Integrated microfluidic systems with sample preparation and nucleic acid amplification
Juxin
Yin
 a,
Yuanjie
Suo
a,
Zheyu
Zou
a,
Jingjing
Sun
a,
Shan
Zhang
a,
Beng
Wang
a,
Yuanjie
Suo
a,
Zheyu
Zou
a,
Jingjing
Sun
a,
Shan
Zhang
a,
Beng
Wang
 bf,
Yawei
Xu
c,
Diane
Darland
*d,
Julia Xiaojun
Zhao
*e and
Ying
Mu
bf,
Yawei
Xu
c,
Diane
Darland
*d,
Julia Xiaojun
Zhao
*e and
Ying
Mu
 *a
*a
aResearch Centre for Analytical Instrumentation, Institute of Cyber-Systems and Control, State Key Laboratory of Industrial Control Technology, Zhejiang University, Hangzhou, 310058, China. E-mail: muying@zju.edu.cn
bCancer Institute (Key Laboratory of Cancer Prevention and Intervention, National Ministry of Education), The Second Affiliated Hospital, School of Medicine, Zhejiang University, Hangzhou, 310009 China
cCollege of Biological and Pharmaceutical Engineering, Jilin Agricultural Science and Technology University, Jilin, 132000 China
dDepartment of Biology, University of North Dakota, USA. E-mail: diane.darland@und.edu
eDepartment of Chemistry, University of North Dakota, USA. E-mail: julia.zhao@und.edu
fInstitute of Translational Medicine, Zhejiang University, Hangzhou, 310029 China
First published on 31st July 2019
Abstract
Rapid, efficient and accurate nucleic acid molecule detection is important in the screening of diseases and pathogens, yet remains a limiting factor at point of care (POC) treatment. Microfluidic systems are characterized by fast, integrated, miniaturized features which provide an effective platform for qualitative and quantitative detection of nucleic acid molecules. The nucleic acid detection process mainly includes sample preparation and target molecule amplification. Given the advancements in theoretical research and technological innovations to date, nucleic acid extraction and amplification integrated with microfluidic systems has advanced rapidly. The primary goal of this review is to outline current approaches used for nucleic acid detection in the context of microfluidic systems. The secondary goal is to identify new approaches that will help shape future trends at the intersection of nucleic acid detection and microfluidics, particularly with regard to increasing disease and pathogen detection for improved diagnosis and treatment.
1. Introduction
Molecular diagnosis is a technique that uses molecular biology methods to detect changes in the structure or level of expression of patient-derived genetic material. This technique involves nucleic acid amplification testing (NAAT) technology. With the high demand for rapid detection tools for diseases and pathogens in the fields of medicine and food safety, the focus has been directed towards molecular diagnostic approaches for point of care (POC). POC is an approach that involves analyzing samples at the site of sample collection, thereby reducing the time required for diagnosis and eliminating (or reducing) the need to transfer specimens to offsite testing laboratories.1–5 The rapid turnaround for POC diagnosis usually does not require professional personnel, such as clinical examiners, for sample processing and analysis. Compared with traditional laboratory detection methods, POC primarily achieves portable in situ detection partially completed by technical staff which streamlines operational procedures, integrates detection devices, and reduces detection costs.6–8 The microfluidic chip technology, also known as the micro total analysis system (μTAS) was first proposed by Manz and colleagues in the 1990s.9 With more than twenty years of development, it has become an independent area of biochemical analysis. The microfluidic chip has features such as rapid and efficient analysis, low consumption, and miniaturization. Moreover, it has the ability to perform nucleic acid extraction and detection on a single chip, thereby providing an excellent platform for POC.10,11Fig. 1 highlights the conceptual intersection of technical improvements for nucleic acid isolation and amplification with the microfluidics platform. Improvement at any of these critical technical points confers additive advancements that directly contribute to improved POC and increased treatment efficiency (Fig. 1).To provide real-time diagnosis of diseases and early screening for diseases,12–14 it is important to develop techniques that allow for accurate and rapid detection of nucleic acids, including deoxyribonucleic acid (DNA) and ribonucleic acid (RNA), which may reveal the contamination of pathogens in food and the environment as well as the emergence of tumor markers using tissue scrapes and biopsies. Indeed, the discovery of circulating nucleic acids has allowed for fast, accurate, and cost-effective detection of nucleic acids, thereby playing an increasingly important role in the early diagnosis of disease.15–17 Traditional nucleic acid detection methods have several challenges, including lengthy preparation and analysis time, vulnerability to contamination, sensitivity limitations, and complex procedures and operation. Collectively, these challenges limit the effectiveness of these methods in POC. Integration of microfluidic technology with nucleic acid extraction helps to address previous technical and analytical limitations associated with nucleic acid detection.
The development of nucleic acid detection technology based on the microfluidic chip has greatly improved the diagnostic efficiency and accuracy of various diseases.18,19 NAAT based on the microfluidic chip has the advantages of being highly sensitive and specific, while still providing a user-friendly workflow with good qualitative and quantitative detection potential.20–22 A specific example of this is the integrated nucleic acid extraction, amplification, and detection (INEAD) system developed by combining microfluidic applications in POC and clinical diagnosis. The chip can be used for sample preparation, nucleic acid signal amplification, and quantification of results on a single device, which actualizes the goal of “sample-in-answer-out”. The entire procedure is performed in a closed environment, which reduces the burden on the operator and addresses the need for rapid detection without time and location constraints.
In this review, we will focus on current methods for extraction of nucleic acids in microfluidic systems and advanced methods for nucleic acid amplification implemented in microfluidic systems. Furthermore, we will introduce current technologies and approaches integrating the INEAD system and provide an outlook for future technological approaches.
