Open Access Article
Open Access ArticleCombining dielectrophoresis and computer vision for precise and fully automated single-cell handling and analysis†
Neus
Godino
,
Felix
Pfisterer
,
Tobias
Gerling
,
Christian
Guernth-Marschner
,
Claus
Duschl
and
Michael
Kirschbaum
 *
*
Fraunhofer IZI-BB, Am Muehlenberg 13, 14476 Potsdam, Germany. E-mail: michael.kirschbaum@izi-bb.fraunhofer.de
First published on 20th November 2019
Abstract
With the advent of single-cell technologies comes the necessity for efficient protocols to process single cells. We combine dielectrophoresis with open source computer vision programming to automatically control the trajectories of single cells inside a microfluidic device. Using real-time image analysis, individual cells are automatically selected, isolated and spatially arranged.
The possibility to study biology at the single-cell level is about to revolutionize our understanding of many fundamental processes in biology. Recent technological advances simplified and economized protocols for the molecular analysis of single cells, therefore providing unprecedented opportunities to analyze the complexities of life's most basic elements. While the genomic and transcriptomic characterization of single cells has been extensively demonstrated, their functional profiling and analysis of cellular reactions to external cues remains difficult at the single-cell level. For many important assays like in situ analysis of cell-to-cell interaction,1 chemical or mechanical stimulations,2 secretion3,4 or paracrine cell signaling studies,5 it is necessary to select and precisely place the cells of interest at defined locations and analyze them over time. Microfluidic platforms, where cells can be manipulated in a controlled and reliable way, have become a powerful alternative to traditional techniques.6 Unfortunately, most of them are of passive (whether stochastic or deterministic) nature and are tailored to a very specific problem or sample which limits their applicability if minor deviations from the assay protocol or another type of sample are to be used.7 This is of particular importance when working with high-quality cell samples, as they result from the pre-enrichment of valuable cells like circulating tumor cells, antigen-specific T-cells or specific subtypes of immune cells with FACS or alternative methods and whose loss or incorrect processing cannot be tolerated. In this case, decision-based protocols that employ (e.g. visual) information about the cells for an active position control are highly valuable because they are more flexible and at the same time accurate and reliable. In contrast to the tedious and low-throughput technologies usually used for active positioning of cells, such as micromanipulator or optical tweezers,8,9 automated flow-through protocols that allow for robust, flexible, efficient and yet precise single-cell handling are strongly desired. A popular technique to selectively pick and place single cells in microfluidic devices is dielectrophoresis (DEP),7,10 where a non-uniform electric field polarizes and exerts force on the cells.11 In combination with image processing this offers exciting opportunities for assay automation and standardization.12–16 In this work, we demonstrate the combination of precise cell manipulation by DEP actuation with computer vision (CV) for active and fully automated single-cell handling in a microfluidic device. As a proof of concept, we demonstrate multi-target color-based cell selection and spatial arrangement of cells on a dielectrophoretic capture array.
We have demonstrated in previous work the effectiveness of DEP for high-precision cell handling,17–19 where different DEP microelectrode configurations were designed to focus, move or trap single cells inside a microfluidic channel. For the exertion of DEP, an alternating voltage generated by a signal generator is applied to microelectrodes in the channel. The relatively simple and economical instrumentation compared to expensive lasers or optical components is suitable for parallelization and automation.20 The DEP electrodes can be arranged to functional elements that allow the execution of particular tasks. According to the user-defined sequence of actions on the cells, each of these functional elements is positioned in the microchannel making DEP actuation a modular and scalable technique.
As a novel approach we now introduce a fully automated control of several of those functional elements (i.e., activation or deactivation of the electric potential applied to the DEP electrodes), by a CV program in order to perform a complex cell handling protocol with high precision, robustness and efficiency. The CV program decides whether to activate or deactivate the electrodes according to the specific selection parameters of the target cells and the previously defined sequence of actions on the cells.
The microscope image is generated by a general purpose commercial camera. For image analysis we employ openCV (open source computer vision) which is a platform-independent open source programming library for vision-based applications. The program analyzes in real-time where the target cells are and controls the instrumentation (i.e., switching on and off the corresponding microelectrodes). The code is written in Python and employs open source packages like cv2 (opencv-python) and imutils for image acquisition and analysis, numpy for the scientific computing, time for reading the necessary time of the different steps and serial for the control the relay board (see below). A USB-controlled camera, GoPro H5Pro modified with C-Mount (RIBCAGE H5PRO, Back-Bone, USA), is attached to the ocular of a fluorescence microscope (IX81, Olympus, Germany) with a magnification of 0.5× to fit the field of view to the size of the CCD chip of the camera.
