polymeric nanoparticles (polystyrene-b-polyethylene glycol) with auto/pair-correlation microscopy. Through comparing the number of moving nanoparticles at the different subcellular compartments/organelles and transit time of the nanoparticles travelling in/between different subcellular compartments, it was found that the nanoparticles with high aspect ratio [5–10 nm (diameter) × 100–300 nm (length); 5–10 nm (diameter) × 400–700 nm (length)] can diffuse into the nucleus more efficiently and have the more accumulation amount in the nucleus than the spherical nanoparticles [20 nm], which provided strong evidence of the highly nuclear targeting efficiency of the rod-shaped nanoparticles. At present, auto/pair-correlation microscopy is compatible with investigating one cell at a time, and it is still a big challenge to apply this technique to analyse nanoparticle movement in multiple cells at the same time.
Summary and future
With nanoparticles that are identical in all aspects except shape, we have learnt that shape alone can influence nanoparticle internalisation. In most cases, rod-shaped nanoparticles exhibit greater amounts of cell internalisation, and higher drug delivery efficiencies, than spherical nanoparticles. There are studies that arrive at the opposite conclusion. However, we feel that with some of these studies there is some uncertainty as to why that might be. One common ambiguity relates to the surface chemistry of the nanoparticles and unequal dimensions being compared (by this we mean that the best comparisons are made when one dimension is the same). Certainly, contradictory results should motivate researchers who work on bio-nano interaction to subscribe to some form of minimum reporting standards to describe the nanoparticles they use in future work.52 The different types of information that different analysis methods provide may also be responsible for some contradictory conclusions as discussed.The cell-based assays rely on the assumption that a cell culture reflects the physiology of a tissue.69,70 Therefore, as powerful as 2D cell culture is, it does not always provide the ideal tissue-specific architecture, mechanical strain, cell–cell communication and nutrient flow found in vivo.71 As such studies are emerging on the use of 3D cell cultures for monitoring drug delivery and the impact of nanoparticle shape. These initial studies also show the same trends that we see in 2D cultures with regards to nanorods more efficiently entering cells relative to spherical nanoparticles. Again, there are some studies that come to the opposite conclusion that spherical nanoparticles are more effectively taken up by cells relative to high aspect ratio nanoparticles in 3D cell cultures.
Among all the analysis methods discussed above, conventional quantification tools, such as mass spectrometry and flow cytometry, can still be used in 3D cell models because in these methods the cells are either destroyed or processed into single-cell suspension before injecting into the instruments. But for 3D live cell imaging, deep penetration into the 3D cell model, high imaging speed and ultra-low intensity of the excitation light are three additional key requirements,71,72 which cannot be realized with conventional microscopes. A lattice light-sheet microscope is a promising imaging tool to satisfy the requirements for 3D cell imaging, and it performs particularly well with long working distance lenses and has a good penetration depth and so millimeter-sized live specimens can be observed in their entirety under high resolution.73–75 Lattice light-sheet microscopy also has the potential to measure the endocytosis lifetime of nanoparticles and track their diffusions in a 3D cellular specimen with the assistance of a high-throughput algorithm.
The study of cellular internalisation of nanoparticles in 2D and 3D cell culture will promote the innovation of nanoparticle-based stimuli-responsive drug delivery systems. Although there have already been a large number of drug delivery systems reported, it is still not clear where and how much of the drug molecules are released from the nanoparticles inside the cells. Along with the understanding of cellular internalisation of the nanoparticles, a specific type of nanoparticle, which is suitable for delivering the drug molecules to the target cell, can be selected out. For this chosen nanoparticle, first, it is necessary to dynamically understand the local physicochemical environment that the chosen nanoparticles will encounter during internalisation, which can be helpful to program the release location of the loading drug molecules. Second, it is necessary to know the intracellular release dynamics of the drug molecules from the nanoparticles, which can be used to calculate the releasing dosage of the drug molecules, evaluate the side effects of the whole drug delivery system and improve the translation of the nanoparticle-based stimuli-responsive drug delivery systems from the experiment bench to the clinic.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
W. W. acknowledges the seed funding from the Australian Centre for Nanomedicine. J.J.G. acknowledges funding from the ARC centre of Excellence in Convergent Bio-Nano Science and Technology (CE140100036), the ARC Laureate Fellowship (FL150100060) program and a National Health and Medical Research Council program grant (1091261). K. G. acknowledges funding from the ARC Centre of Excellence in Advanced Molecular Imaging (CE140100011), the Australian Research Council (LP140100967 and DP130100269) and National Health and Medical Research Council of Australia (1059278 and 1037320). R. D. T. acknowledges funding from the Australian Research Council (LP150101014 and DP190102659).References
- T. Jiang, R. Mo, A. Bellotti, J. Zhou and Z. Gu, Adv. Funct. Mater., 2014, 24, 2295–2304 CrossRef CAS.