2. Current methods in nucleic acid isolation and purification
Nucleic acid extraction is the “first step” in molecular diagnostics and nucleic acid detection for research applications. The extraction of nucleic acids mainly refers to the separation of nucleic acids from biological macromolecules such as proteins, polysaccharides, and fats. The following target goals are associated with nucleic acid extraction: to ensure the integrity of the primary structure of nucleic acid molecules is preserved; to exclude other molecular contaminants; and to optimize yield. Sample lysis (whether from whole tissue, cells, or blood samples) and nucleic acid adsorption are necessary steps for nucleic acid extraction. The quality and efficiency of nucleic acid extraction directly influence the research results or diagnostic output. In many situations, nucleic acid extraction time accounts for a higher fraction of the total time spent during the detection process, and hence greatly affects the efficiency of the entire detection process. Any mistakes during the extraction process may inhibit the downstream applications such as polymerase chain reaction (PCR) or alternative amplification approaches as well as the purity of the end product.23 This also poses a huge challenge to quality and consistency. Application of microfluidic systems overcomes this limitation and improves nucleic acid extraction efficiency through the precise design, automation, and single system approach.24–26 Here we highlight nucleic acid extraction methods that are based on microfluidic systems, and identify their advantages and limitations.2.1 Isolation of nucleic acids by magnetic beads
Magnetic beads are formed by coating a core of Fe3O4 with an active group that can be adsorbed by a magnet and then bind to nucleic acids in a cell or tissue lysate.21,27,28 The combination of beads with nucleic acids is controlled by adjusting the pH or the salt concentration.29 High flux, automated extraction of nucleic acids is achieved by applying magnetic beads in synergy with other bead compositions, such as silicon.30 Their compact size, ease of handling, and high efficiency when used to isolate nucleic acids make magnetic beads suitable for microfluidic platform applications.27,31–33 The following factors contribute to the efficiency of magnetic beads for nucleic acid extraction. (1) The magnetic force on the surface of the magnetic beads. (2) The strength of the magnetic field. It should be emphasized here that the force is proportional to the magnetic field gradient and not the magnetic field intensity. (3) The binding capacity of the magnetic beads with the nucleic acid. Prior studies have demonstrated the importance of magnetic beads in isolating RNA. For example, Shi et al. proposed a total RNA extraction droplet array (TREDA) system for RNA purification. The authors have developed a system that uses hydrophilic spots to hold stationary droplets and uses magnets to control the movement of magnetic beads between droplets (Fig. 2A). This approach can extract total RNA in a low cell concentration medium in less than 5 minutes and the product can be used readily for quantitative real-time PCR (qPCR) platforms.34 This approach is, therefore, highly amenable to simultaneous extraction of multiple samples and the extraction of trace samples from complex sources.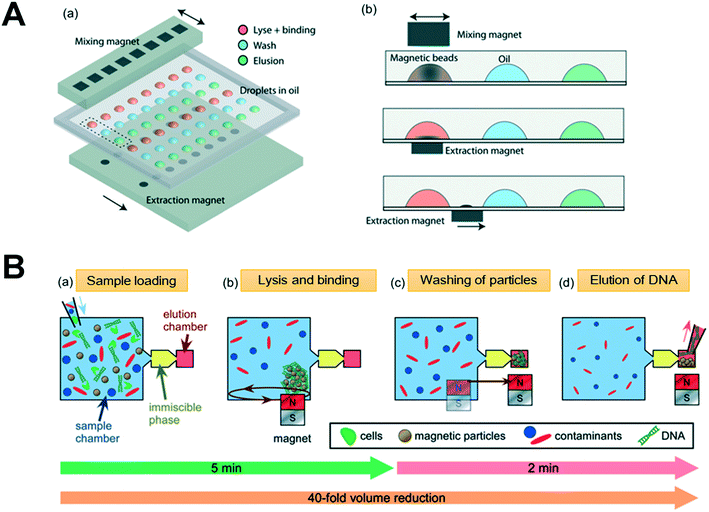 | ||
| Fig. 2 Magnetic beads-based microfluidic nucleic acid extraction chip. (A) Overview of the TREDA system (Shi et al., 2015) (a) schematic of the TREDA chip where (b) controls the dispersion, aggregation, and movement of magnetic beads. Adapted from ref. 34 with permission from the Royal Society of Chemistry. (B) Schematic of the DNA extraction process showing (Mosley et al., 2016) (a) sample loading and lysis. (b) Mixing of superparamagnetic particles (PMPs) to combine with DNA. During the process, the magnet can control the movement of the magnetic beads, so the magnetic beads are in an aggregate state. This process can be accelerated by controlling the movement of the magnetic beads. (c) Transfer of PMPs through the immiscible phase for washing, (d) elution of DNA from the PMPs and collection of the nucleic acid for off-chip analysis is shown. Adapted from ref. 38 with permission from the Royal Society of Chemistry. | ||
Surface tension is an important parameter in the application of magnetic beads in microfluidic chips since it can achieve the separation of different extraction components at the microscopic scale.35–37 Mosley et al. revealed the potential of magnetic beads to extract nucleic acids from complex samples using a microfluidic system (Fig. 2B). They leveraged surface tension to separate reagents and employed an external magnetic field to control the movement of magnetic beads to achieve DNA extraction from stool samples. In this study, the authors used mineral oils with better biocompatibility to separate different components, and chose the appropriate lysis buffer (5M guanidinium hydrochloride, GuHCl) optimized to the amount of magnetic beads for efficient nucleic acid extraction.38 Shu et al.39 proposed an active droplet-array (ADA) approach in which the reagents are pre-stored in water-in-oil form and the oil phase infiltrates the micro-cells and slits while the aqueous phase is excluded due to surface tension. Meanwhile, lysis, washing, elution, and detection are accomplished by automatically controlling the movement of beads in the droplet array. The latter system provides the magnetic beads with automation control and reduces handling-related contamination issues.
The magnetic bead method is suitable for automated extraction and many commercial automatic nucleic acid extractors are based on this method such as MagNA Pure96 (Roche™), Smart LabAssist-32 (TAN Bead™), BioRobot MDx (QIAGEN), and SPRI-TE (Beckman). The magnetic bead-based nucleic acids extraction process is rapid and results in high-efficiency extraction when integrated with microfluidic platforms. The advantage of magnetic beads combined with microfluidic platforms in automated extraction of nucleic acids is likely to promote the development of molecular diagnostics in the future. However, the control of magnetic beads is still a difficult challenge and requires a complicated control system to achieve automation. This technical limitation renders most of the current products to be bulky and costly, which limits its application in POC and other clinical applications. It is also important to note that nucleic acid extraction should be performed in an absolutely closed environment to prevent exogenous contamination. At present, although certain measures have been developed to prevent contamination, some products are still vulnerable to contamination.