The DEP microfluidic chips were previously described.17–19 They were manufactured by sandwiching a 35 μm thick SU8 polymer spacer between two glass slides which contain 15 μm wide Pt microelectrodes (GeSiM GmbH, Germany). For all the results shown in the present work, the voltage used for DEP is 3 V peak to peak at a frequency of 1 MHz applied by a radio frequency generator (Cytocon 400, Evotec Technologies, Germany). To switch on and off the voltage on the electrodes, a computer-controlled relay board (8× relay card 24V/7A, Conrad Electronics SE, Germany) is placed between the generator and the chip.
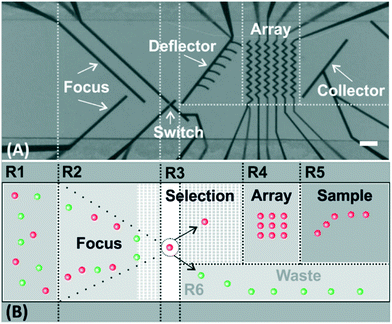
Fig. 1A shows a micrograph of the top view of the microchannel with the most relevant functional elements highlighted. The hydrodynamic forces (the fluid flow is going from left to right) in combination with the repulsive dielectrophoretic force (i.e., negative DEP) exerted on the cells at the microelectrodes determine the cell trajectories and allow us to precisely control their position in the microchannel (Fig. 1B, for a detailed description of the used concept, see ref. 21 and 22). According to the arrangement of these elements in the microchannel, the channel can be sectioned into 6 functional regions, R1–6. Initially, the cells are flowing randomly in the microchannel (Fig. 1B-R1). Afterwards, the cells are focused to the center of the microchannel (Fig. 1B-R2) following the trajectories defined by the focusing electrodes. The target cells are automatically identified and isolated at the selection element (Fig. 1B-R3). The selection element comprises three electrodes. Depending on the activation/deactivation of these electrodes (see ref. 16 for further details), the cells are either selected, so driven to the deflector on their way to the array, or discarded towards the waste outlet (Fig. 1B-R6). The selected cells are guided to a precise position in the array, where each cell can be individually identified and addressed (Fig. 1B-R4). The microelectrodes in the array have a “zig-zag”-shaped configuration, which allows stopping and trapping of cells in each pocket of the zig-zag.17–19 Once the cells are placed in the array, they keep trapped at the zig-zag electrode allowing prolonged examination of cellular reactions to external stimuli. By consecutively repeating the whole process, up to forty two cells can be allocated to each trapping position of the array for further analysis (e.g., cell-to-cell interaction studies). Afterwards, the cells can be sampled out (Fig. 1B-R5).17 The number of trapping positions can be easily increased by adding more zig-zag electrodes or those with more turns. Although the microfluidic device is placed under a microscope for the image-based control of the cell movement, the actuation of the electrodes (and, thus, control of the movement of the cells) is independent of the field of view, which is in contrast to optical actuation.
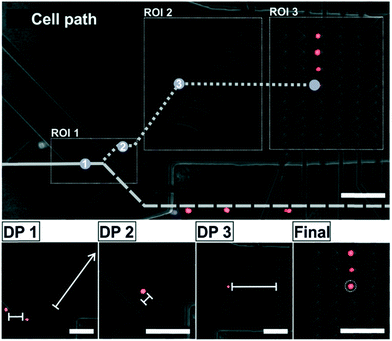
The cell handling process starts with placing and focusing the chip under the microscope and starting video acquisition. In a first step, a pattern recognition function defined in cv2 identifies the position of the microelectrodes in the image. On that basis three regions of interest (ROI) in the microchannel are defined, as depicted in Fig. 2. They are continuously analyzed for the presence or position of cells in order to perform different actions on the cells: ROI1: selection, ROI2: transport and ROI3: arrangement. For this, there are three relevant decision points (DP1–3) along the path of the cells to the array (see Fig. 2). They are explained in the following. Initially, all the cells are freely flowing along the microchannel. They are focused by the focus element and forwarded to DP1 in ROI1. At this point it is decided whether a cell is selected or discarded. In order to select a target cell, several preconditions must be met: (i) in ROI1 there is just one single event detected (ii) size and shape of this event correspond to the size and shape of one single cell (doublets or cell agglomerations are excluded) (iii) the event is of the target color, and (iv) ROI2 is free of cells. Immediately after the switch element allows the cell to pass, the deflector element is activated and a control timer starts counting. If the target cell is detected within a critical time frame at DP2, it means that the cell is out of the influence of the switch element, so it is set back to its initial state and, thus, prevents any other cell to pass. In contrast, if no cell is detected during this time frame at DP2 (i.e., the cell is lost), both electrodes switch back to their initial state being ready for the selection of a new target cell in ROI1. In the first case, the selected target cell leaves ROI1 and enters ROI2 where the deflector element drives the cell along the microchannel. When the target cell reaches the required height in relation to its intended position in the array, the deflector element is switched OFF and the corresponding zig-zag column is activated. As a consequence, the flow carries the cell towards its final position in the array. As a safety mechanism, there is also a control timer between DP2 and DP3 in order to terminate the waiting phase and recover the initial status of the electrodes, if the cell is lost caused by an unpredictable event like an air bubble or a mechanical shock (which did not happen in our experiments). Due to the electrode design, the deflector element can handle only one cell at a time in our set-up, which is the reason for what ROI2 needs to be free of cells before selecting a new target cell. Now, the target cell is in ROI3. When the cell reaches its corresponding position at the zig-zag array, the column is kept activated to prevent the cell to leave with the flow. The sequence to fill the full array is from right (distant electrodes) to left (close electrodes). The actions of the electrodes at the various decision points during the process of selection and arrangement are shown in the micrographs in Fig. 2.