- S. Mura, J. Nicolas and P. Couvreur, Nat. Mater., 2013, 12, 991–1003 CrossRef CAS PubMed.
- W. Wang, Y. Wen, L. Xu, H. Du, Y. Zhou and X. Zhang, Chem. – Eur. J., 2014, 20, 7796–7802 CrossRef CAS PubMed.
- A. Verma, O. Uzun, Y. H. Hu, Y. Hu, H. S. Han, N. Watson, S. L. Chen, D. J. Irvine and F. Stellacci, Nat. Mater., 2008, 7, 588–595 CrossRef CAS PubMed.
- K. Kostarelos, L. Lacerda, G. Pastorin, W. Wu, S. Wieckowski, J. Luangsivilay, S. Godefroy, D. Pantarotto, J.-P. Briand, S. Muller, M. Prato and A. Bianco, Nat. Nanotechnol., 2007, 2, 108–113 CrossRef CAS PubMed.
- R. A. Petros and J. M. DeSimone, Nat. Rev. Drug Discovery, 2010, 9, 615–627 CrossRef CAS PubMed.
- I. Canton and G. Battaglia, Chem. Soc. Rev., 2012, 41, 2718–2739 RSC.
- F. Zhao, Y. Zhao, Y. Liu, X. L. Chang, C. Y. Chen and Y. L. Zhao, Small, 2011, 7, 1322–1337 CrossRef CAS PubMed.
- Y. Mo and L.-Y. Lim, J. Pharm. Sci., 2004, 93, 20–28 CrossRef CAS PubMed.
- K. C. Partlow, G. M. Lanza and S. A. Wickline, Biomaterials, 2008, 29, 3367–3375 CrossRef CAS PubMed.
- K. Kettler, K. Veltman, D. van de Meent, A. van Wezel and A. J. Hendriks, Environ. Toxicol. Chem., 2014, 33, 481–492 CrossRef CAS PubMed.
- A. Albanese, P. S. Tang and W. C. W. Chan, Annu. Rev. Biomed. Eng., 2012, 14, 1–16 CrossRef CAS PubMed.
- M. T. Zhu, G. J. Nie, H. Meng, T. Xia, A. Nel and Y. L. Zhao, Acc. Chem. Res., 2013, 46, 622–631 CrossRef CAS PubMed.
- C. He, Y. Hu, L. Yin, C. Tang and C. Yin, Biomaterials, 2010, 31, 3657–3666 CrossRef CAS PubMed.
- V. T. Cong, K. Gaus, R. D. Tilley and J. J. Gooding, Expert Opin. Drug Delivery, 2018, 15, 881–892 CrossRef CAS PubMed.
- N. P. Truong, M. R. Whittaker, C. W. Mak and T. P. Davis, Expert Opin. Drug Delivery, 2015, 12, 129–142 CrossRef CAS PubMed.
- C. Kinnear, T. L. Moore, L. Rodriguez-Lorenzo, B. Rothen-Rutishauser and A. Petri-Fink, Chem. Rev., 2017, 117, 11476–11521 CrossRef CAS PubMed.
- M. Björnmalm, M. Faria, X. Chen, J. Cui and F. Caruso, Langmuir, 2016, 32, 10995–11001 CrossRef PubMed.