2.2 Silicon-based method of nucleic acid isolation
Silica is widely used in DNA extraction because of its stability, biocompatibility, and easy modification properties, especially in solid phase extraction (SPE).40,41 Being an important sample pretreatment and enrichment technology, the application of SPE in microfluidic chips has attracted much attention and is an effective strategy for integrating nucleic acids in microfluidic devices.42–44 The target nucleic acid molecule combines with the silica-based material that is modified on the microfluidic chip in the presence of high chaotropic salt concentration.45–47 The bound nucleic acids can be washed with the appropriate solution, such as diluted ethanol or isopropanol, and then quickly eluted by a small amount of solvent to extract the target nucleic acid.48 Different forms of silicon have been utilized in the microfluidic system for nucleic acid extraction. In 1999, Christel et al.49 demonstrated that silicon fluidic microchips could be used for DNA extraction. In this study, thousands of micropillars with a silica surface were fabricated by deep reactive ion etching on a silicon wafer, which increased the internal surface area of the chip. Ramsey's group50 applied silica membranes for the concentration of DNA on the microfluidics which allowed for DNA fragments to be concentrated up to 2 orders of magnitude. Petralia et al. used silica-coated pillar arrays on microchips to extract DNA (Fig. 3A) and reported that elution efficiency strongly depended on the geometrical dimensions of the pillars and efficiency increased with the surface/volume ratio.51 Park et al.48 developed a complete rotating microfluidic system using silicon beads as a material for DNA extraction (Fig. 3B). The authors applied the geometrical depth on the chip so that reaction liquid was stored in the chip, and the precise design of the microfluidic system and rotational speed control could ensure that each step progressed smoothly. Based on this method, they integrated DNA extraction and amplification into a centrifugal chip and detected 50 colony forming units (CFU) of pathogens within 80 minutes. Gunal et al.52 used monodisperse-porous silica microspheres on the microfluidic system to isolate 14 ng DNA from 10 μL of whole blood lysate, further demonstrating the value of silicon-based approaches in nucleic acid isolation.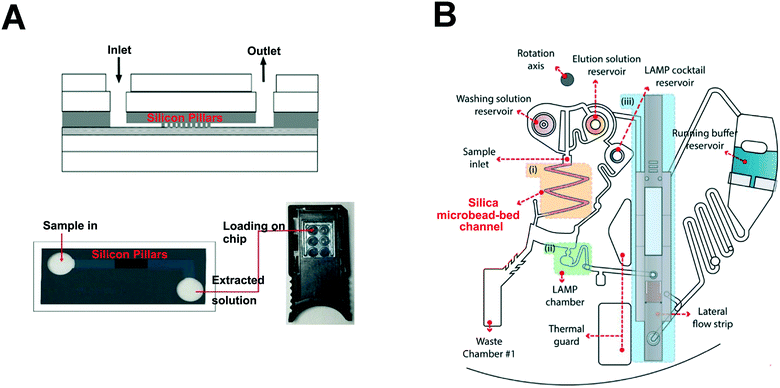 | ||
| Fig. 3 Silica pillar-based nucleic acid extraction method. (A) Silica-coated pillar arrays on microchips for DNA extraction (Petralia et al., 2017). The chip is composed of a 6-layer structure and the size of silicon pillars array is 5 × 1.8 mm2. Adapted from ref. 51 with permission from the Royal Society of Chemistry. (B) Silicon bead-silicon beads nucleic acid extraction method. Schematic illustration of the integrated rotary microdevice for the DNA extraction, the loop-mediated isothermal amplification (LAMP) reaction, and the lateral flow strip detection are shown (Park et al., 2017). Nucleic acid extraction is based on the silica microbead-bed channel, which serves as a solid phase matrix. DNA extraction is achieved by controlling different speeds and the extraction efficiency can be up to 80% in the microdevice. Adapted from ref. 48 with permission from Elsevier. | ||
On-chip silicon solid-phase extraction technology not only reduces the complexity of the analysis and the time required, but also promotes the integration and miniaturization of analytical instruments. However, nucleic acid extraction based on this method produces more waste liquid; therefore, a suitable waste liquid storage area has to be designed on the microfluidic chip to prevent or reduce pollution. Moreover, microfluidic nucleic acid extraction based on this method requires relatively complex equipment such as those required for manufacturing of the chips, which limits the application of this method.
2.3 Paper-based method of nucleic acid isolation
Paper, a new branch of microfluidic technology, is an attractive and inexpensive platform (Lab-on-a-paper) for nucleic acid extraction due to its inherent advantages such as biocompatibility, high surface area, and absorptive nature.53–55 A filter paper consisting of lysis reagent and protein denaturation reagent can be used to store reagents in a dried form for long periods without refrigeration.19,56,57 Commercial filter papers have been launched in the market, including products such as 903® paper and a Fast Technology Analysis (FTA)® card.58 The pores of filter paper not only affect the binding of nucleic acids (such as DNA) but also influence the post-preparation processes such as PCR efficiency and stability. These are the two major challenges of using paper-based microfluidic chips for nucleic acid extraction.59 One of the common methods of nucleic acid extraction by paper-based microfluidic chips involves the use of lysis buffer and extraction buffer in the reservoir module.60,61 Chitosan is used to prevent the inhibitory effects of guanidine and isopropanol on PCR and it can be applied in nucleic acid extraction with paper-based microfluidic chips.62 Gan et al. (Fig. 4) developed a chitosan-modified Fusion 5 filter paper by embedding it in a thermoplastic microchip. This highly efficient approach leveraged the principle of the entanglement of DNA with fibers and the electrostatic adsorption of DNA to chitosan polymers.63 This idea is similar in principle to the approach proposed by Byrnes et al., who developed a porous chitosan membrane that could purify and concentrate DNA from complex samples.64 To extract RNA, Rodriguez et al.65 proposed an alcohol precipitation method based on a polyethersulfone (PES) filter paper. The filter paper was used to detect H1N1 from human clinical nasopharyngeal specimens. The fastest nucleic acid extraction has been achieved using a filter paper (Whatman No. 1). Filter paper allows rapid extraction and purification of nucleic acids from plants, animals and microorganisms within 30 seconds at a very low cost.66 As a result, this technology can be used to extract DNA and RNA without specialized equipment.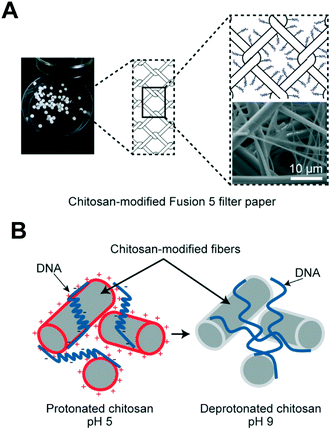 | ||
| Fig. 4 Chitosan-modified Fusion 5 filter paper and DNA capture mechanism (Gan et al., 2017). (A) 3 mm-diameter discs of chitosan-modified Fusion 5 filter paper. Schematic and scanning electron microscope image of the fiber matrix coated with chitosan polymers. (B) Schematic of the DNA capture mechanism. At a pH around 5, DNA molecules are “actively” adsorbed onto the chitosan-modified fibers. Once DNA is on the fibers, the physical entanglement of the long-chain molecules with the fiber matrix can also assist the capture. At a pH of 9, although DNA is not “actively” absorbed onto the fiber, DNA molecules remain bound due to the physical trapping of these long-chain DNA molecules within the fiber matrix against washing and elution. Adapted from ref. 63 with permission from ACS Publications. | ||
Over the years, researchers in different fields have been using several methods for nucleic acid extraction from paper technology. This technology can extract nucleic acids from samples with different viscosities such as raw viscous samples,67 urine,68 and bacteria solutions.69 Optimization of paper-based microfluidic chips will help increase the efficiency and decrease the cost of nucleic acid extraction. In addition, several studies have revealed that nucleic acid amplification methods (such as PCR, LAMP) can be conducted directly within a paper membrane,70–72 making the paper method suitable for nucleic acid extraction. Although the paper-based microfluidic chip has its unique advantages, it still needs to overcome the challenge in NAAT such as reagent storage, and automation.55,73 In addition, paper-based nucleic acid extraction is performed in an open environment and it is susceptible to contamination during the extraction process. Therefore, it is likely that a combination of paper extraction methods with microfluidics will overcome these challenges and broaden the scope of application for this cost-effective approach.