Based on the previous conditions, we set-up a protocol for the classification and isolation of single cells from a sample containing a mixture of differently labeled cells and their automated arrangement as a group of 3 × 4 cells, as depicted in Fig. 3A. We chose this assay as an example, because color staining and fluorescent labelling are standard processes in cell biology to analyze or identify cells of interest in a heterogeneous cell sample. An image- and color-based cell processing has therefore multiple applications in biomedicine and biotechnology (e.g., analysis and characterization of circulating tumor cells,4 T cell receptor binding kinetics of antigen-specific immune cells23 or the dynamic analysis of immune cell interactions24). We used myeloma cells (Sp2/01-Ag, ATTC CRL-8006), with a diameter of approximately 10 to 12 μm. They are injected in the chip using a syringe pump (SP230IWZ, WPI, USA) at 7 μl h−1. The cell sample consisted of a mixture of non-stained, red stained (CellTrace Calcein Red-Orange AM, Invitrogen, Thermo Fisher Scientific, USA) and blue stained cells (CellTrace Calcein Violet AM, Invitrogen, Thermo Fisher Scientific, USA). After the staining process, the cells are suspended in PBS (phosphate buffered saline) with a conductivity around 1.4 S m−1.
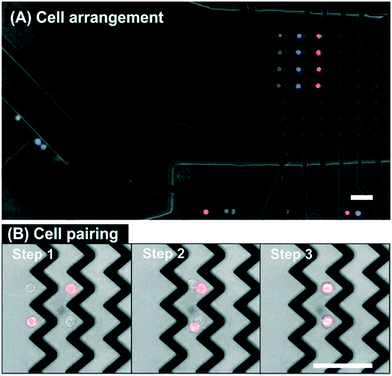
On the basis of color selection, we automatically and precisely isolated and positioned the target cells at specific locations in the microchannel (see Fig. 3A). The complete sequence of the experiment is shown in Video S1 (ESI†). First, four red stained cells are successfully isolated and arranged, then four blue stained cells, and finally four non-stained cells. Apart from the handling of the selected cells, it is also interesting to emphasize examples of cell exclusion found in Video S1† and in order to corroborate the correct performing of the protocol. At time 6 s the potentially selectable red stained cell is excluded because the previous selected cells are still under the influence of the deflector element. Another exclusion occurs at time 32 s where a doublet of red stained cells is discarded. At time 52 s the upcoming blue stained cell is not selected because the distance to the following cell is too close. Following the previously described exclusion rules, Video S1† shows how it is possible to automatically select and arrange by color a total of twelve cells in two minutes.
In another assay format, and to highlight how accurate and versatile the DEP cell handling technology is, two different cell types are put into contact by guiding them into the same trapping position of the zig-zag array. Fig. 3B shows the final sequence after selecting four cells of different colors, positioning them in two columns and two rows (2 × 2) and overlapping them in one single column. So the cells are selected and arranged in two different zig-zag columns, then those on the left are switched OFF and the flow carries the cells from the previous column to the next downstream zig-zag column where they are trapped. Now there are two cells at each of the traps of the zig-zag and they both arrange in their equilibrium position where the DEP forces and the hydrodynamic forces compensate each other: one cell is on top of the other and in direct contact. Direct contact between the cell membranes is relevant for studies of in situ cell-to-cell interaction, for instance, for immune cell activation studies or the development of chimeric antigen receptor (CAR) T-cell immunotherapies.25 The video containing the complete sequence is in ESI† (Video S2).