- R. Agarwal, V. Singh, P. Jurney, L. Shi, S. V. Sreenivasan and K. Roy, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 17247–17252 CrossRef CAS PubMed.
- D. Sen Karaman, D. Desai, R. Senthilkumar, E. M. Johansson, N. Ratts, M. Oden, J. E. Eriksson, C. Sahlgren, D. M. Toivola and J. M. Rosenholm, Nanoscale Res. Lett., 2012, 7, 358 CrossRef PubMed.
- H. Meng, S. Yang, Z. X. Li, T. Xia, J. Chen, Z. X. Ji, H. Y. Zhang, X. Wang, S. J. Lin, C. Huang, Z. H. Zhou, J. I. Zink and A. E. Nel, ACS Nano, 2011, 5, 4434–4447 CrossRef CAS PubMed.
- S. E. A. Gratton, P. A. Ropp, P. D. Pohlhaus, J. C. Luft, V. J. Madden, M. E. Napier and J. M. DeSimone, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 11613–11618 CrossRef CAS PubMed.
- P. Zhang, B. B. Li, J. W. Du and Y. X. Wang, Colloids Surf., B, 2017, 157, 18–25 CrossRef CAS PubMed.
- A. Banerjee, J. P. Qi, R. Gogoi, J. Wong and S. Mitragotri, J. Controlled Release, 2016, 238, 176–185 CrossRef CAS PubMed.
- N. J. Hao, L. L. Li, Q. Zhang, X. L. Huang, X. W. Meng, Y. Q. Zhang, D. Chen, F. Q. Tang and L. F. Li, Microporous Mesoporous Mater., 2012, 162, 14–23 CrossRef CAS.
- B. G. Trewyn, J. A. Nieweg, Y. Zhao and V. S. Y. Lin, Chem. Eng. J., 2008, 137, 23–29 CrossRef CAS.
- K. Zhang, H. F. Fang, Z. Y. Chen, J. S. A. Taylor and K. L. Wooley, Bioconjugate Chem., 2008, 19, 1880–1887 CrossRef CAS PubMed.
- Z. Q. Chu, S. L. Zhang, B. K. Zhang, C. Y. Zhang, C. Y. Fang, I. Rehor, P. Cigler, H. C. Chang, G. Lin, R. B. Liu and Q. Li, Sci. Rep., 2014, 4, 4495 CrossRef PubMed.
- N. Doshi and S. Mitragotri, J. R. Soc., Interface, 2010, 7, S403–S410 CrossRef CAS PubMed.
- P. M. Favi, M. Gao, L. J. S. Arango, S. P. Ospina, M. Morales, J. J. Pavon and T. J. Webster, J. Biomed. Mater. Res., Part A, 2015, 103, 3449–3462 CrossRef CAS PubMed.
- E. Hinde, K. Thammasiraphop, H. T. Duong, J. Yeow, B. Karagoz, C. Boyer, J. J. Gooding and K. Gaus, Nat. Nanotechnol., 2017, 12, 81–89 CrossRef CAS PubMed.
- A. K. Salem, P. C. Searson and K. W. Leong, Nat. Mater., 2003, 2, 668–671 CrossRef CAS PubMed.
- M.-M. Fan, W.-Z. Zhang, C. Cheng, Y. Liu, B.-J. Li, X. Sun and S. Zhang, Part. Part. Syst. Charact., 2014, 31, 994–1000 CrossRef CAS.
- U. Kubitscheck, D. Grunwald, A. Hoekstra, D. Rohleder, T. Kues, J. P. Siebrasse and R. Peters, J. Cell Biol., 2005, 168, 233–243 CrossRef PubMed.
- K. E. Knockenhauer and T. U. Schwartz, Cell, 2016, 164, 1162–1171 CrossRef CAS PubMed.
- N. Panté and M. Kann, Mol. Biol. Cell, 2002, 13, 425–434 CrossRef PubMed.
- S. B. Lowe, V. T. G. Tan, A. H. Soeriyadi, T. P. Davis and J. J. Gooding, Bioconjugate Chem., 2014, 25, 1581–1601 CrossRef CAS PubMed.