2.4 Alternative nucleic acid purification methods based on microfluidics
In addition to the methods mentioned above, several simple and efficient nucleic acid extraction methods based on microfluidic systems have been developed for nucleic acid detection. These methods play a positive role in the development of microfluidic systems and increase the efficiency of nucleic acid extraction using microfluidics platforms. Appropriate processing and modification of existing materials could allow extraction of nucleic acids using simple and common materials, thereby making these approaches more accessible. Fu et al. used poly-diallyl-dimethylammonium-chloride (PDDA), which has a large amount of positively charged quaternary ammonium groups, to modify the internal surface of a capillary making it able to attract negatively charged DNA or RNA.74 Organic solvents can also be applied to microfluidic systems for nucleic acid extraction. Morales et al. developed a dual inlet-dual output serpentine device that allows DNA isolation in the aqueous phases under stratified flow and droplet-based flow conditions.75 In a recent study, a novel microfluidic liquid phase nucleic acid purification chip was developed by Zhang et al. The chip was designed to selectively isolate DNA or RNA from low copy/single bacterial cells in the range of 5000 down to a single cell in a sample volume of 1 μL or 125 nL. The on-chip liquid phase nucleic acid purification was 10-fold higher than the conventional column-based solid phase nucleic acid extraction methods with the added advantage of small volume handling capacity.76Application of microelectronic components to a microfluidic system has also been found to be effective in nucleic acid extraction. Marshall et al. proposed the use of integrated heaters in combination with isotachophoresis to lyse the sample and isolate nucleic acids.77 The authors used this system to process nucleic acids from clinically-derived blood samples. Han et al. invented a self-powered switch-controlled system (SSNES) which has two disposable syringes and a switchgear made of polymethyl methacrylate (PMMA) blocks and an O-ring for the nucleic acid extraction. In this system, the authors use a dimethyl adipimidate (DMA)-based extraction method with the microfluidic system.78 Yoon et al.79 also reported a DMA chip for RNA extraction from different cell lines with rapid processing time and high product purity (Fig. 5A).
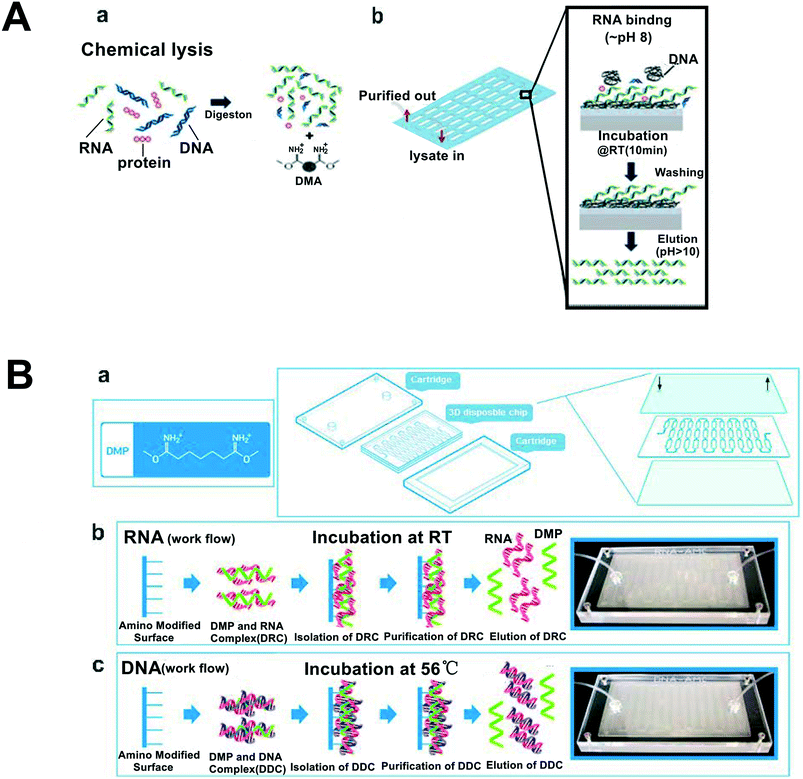 | ||
| Fig. 5 Schematic of DMA chip and DMP chip for nucleic acid extraction. (A) DMA chip for RNA isolation. a: Cell lysis. Different components including DNA, RNA and protein are released. b: On-chip RNA isolation. RNA is bound and eluted by controlling pH. Adapted from ref. 79 with permission from the Elsevier. (B) DMP chip for DNA and RNA isolation. (a) Chemical structure of DMP and schematic drawing for assembling of a plastic type microfluidic cartridge with a 3D disposable chip. (b) Schematic and photograph workflows for the DMP system for RNA (b) and DNA (c) extraction. Adapted from ref. 20 with permission from ACS Publications. | ||
Different chemicals that bind nucleic acids, can be applied to microfluidic systems for efficient nucleic acid extraction. Jin et al. incorporated dimethyl pimelimidate (DMP) into the microfluidic system which led to efficient extraction of nucleic acids (DNA and RNA) from various samples such as mammalian cells, bacterial cells, and viruses from human disease (Fig. 5B). The DMP captures the nucleic acid and the system does not need instruments or electricity.20 Since plasma is an important sample source for clinical testing, Liu et al. proposed a simple and highly efficient plasma separator that does not need centrifugation. Compared with other POC methods, this separator has higher DNA recovery efficiency, reaching up to 84.5%.80 Adjusting the properties of the material and the charge of the nucleic acid allows for a greater range of isolation methods to be integrated with microfluidic platforms. Although the methods mentioned above apply different principles for nucleic acid extraction using microfluidic systems with unique advantages, their efficiency, affordability, security, and simplicity need to be further optimized for broader application and accessibility. A key area that need to be improved is the integration of nucleic acid extraction with downstream reactions. Several integration approaches have been identified to date that can contribute to POC and increased treatment efficiency,81–84 but opportunities remain to build on these technologies. Regardless of the isolation method utilized, certain measures should be taken to prevent nucleic acid contamination and ensure accuracy of the results and quality of the product.