The combination of DEP with CV is a precise, effective and selective way to select and position single cells in a microfluidic device. Nevertheless, considerations about throughput are important when it comes to cell processing techniques. The throughput of our approach is limited by the time each target cell needs to be selected, transported and trapped for their arrangement in the array.
It is easy to accept that the time required for this is inversely proportional to the flow velocity in the microchannel. Higher flow velocities require higher DEP forces for cell manipulation, which scale as the square value of voltage applied to the electrodes. However, as the applied voltage contributes to temperature elevation inside the microchannel and, thus, affecting the viability of the cells, it is important to keep it at reasonably low values.26 In our configuration, with an applied voltage of 3 V, the maximum flow velocity is around 120 μm s−1 (for a microchannel of 35 μm height and 700 μm width this corresponds to a flow of 7 μl h−1). One of the most critical points is the time needed for a cell to cross the switch electrode which is less than 0.5 s, limiting the maximum throughput to approximately 2 cells per second. However, in order to process several thousands of cells, the system must be automatically running for approximately half an hour. Furthermore, parallelization of several microchannels or modification of the geometry of the electrodes (e.g., flatter electrode angles) are also feasible alternative to increase throughput.
Conclusions
This work presents the combination of a precise single-cell manipulation technique (i.e., DEP) with a CV program as a powerful tool to achieve fully automated and highly controlled handling of single cells in a microfluidic device. We have designed several electrode configurations which automatically and robustly perform specific actions on the cells, like: focusing, isolating, moving and trapping. DEP handling is a customizable and modular technique for single-cell manipulation. As an example, we describe here a color-based cell selection, where the target cells are automatically classified and isolated into a DEP-array, based on a CV program written in Python and using open source openCV functions. We also show the possibility of placing two cells at the same position of the array for in situ cell-to-cell interaction studies. We believe in the enormous potential of the combination of DEP with CV programming as a tool for the development of more specific and selective single-cell selection and processing protocols.Author contributions
Concept: MK, NG. Methodology: CGM, MK, CD. Software: NG. Validation: FP, TG, CGM, NG, MK. Writing and review: NG, MK, CD.Conflicts of interest
The authors ensure that there are no conflicts to declare.Acknowledgements
We acknowledge financial support by the Federal Ministry of Education and Research of Germany (project numbers 13GW0283A and 031A495B).Notes and references
- B. Dura, S. K. Dougan, M. Barisa, M. M. Hoehl, C. T. Lo, H. L. Ploegh and J. Voldman, Nat. Commun., 2015, 6, 5940 CrossRef CAS PubMed.
- M. Poudineh, M. Labib, S. Ahmed, L. N. M. Nguyen, L. Kermanshah, R. M. Mohamadi, E. H. Sargent and S. O. Kelley, Angew. Chem., Int. Ed., 2017, 56, 163–168 CrossRef CAS PubMed.
- Y. Deng, Y. Zhang, S. Sun, Z. Wang, M. Wang, B. Yu, D. M. Czajkowsky, B. Liu, Y. Li, W. Wei and Q. Shi, Sci. Rep., 2014, 4, 7499 CrossRef CAS PubMed.
- M. Dhar, J. N. Lam, T. Walser, S. M. Dubinett, M. B. Rettig and D. Di Carlo, Proc. Natl. Acad. Sci. U. S. A., 2018, 115, 9986–9991 CrossRef CAS PubMed.
- F. Wimmers, N. Subedi, N. van Buuringen, D. Heister, J. Vivié, I. Beeren-Reinieren, R. Woestenenk, H. Dolstra, A. Piruska, J. F. M. Jacobs, A. van Oudenaarden, C. G. Figdor, W. T. S. Huck, I. J. M. de Vries and J. Tel, Nat. Commun., 2018, 9, 3317 CrossRef PubMed.
- A. Gross, J. Schoendube, S. Zimmermann, M. Steeb, R. Zengerle and P. Koltay, Int. J. Mol. Sci., 2015, 16, 16897–16919 CrossRef CAS PubMed.
- M. Li and R. K. Anand, Anal. Bioanal. Chem., 2018, 410, 2499–2515 CrossRef CAS PubMed.
- E. Sinkala, E. Sollier-Christen, C. Renier, E. Rosàs-Canyelles, J. Che, K. Heirich, T. A. Duncombe, J. Vlassakis, K. A. Yamauchi, H. Huang, S. S. Jeffrey and A. E. Herr, Nat. Commun., 2017, 8, 14622 CrossRef CAS PubMed.