- R. Agarwal, P. Jurney, M. Raythatha, V. Singh, S. V. Sreenivasan, L. Shi and K. Roy, Adv. Healthcare Mater., 2015, 4, 2269–2280 CrossRef CAS PubMed.
- R. Fernandes, N. R. Smyth, O. L. Muskens, S. Nitti, A. Heuer-Jungemann, M. R. Ardern-Jones and A. G. Kanaras, Small, 2015, 11, 713–721 CrossRef CAS PubMed.
- K. C. L. Black, Y. Wang, H. P. Luehmann, X. Cai, W. Xing, B. Pang, Y. Zhao, C. S. Cutler, L. V. Wang, Y. Liu and Y. Xia, ACS Nano, 2014, 8, 4385–4394 CrossRef CAS PubMed.
- J. Shi, J. L. Choi, B. Chou, R. N. Johnson, J. G. Schellinger and S. H. Pun, ACS Nano, 2013, 7, 10612–10620 CrossRef CAS PubMed.
- O. Shimoni, Y. Yan, Y. J. Wang and F. Caruso, ACS Nano, 2013, 7, 522–530 CrossRef CAS PubMed.
- L. Florez, C. Herrmann, J. M. Cramer, C. P. Hauser, K. Koynov, K. Landfester, D. Crespy and V. Mailander, Small, 2012, 8, 2222–2230 CrossRef CAS PubMed.
- B. D. Chithrani, A. A. Ghazani and W. C. W. Chan, Nano Lett., 2006, 6, 662–668 CrossRef CAS PubMed.
- T. Yu, A. Malugin and H. Ghandehari, ACS Nano, 2011, 5, 5717–5728 CrossRef CAS PubMed.
- J. Zhao, H. Lu, S. Wong, M. Lu, P. Xiao and M. H. Stenzel, Polym. Chem., 2017, 8, 3317–3326 RSC.
- J. Zhao, H. Lu, P. Xiao and M. H. Stenzel, ACS Appl. Mater. Interfaces, 2016, 8, 16622–16630 CrossRef CAS PubMed.
- D. R. Dias, A. F. Moreira and I. J. Correia, J. Mater. Chem. B, 2016, 4, 7630–7640 RSC.
- Y. Geng, P. Dalhaimer, S. S. Cai, R. Tsai, M. Tewari, T. Minko and D. E. Discher, Nat. Nanotechnol., 2007, 2, 249–255 CrossRef CAS PubMed.
- Arnida, M. M. Janát-Amsbury, A. Ray, C. M. Peterson and H. Ghandehari, Eur. J. Pharm. Biopharm., 2011, 77, 417–423 CrossRef CAS PubMed.
- Y. Qiu, Y. Liu, L. Wang, L. Xu, R. Bai, Y. Ji, X. Wu, Y. Zhao, Y. Li and C. Chen, Biomaterials, 2010, 31, 7606–7619 CrossRef CAS PubMed.
- M. Faria, M. Björnmalm, K. J. Thurecht, S. J. Kent, R. G. Parton, M. Kavallaris, A. P. R. Johnston, J. J. Gooding, S. R. Corrie, B. J. Boyd, P. Thordarson, A. K. Whittaker, M. M. Stevens, C. A. Prestidge, C. J. H. Porter, W. J. Parak, T. P. Davis, E. J. Crampin and F. Caruso, Nat. Nanotechnol., 2018, 13, 777–785 CrossRef CAS PubMed.
- S. Hou, K. N. Sikora, R. Tang, Y. Liu, Y.-W. Lee, S. T. Kim, Z. Jiang, R. W. Vachet and V. M. Rotello, ACS Nano, 2016, 10, 6731–6736 CrossRef CAS PubMed.
- R. Toy, P. M. Peiris, K. B. Ghaghada and E. Karathanasis, Nanomedicine, 2014, 9, 121–134 CrossRef CAS PubMed.
- K. Yang and Y. Q. Ma, Nat. Nanotechnol., 2010, 5, 579–583 CrossRef CAS PubMed.