3. Nucleic acid amplification using microfluidics approaches
Microfluidics-based approaches provide an ideal platform for nucleic acid amplification and rely mainly on fast prototyping by soft lithography in polydimethylsiloxane (PDMS) or glass etching.85,86 Although PDMS has been applied in many studies, the issue of evaporation limits its practical application due to gas permeability. In addition to this, injection molding,87,88 thermal bonding and hot embossing89,90 along with changes in solvent use91–93 are important alternatives which can improve nucleic acid amplification on a microfluidics system. Nucleic acid amplification on chips also confers the advantages of amplification speed, detection limit, sample demand, and detection accuracy.42,94 In recent years, many kinds of nucleic acid amplification approaches such as PCR and isothermal amplification have been performed on the microfluidic system, with the achievement of fast and accurate detection. This section will summarize several promising techniques which have been used on microfluidic systems.3.1 Digital PCR (dPCR) amplification of nucleic acids
qPCR technology is used to quantify nucleic acids and is the gold standard for the diagnosis of infectious diseases.95–97 However, it is a relative quantification technique that relies on a standard curve or reference gene assessment to determine the amount of target nucleic acid. For low copy number target DNA, issues such as primer efficiency and differences in template concentrations can markedly affect the detection sensitivity and accuracy at the end-stage.18,19,98,99 dPCR is a third generation PCR technology which can provide a direct count of target molecules without relying on any calibrators or external standards to determine the absolute number of molecules as low as a single copy.66 The concept behind digital PCR was first proposed by Sykes et al. in 1992.100 Although this method was not referred to as “digital PCR” at the time, the basic experimental procedure for digital PCR was established. Moreover, the fundamental principle of digital PCR detection provided an all-or-none for the assay.101 In 1999, Vogelstein and Kinzler performed dPCR reactions using microplates for the first time, which allowed for a high throughput strategy.102 This approach allows for large-scale multiple dilution and liquid separation until the number of molecules to be detected does not exceed 1 (0 or 1) in each subdivided sample. All subdivided samples were subjected to PCR amplification under the same conditions, and the target molecules are amplified to generate a very strong fluorescence source. The number of samples and total number of microwells are counted and the Poisson distribution is used to predict the initial concentration of the target sequence.103,104 dPCR can be further divided into droplet digital PCR (ddPCR) and integrated flow circuit (IFC) chip dPCR. Studies have demonstrated that ddPCR and IFC dPCR are more robust than standard qPCR approaches for detecting trace samples101,105,106 and have been applied in gene mutation analysis,47,107 prenatal diagnosis of chromosome abnormality,46,108 DNA copy number determination,50,107 pathogen detection,78,109 transgenic detection,55,68 among other applications. Modifications to dPCR chip have been developed in recent years that have advanced the application of this innovative approach for detection and quantification of nucleic acids. Microfluidic technology in combination with dPCR offers greater throughput, high sensitivity, and improved accuracy over more traditional PCR-based methods.Researchers have broken through many technical bottlenecks with the development of micro-electro-mechanical system (MEMS) and microfluidic technology.73 Hansen's group developed an ultra-high density megapixel digital PCR chip with 1 million cells. The chip can have a density of up to 440![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 reactors per square centimeter and a reaction chamber size of 20 μm × 20 μm × 25 μm, which offers a dynamic range of 107.106 Shen et al.110 established a slipchip consisting of two glass sheets, one etched with a reaction cell and the other linked to a channel. When the two lamellae are combined with the channel, a continuous flow path is formed, and the reaction reagent can enter the reaction chamber from the inlet to reach the outlet. When the two chips slide, the channels and the ridges are misaligned and the flow path is cut off. In recent years, our group developed an integrated temporary negative pressure-assisted microfluidic chip to combine DNA isolation and digital PCR detection in one chip. This research laid the foundation for the connection between different functional areas.111 Our team has developed a series of self-priming, liquid-divided, integrated flow path chips. An integrated on-chip, valve-free and power-free microfluidic dPCR device has been developed based on a novel self-priming compartmentalization for the first time.112 In 2017, our team reported a dPCR chip with a scalable branch network structure (Fig. 6). The chip is a fully-encapsulated dPCR chip that has no waste or cross-contamination from the external environment. In addition, a negative pressure liquid separation method has been adopted in which the uniform dispersion of the sample can be accomplished without the aid of an external device. This method is more flexible and is suitable for rapid isolation.113 Ning et al.114 proposed a digital PCR chip with zero-water-loss. In this low-cost chip, the authors integrated a void in the space beneath the microwells, which provided power and water storage during the isolation procedure.
000 reactors per square centimeter and a reaction chamber size of 20 μm × 20 μm × 25 μm, which offers a dynamic range of 107.106 Shen et al.110 established a slipchip consisting of two glass sheets, one etched with a reaction cell and the other linked to a channel. When the two lamellae are combined with the channel, a continuous flow path is formed, and the reaction reagent can enter the reaction chamber from the inlet to reach the outlet. When the two chips slide, the channels and the ridges are misaligned and the flow path is cut off. In recent years, our group developed an integrated temporary negative pressure-assisted microfluidic chip to combine DNA isolation and digital PCR detection in one chip. This research laid the foundation for the connection between different functional areas.111 Our team has developed a series of self-priming, liquid-divided, integrated flow path chips. An integrated on-chip, valve-free and power-free microfluidic dPCR device has been developed based on a novel self-priming compartmentalization for the first time.112 In 2017, our team reported a dPCR chip with a scalable branch network structure (Fig. 6). The chip is a fully-encapsulated dPCR chip that has no waste or cross-contamination from the external environment. In addition, a negative pressure liquid separation method has been adopted in which the uniform dispersion of the sample can be accomplished without the aid of an external device. This method is more flexible and is suitable for rapid isolation.113 Ning et al.114 proposed a digital PCR chip with zero-water-loss. In this low-cost chip, the authors integrated a void in the space beneath the microwells, which provided power and water storage during the isolation procedure.
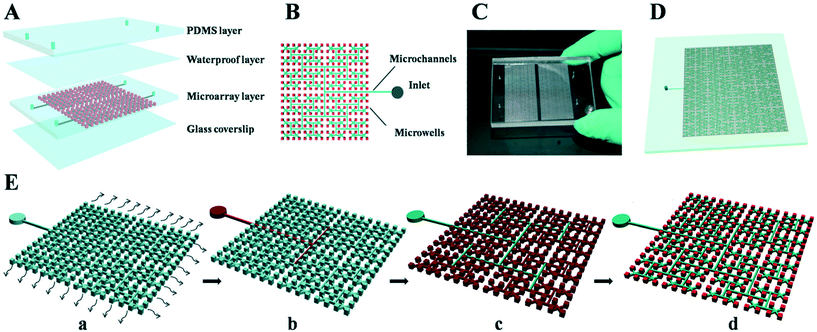 | ||
Fig. 6 Schematic drawing of the fractal branching microchannel net chip (Zhu et al., 2017). (A) Schematic diagram of the chip that has 4096 microwells for dPCR reaction. (B) Diagram of the details of the chip design. (C) Photograph of the chip. (D) The scalability of the chip with 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 384 microwells in each reaction panel. (E) The principle and operation procedure of the microfluidic device: (a) the chip is degassed in a vacuum pump and then adhesive tape is attached to seal the top surface of the chip after the degassing step; (b) the adhesive tape is punctured, and the reagent can be dispensed into the inlet, while the degassing-drive flow primes the sample into the microwells quickly; (c) the oil is then dispensed into the inlet, and the oil phase is self-primed into the channels; (d) all the sample solutions are partitioned into each microwell by the oil, and no sample is wasted. Finally, the chip is sealed using a coverslip to run PCR amplification. Adapted from ref. 113 with permission from the Royal Society of Chemistry. 384 microwells in each reaction panel. (E) The principle and operation procedure of the microfluidic device: (a) the chip is degassed in a vacuum pump and then adhesive tape is attached to seal the top surface of the chip after the degassing step; (b) the adhesive tape is punctured, and the reagent can be dispensed into the inlet, while the degassing-drive flow primes the sample into the microwells quickly; (c) the oil is then dispensed into the inlet, and the oil phase is self-primed into the channels; (d) all the sample solutions are partitioned into each microwell by the oil, and no sample is wasted. Finally, the chip is sealed using a coverslip to run PCR amplification. Adapted from ref. 113 with permission from the Royal Society of Chemistry. | ||
In recent years, several techniques and instruments based on digital drop PCR (ddPCR) have been developed. Compared to microarray chips, the processing of ddPCR chips is simpler and the number of droplets is more flexible than in previous iterations of the technology.72,115 Chen et al. reported a capillary-based droplet dPCR chip, which integrates droplet generation, PCR reaction, and flow detection in a single system. This approach avoids the fusion of droplets in the reaction chamber and sample loss caused by transfer between instruments.116 An ideal ddPCR assay should be easy and fast to operate in most laboratories. Huang et al. developed an off-chip monodispersion droplet generation method that can efficiently produce results without an emulsion step using a highly precise micro-channel array and a bench-top centrifuge machine. The results were robust and the cost of dPCR was reduced, supporting the idea that dPCR could be adopted in standard molecular laboratories.117
Compared to traditional quantitative PCR techniques, dPCR achieves absolute quantification by measuring the copy number of a specific gene. However, dPCR cannot be used, at this time, to perform large-scale gene sequencing and can only be used for quantitative detection of specific known genes. Multiplex PCR detection is likely to be an attractive development direction for this technology and will promote the application of dPCR in mutation analysis96 and genotyping,105 for example. In combination with different fluorescent probes or different intensities of fluorescence, dPCR can be applied in multiple detection systems.105,118–120 Didelot et al.121 used a multiplex picoliter droplet-based digital PCR method to detect human genomic DNA samples of 4 lengths (78, 159, 197, and 550 bp) and the results were consistent with the sequencing data run in parallel. Zhong et al.122 reported a novel and easy method which is based on the singular nature of amplifications at terminal dilution for multiplexing dPCR in picolitre droplets. The authors theoretically performed 10 multiple detections by this approach. To achieve these results, they used 5 multiple detection assays for spinal muscular atrophy with just two fluorophores.