- M. Werner, F. Merenda, J. Piguet, R.-P. Salathé and H. Vogel, Lab Chip, 2011, 11, 2432–2439 RSC.
- Z. R. Gagnon, Electrophoresis, 2011, 32, 2466–2487 CrossRef CAS PubMed.
- A. Ramos, H. Morgan, N. G. Green and A. Castellanos, J. Phys. D: Appl. Phys., 1998, 31, 2338–2353 CrossRef CAS.
- M. Doan and A. E. Carpenter, Nat. Mater., 2019, 18, 414 CrossRef CAS PubMed.
- X. Li, W. Chen, Z. Li, L. Li, H. Gu and J. Fu, Trends Biotechnol., 2014, 32, 586–594 CrossRef CAS PubMed.
- J. Riordon, D. Sovilj, S. Sanner, D. Sinton and E. W. K. Young, Trends Biotechnol., 2019, 37, 310–324 CrossRef CAS PubMed.
- R. S. W. Thomas, P. D. Mitchell, R. O. C. Oreffo and H. Morgan, Biomicrofluidics, 2010, 4, 022806 CrossRef PubMed.
- N. Nitta, T. Sugimura, A. Isozaki, H. Mikami, K. Hiraki, S. Sakuma, T. Iino, F. Arai, T. Endo, Y. Fujiwaki, H. Fukuzawa, M. Hase, T. Hayakawa, K. Hiramatsu, Y. Hoshino, M. Inaba, T. Ito, H. Karakawa, Y. Kasai, K. Koizumi, S. Lee, C. Lei, M. Li, T. Maeno, S. Matsusaka, D. Murakami, A. Nakagawa, Y. Oguchi, M. Oikawa, T. Ota, K. Shiba, H. Shintaku, Y. Shirasaki, K. Suga, Y. Suzuki, N. Suzuki, Y. Tanaka, H. Tezuka, C. Toyokawa, Y. Yalikun, M. Yamada, M. Yamagishi, T. Yamano, A. Yasumoto, Y. Yatomi, M. Yazawa, D. Di Carlo, Y. Hosokawa, S. Uemura, Y. Ozeki and K. Goda, Cell, 2018, 175, 266–276.e13 CrossRef CAS PubMed.
- M. Kirschbaum, M. S. Jaeger, T. Schenkel, T. Breinig, A. Meyerhans and C. Duschl, J. Chromatogr. A, 2008, 1202, 83–89 CrossRef CAS PubMed.
- M. Kirschbaum, C. R. Guernth-Marschner, S. Cherré, A. de P. Peña, M. S. Jaeger, R. A. Kroczek, T. Schnelle, T. Mueller and C. Duschl, Lab Chip, 2012, 12, 443–450 RSC.
- M. Kirschbaum, M. S. Jaeger and C. Duschl, Lab Chip, 2009, 9, 3517–3525 RSC.
- Z. Hu and L. Kuo-Kang, J. R. Soc., Interface, 2008, 5, 671–690 CrossRef PubMed.
- M. Dürr, J. Kentsch, T. Müller, T. Schnelle and M. Stelzle, Electrophoresis, 2003, 24, 722–731 CrossRef PubMed.
- T. Muller, A. Pfennig, P. Klein, G. Gradl, M. Jager and T. Schnelle, IEEE Eng. Med. Biol. Mag., 2003, 22, 51–61 CrossRef PubMed.
- M. Nauerth, B. Weißbrich, R. Knall, T. Franz, G. Dössinger, J. Bet, P. J. Paszkiewicz, L. Pfeifer, M. Bunse, W. Uckert, R. Holtappels, D. Gillert-Marien, M. Neuenhahn, A. Krackhardt, M. J. Reddehase, S. R. Riddell and D. H. Busch, Sci. Transl. Med., 2013, 5, 192ra87 Search PubMed.
- J. H. Jang, Y. Huang, P. Zheng, M. C. Jo, G. Bertolet, M. X. Zhu, L. Qin and D. Liu, J. Immunol., 2015, 195, 1320–1330 CrossRef CAS PubMed.
- M. Sadelain, R. Brentjens and I. Rivière, Cancer Discovery, 2013, 3, 388–398 CrossRef CAS PubMed.
- M. S. Jaeger, T. Mueller and T. Schnelle, J. Phys. D: Appl. Phys., 2006, 40, 95–105 CrossRef.
Footnote |
| † Electronic supplementary information (ESI) available: Video1.avi (cell selection, handling and trapping) and Video2.avi (cell pairing). See DOI: 10.1039/c9lc00800d |
| This journal is © The Royal Society of Chemistry 2019 |