- S. Muro, C. Garnacho, J. A. Champion, J. Leferovich, C. Gajewski, E. H. Schuchman, S. Mitragotri and V. R. Muzykantov, Mol. Ther., 2008, 16, 1450–1458 CrossRef CAS PubMed.
- J. A. Champion and S. Mitragotri, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 4930–4934 CrossRef CAS PubMed.
- Y. Han, X. Wang, H. Dai and S. Li, ACS Appl. Mater. Interfaces, 2012, 4, 4616–4622 CrossRef CAS PubMed.
- N. de Jonge and D. B. Peckys, ACS Nano, 2016, 10, 9061–9063 CrossRef CAS PubMed.
- H. Y. Nam, S. M. Kwon, H. Chung, S. Y. Lee, S. H. Kwon, H. Jeon, Y. Kim, J. H. Park, J. Kim, S. Her, Y. K. Oh, I. C. Kwon, K. Kim and S. Y. Jeong, J. Controlled Release, 2009, 135, 259–267 CrossRef CAS PubMed.
- B. R. Smith, P. Kempen, D. Bouley, A. Xu, Z. Liu, N. Melosh, H. J. Dai, R. Sinclair and S. S. Gambhir, Nano Lett., 2012, 12, 3369–3377 CrossRef CAS PubMed.
- D. A. Christian, S. Cai, O. B. Garbuzenko, T. Harada, A. L. Zajac, T. Minko and D. E. Discher, Mol. Pharmaceutics, 2009, 6, 1343–1352 CrossRef CAS PubMed.
- A. P. R. Johnston, ACS Sens., 2017, 2, 4–9 CrossRef CAS PubMed.
- A. von Diezmann, Y. Shechtman and W. E. Moerner, Chem. Rev., 2017, 117, 7244–7275 CrossRef CAS PubMed.
- D. D. Chen, N. A. Monteiro-Riviere and L. S. W. Zhang, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2017, 9, e1419 Search PubMed.
- M. A. Digman and E. Gratton, Annu. Rev. Phys. Chem., 2011, 62, 645–668 CrossRef CAS PubMed.
- M. Baum, F. Erdel, M. Wachsmuth and K. Rippe, Nat. Commun., 2014, 5, 4494 CrossRef CAS PubMed.
- M. A. Digman, R. Dalal, A. F. Horwitz and E. Gratton, Biophys. J., 2008, 94, 2320–2332 CrossRef CAS PubMed.
- L. G. Griffith and M. A. Swartz, Nat. Rev. Mol. Cell Biol., 2006, 7, 211–224 CrossRef CAS PubMed.
- J. E. Cohen, PLoS Biol., 2004, 2, e439 CrossRef PubMed.
- F. Pampaloni, N. Ansari and E. H. Stelzer, Cell Tissue Res., 2013, 352, 161–177 CrossRef PubMed.
- T. Bruns, S. Schickinger, R. Wittig and H. Schneckenburger, J. Biomed. Opt., 2012, 17, 101518 CrossRef PubMed.
- K. He, R. Marsland, III, S. Upadhyayula, E. Song, S. Dang, B. R. Capraro, W. Wang, W. Skillern, R. Gaudin, M. Ma and T. Kirchhausen, Nature, 2017, 552, 410–414 CrossRef CAS PubMed.
- T. L. Liu, S. Upadhyayula, D. E. Milkie, V. Singh, K. Wang, I. A. Swinburne, K. R. Mosaliganti, Z. M. Collins, T. W. Hiscock, J. Shea, A. Q. Kohrman, T. N. Medwig, D. Dambournet, R. Forster, B. Cunniff, Y. Ruan, H. Yashiro, S. Scholpp, E. M. Meyerowitz, D. Hockemeyer, D. G. Drubin, B. L. Martin, D. Q. Matus, M. Koyama, S. G. Megason, T. Kirchhausen and E. Betzig, Science, 2018, 360, eaaq1392 CrossRef PubMed.
- P. de Boer, J. P. Hoogenboom and B. N. Giepmans, Nat. Methods, 2015, 12, 503–513 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2019 |