In addition, it is worth mentioning that the detection method of digital PCR results mainly relies on the quality and intensity of the fluorescent signals (fluorescent dyes and probes) which can increase the cost of digital PCR and limit its application in POC diagnosis. Therefore, it is necessary to develop low cost detection devices or other signal readout methods to promote its wider application. The technology of dPCR is rapidly growing and its performance in different fields is expected to eventually outperform qPCR. The Minimum Information for Publication of Quantitative Real-Time PCR Experiments (MIQE) Guidelines for dPCR were proposed by Huggett et al.123 Similar to the MIQE for qPCR,124 the MIQE for dPCR will help to standardize experimental protocols, maximize efficient utilization of resources, and enhance the impact of this promising new technology.
3.2 Isothermal quantification of nucleic acids
Since qPCR requires a thermocycler and a 2–3 hour amplification time, it is not suitable for POC in all cases. Given the developments of nucleic acid amplification technology in recent years, nucleic acid isothermal amplification technology has been increasingly applied in POC systems. The nucleic acid isothermal amplification technology does not require different temperature cycles to generate new product from the nucleic acid template. It does not depend on sophisticated equipment and has shown good application prospects in POC. Among the isothermal amplification techniques, loop-mediated isothermal amplification (LAMP) has relatively wider application than other technologies. Recombinase polymerase amplification (RPA) is a nucleic acid detection technology that has also shown broad application prospects.125–127 Both approaches offer alternatives for nucleic acid amplification that may be amenable for use in POC.In many cases, only the presence or absence of target is required in POC. Therefore, the application of LAMP in POC does not necessarily require absolute quantification. Yuan et al. developed a disc chip using colorimetric LAMP for the detection of peanut, sesame and soybean allergens. The authors found that the detection limit was as low as 0.4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ng μL−1.130 An automated approach using the lab-on-a-disc and a miniaturized rotary instrument equipped with three heating blocks was developed by Oh et al.135 The authors demonstrated that a milk sample contaminated with foodborne pathogens could be automatically run on the centrifugal disc by LAMP with a detection level as low as 10 bacteria within 65 minutes. A LAMP-based approach using on-capillary array technology was developed by Shao et al.136 This microarray performs hydrophobic processing on capillaries and immobilizes primers into capillaries via chitosan to process LAMP with a detection sensitivity as low as 50 copies/capillary. Trinh et al.137 have designed a multiplex LAMP plastic microdevice chip that is eco-friendly, and can rapidly detect four pathogens within a concentration range of 0.12–0.13
ng μL−1.130 An automated approach using the lab-on-a-disc and a miniaturized rotary instrument equipped with three heating blocks was developed by Oh et al.135 The authors demonstrated that a milk sample contaminated with foodborne pathogens could be automatically run on the centrifugal disc by LAMP with a detection level as low as 10 bacteria within 65 minutes. A LAMP-based approach using on-capillary array technology was developed by Shao et al.136 This microarray performs hydrophobic processing on capillaries and immobilizes primers into capillaries via chitosan to process LAMP with a detection sensitivity as low as 50 copies/capillary. Trinh et al.137 have designed a multiplex LAMP plastic microdevice chip that is eco-friendly, and can rapidly detect four pathogens within a concentration range of 0.12–0.13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ng μL−1.
ng μL−1.
Since the amplification product of LAMP contains large amounts of pyrophosphate in addition to the double-stranded nucleic acid, the signal readout of LAMP is more flexible than that of traditional PCR. In recent years, fluorescence signals,138,139 electrochemical signals,140,141 and visual detection142,143 readout methods have been applied successfully to microfluidic platforms to promote the application of LAMP in pathogen detection and clinical diagnosis. The microfluidic-based LAMP is effective in achieving rapid results and miniaturized detection. However, due to the LAMP reaction conditions, further efforts are required to improve detection specificity, primer design, and temperature control for optimization of LAMP. Moreover, chip designs that implement multiple target detection on one chip would be of significant value and need to be developed.
In recent years, several researchers have adopted this approach to incorporate microfluidics in order to improve on the rapidity and accuracy of RPA.125,127,145 Li et al. designed a picoliter well array chip for dRPA which could function at 39 °C in 20 minutes.26 The chip has 27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 picoliter wells (314 pL) and cross-contamination between microwells is avoided through silanization modification by methoxy-PEG-silane. Schuler et al.25 performed a digital droplet RPA by the centrifugal step emulsification method in order to detect Listeria monocytogenes DNA. Shen et al.23 developed a digital RPA Slip Chip, which can be used for absolute quantification of target nucleic acid molecules. Multiple RPA-based detections can also be implemented on a microfluidic platform as demonstrated recently by Song et al.146 The authors designed a high-level multiplexing microfluidics chip dubbed rapid amplification (RAMP), which consists of a first-stage RPA and second-stage LAMP. The approach was rapid, requiring only 40 minutes, and provided high sensitivity and specificity for target detection. Kersting et al. developed an on-chip multiplex RPA approach which is performed in a programmable hybridization chamber and can detect 10 CFU of methicillin-resistant Staphylococcus aureus and Salmonella enterica and 100 CFU for Neisseria gonorrhoeae in <20 minutes.147 Choi et al. described a direct RPA strategy on a disc chip. This strategy can simultaneously detect Salmonella enterica, Escherichia coli O157:H7, and Vibrio parahaemolyticus in 30 minutes from milk samples without DNA extraction.94 Chen et al.24 also developed a disc RPA chip which can simultaneously detect five distinct pathogens from urine samples within 40 minutes. Because RPA has a unique amplification principle and does not require a cycle temperature similar to PCR, the RPA can be combined with other technologies to achieve rapid detection on microfluidics. A new technique for isothermal solid-state amplification/detection (ISAD) was proposed to rapidly detect single point mutations in cancer.148 This is a label-free, real-time detection technique that combines RPA with a solid-phase amplification based on silicon microrings. In this study, ISAD was used to detect single mutations in the Harvey RAS gene, and this approach yielded an amplification product efficiency 100-times greater than that of RPA and conventional PCR methods, alone, in terms of sensitivity. To detect RPA amplification products, real-time fluorescent detection94,149 or lateral flow detection150,151 are mainly used on the microfluidics system. Additional product detection methods should be developed to promote the application of RPA. In recent years, RPA technology based on microfluidics has witnessed rapid advancements. To increase accessibility of this technique, the production cost of the chip and the reaction cost of the RPA should be reduced. Moreover, due to the high sensitivity of RPA, one of the limitations for its use is related to non-specific product amplification and contamination. These latter concerns should be considered with regard to the use of RPA in combination with microfluidic platforms. Further optimization of primers and probe sequences for different genes will increase the feasibility of using RPA-based microfluidic approaches in POC systems.
000 picoliter wells (314 pL) and cross-contamination between microwells is avoided through silanization modification by methoxy-PEG-silane. Schuler et al.25 performed a digital droplet RPA by the centrifugal step emulsification method in order to detect Listeria monocytogenes DNA. Shen et al.23 developed a digital RPA Slip Chip, which can be used for absolute quantification of target nucleic acid molecules. Multiple RPA-based detections can also be implemented on a microfluidic platform as demonstrated recently by Song et al.146 The authors designed a high-level multiplexing microfluidics chip dubbed rapid amplification (RAMP), which consists of a first-stage RPA and second-stage LAMP. The approach was rapid, requiring only 40 minutes, and provided high sensitivity and specificity for target detection. Kersting et al. developed an on-chip multiplex RPA approach which is performed in a programmable hybridization chamber and can detect 10 CFU of methicillin-resistant Staphylococcus aureus and Salmonella enterica and 100 CFU for Neisseria gonorrhoeae in <20 minutes.147 Choi et al. described a direct RPA strategy on a disc chip. This strategy can simultaneously detect Salmonella enterica, Escherichia coli O157:H7, and Vibrio parahaemolyticus in 30 minutes from milk samples without DNA extraction.94 Chen et al.24 also developed a disc RPA chip which can simultaneously detect five distinct pathogens from urine samples within 40 minutes. Because RPA has a unique amplification principle and does not require a cycle temperature similar to PCR, the RPA can be combined with other technologies to achieve rapid detection on microfluidics. A new technique for isothermal solid-state amplification/detection (ISAD) was proposed to rapidly detect single point mutations in cancer.148 This is a label-free, real-time detection technique that combines RPA with a solid-phase amplification based on silicon microrings. In this study, ISAD was used to detect single mutations in the Harvey RAS gene, and this approach yielded an amplification product efficiency 100-times greater than that of RPA and conventional PCR methods, alone, in terms of sensitivity. To detect RPA amplification products, real-time fluorescent detection94,149 or lateral flow detection150,151 are mainly used on the microfluidics system. Additional product detection methods should be developed to promote the application of RPA. In recent years, RPA technology based on microfluidics has witnessed rapid advancements. To increase accessibility of this technique, the production cost of the chip and the reaction cost of the RPA should be reduced. Moreover, due to the high sensitivity of RPA, one of the limitations for its use is related to non-specific product amplification and contamination. These latter concerns should be considered with regard to the use of RPA in combination with microfluidic platforms. Further optimization of primers and probe sequences for different genes will increase the feasibility of using RPA-based microfluidic approaches in POC systems.
4. INEAD system and clinical application
4.1 INEAD systems
The integration of sample preparation and amplification on a microfluidics chip system can prevent contamination, reduce sample loss, and reduce detection time. Thus, the INEAD system has the potential to produce more accurate and robust detection results with reduced time cost. Over the years, various strategies have been tested to optimize integration and automation of the INEAD system and this section introduces some approaches for implementing the INEAD system.The centrifugal microfluidic chip could pre-embed the reagents required for the extraction and amplification processes on a CD-shaped chip. The centrifugal force serves as the driving force for the liquid flow.41,152,153 Jung et al.154 developed a microfluidic chip integrated with RNA extraction and RT-LAMP to extract influenza virus RNA. The chip contains four reservoirs: a viral RNA sample pool, a wash solution pool, an eluent pool, and an RT-LAMP reagent pool. It also contains two chambers: the waste reservoir and the RT-LAMP reaction chamber. By controlling the rotational speed of the chip, viral RNA samples, washings, eluents, and RT-LAMP reagents are sequentially passed through the microbeads by different centrifugal forces to control adsorption and subsequent washing and elution of nucleic acid product. Loo et al.152 introduced a centrifugal chip integrating DNA extraction and LAMP that can be used to quantify the number of target bacteria by fluorescence intensity from only a small amount of body fluid sample (Fig. 7B). Zhang et al.155 reported a disc chip in which the nucleic acid purification was integrated with the LAMP approach. The chip can be used to detect six kinds of pathogens simultaneously in an electricity-free manner. This latter feature demonstrates that this approach may have great potential for application in POC systems. As a simple way to achieve integration, in recent years, many studies have been based on centrifugal force24,156–158 and commercial applications have been implemented that point to this technique as having increasingly broad application.
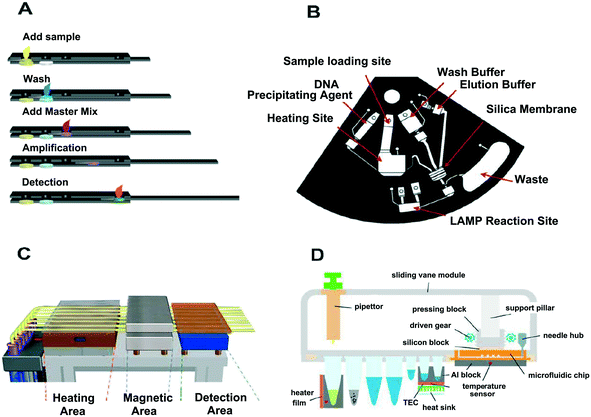 | ||
| Fig. 7 Technical variance for the INEAD and SIDAO systems. (A) The paper-based INEAD system (Connelly et al., 2015) is shown for comparison with the (B) centrifugal microfluidic that integrates the nucleic acid extraction with LAMP (Loo et al., 2016) and (C) the capillary-based INEAD system (Liu et al., 2013). “A” has the advantage of low cost while “B” and “C” can be automated. Despite the different integration options of the microfluidic chip, the systems can be simple and fast to achieve “sample-in-answer-out”. (D) The magnetic bead-based system combines nucleic acid extraction with a digital Recombinase polymerase amplification (RPA) chip (Yang et al., 2018). “D” can automatically achieve “sample-in-digital-answer-out”. Fig. A and C are adapted from ref. 53 and 159 with permission from ACS Publications, Fig. B is adapted from ref. 152 with permission from Elsevier, and Fig. D is adapted from ref. 160 with permission from Springer. | ||
Paper microfluidics is another ideal platform for integration which could process the fluid in the reaction by introducing buffers, samples and reagents. Connelly et al.53 developed an integrated system containing sample preparation and LAMP (Fig. 7A). The paper-based chip has a multi-layered structure and slides in and out of the paper chip to introduce a sample and a reaction reagent. Finally, LAMP's endpoint detection is simplified through use of a handheld UV power and camera. While this will reduce the automation aspect of this particular approach, it will allow for a low tech option in cases where POC sites have limited technical resources. Tang et al. also designed an integrated paper-based system in which the authors used a sponge-based reservoir and a paper-based valve for nucleic acid extraction. The subsequent isothermal reaction was achieved with micro-heated components and pre-embedded reagents. The system could detect Salmonella typhimurium in food samples within 1 h with a detection limit as low as 10−2–10−3 bacteria.60
Capillaries are also an effective tool for integration in many nucleic acid purification and amplification approaches. Liu et al.159 developed an excellent approach for integrating capillary-array microsystems that includes a heating block, a multichannel syringe pump, a bidirectional magnetic force controller, and a fluorescence detection module (Fig. 7C). In this system, DNA extraction and LAMP products detection were in the capillary-array with the capacity to process 10 samples simultaneously. Fu et al. also reported an integrated capillary system which could allow on-site qPCR.74 In this method, nucleic acid extraction was achieved through a PDDA modified capillary followed by temperature control of the PCR through water circulation pumps. This system achieved the steps (including extraction, amplification, and detection) within 40 minutes.
Some industries have launched fully automated nucleic acid diagnostic products such as GeneXpert®, Filmarray®, and Atlas Genetics io®. GeneXpert is a fully automated molecular diagnostics platform developed by Cepheid™. The GeneXpert reduces user handling of samples by integrating sample preparation steps with PCR amplification and real-time fluorescence detection in the same cartridge. This is a closed system that relies on valve actuation and hydraulic control. The PCR process is fully automated, with simple operation and fast results. The Cartridge of GeneXpert is used for quantitative detection of multiple samples. Since it involves nucleic acid extraction, amplification, and monitoring, the cartridge structure is complex and costly. Filmarray is a fully automated molecular diagnostics platform based on microfluidic technology that provides qualitative target detection and is developed by BioFire™. This approach utilizes a test strip that contains all reagents needed for nucleic acid extraction and amplification. The product uses multiplex PCR analysis technology to perform the detection of up to 24 targets at a time on the same sample within an hour and is ideally suited for early rapid screening of multiple infectious diseases. Atlas Genetics is a microfluidic-based POC product developed by Atlas Genetics™. Considering that electrochemical detection is employed in this product, no complicated optics are needed in the instrument, and the instrument could be miniaturized for portability making it ideal for POC sites. In 2016, Bohui Company™ successfully developed a fully automatic genotyping detection chip for human papilloma virus (HPV) that integrates sample processing, nucleic acid amplification and multiple interpretations. One major advantage for this technology is the rapidity since only 2 minutes are required to complete sample processing and detection. The detection requires a chip controller only, which contributes to the automation of the whole process from nucleic acid extraction, PCR amplification, reverse hybridization to results interpretation. Another advantage of this approach is that the entire experiment can be conducted in a sealed chip, which not only avoids DNA contamination, but also eliminates human error introduced by manual operation.
The integrated microfluidic POC system has been flourishing as a means to conduct fast, accurate nucleic acid detection. These systems are often pre-packaged, with a predetermined reagent solution in the chip or with the reagent in the reservoir. Indeed, nucleic acid extraction coupled with “in situ PCR” has been found to have great application in INEAD.63 However, there are not many researches on digital nucleic acid detection integrated with sample preparation to achieve “sample-in-digital-answer out (SIDAO)”. Based on the air permeability of PDMS, a microfluidic chip which used the negative pressure provided by a syringe to achieve sample injection and enable digital PCR reactions was developed. The systems were successful in isolating and amplifying nucleic acid from a tissue source.111 This approach is convenient and rapid, cost-effective, accurate, and suitable for use in a standard molecular laboratory. Yang et al.160 also reported a system that integrated the DNA extraction and digital RPA (Fig. 7D). The system had an automated module to allow liquid transfer and the reaction buffer was injected into the digital RPA chip through the mechanical pressure. Using this system, they successfully performed absolute quantification of a Mycobacterium tuberculosis gene from saliva and serum.
4.2 Clinical application of the INEAD system
As medical care advances, the demand for cost-effective, rapid and accurate nucleic acid detection has pushed technological advances that promote the application of INEAD system in clinical practice. Many commercial INEAD systems have been clinically applied in point-of-care testing (POCT) of tuberculosis,161,162 HIV,163 methicillin-resistant Staphylococcus aureus,164 and Chlamydia trachomatis.165 The INEAD approaches used were found to give accurate results in a short time frame, which helped to promote medical development and support in remote areas. Indeed, some of the approaches have been endorsed by the World Health Organization (WHO).166 In a recent study, Ye et al.156 test for the presence of the rotavirus A from 48 clinical stool samples using the INEAD system and found that the sensitivity and specificity were both 100%. Wang et al.167 detected the S. pneumoniae and M. pneumoniae from 63 clinical specimens (oropharyngeal swabs and bronchoalveolar lavage fluid), which further supported the value of using INEAD systems for the analysis of clinical specimens. The clinical application value of the INEAD system was also demonstrated by Choi et al., in the positive test of 16 HBV-clinical samples.168 While quite a bit of recent progress has been made, it is important to recognize the value of further research involving INEAD-based approaches for accurate, rapid detection of virus, pathogen, or immunogen in patient samples.5. Summary and conclusions
In the current review, the history and development of the microfluidics field was captured to provide an assessment of past and current techniques in terms of accuracy, reliability, cost and speed, particularly in the context of POC. Further, this study offered the current perspective on the state of this field in terms of areas for future growth and technological advancement. This work compared methods for nucleic acid extraction on microfluidic systems, methods for nucleic acid amplification, and integrated microfluidic chips for sample preparation combined with nucleic acid amplification.In terms of sample preparation, this study offered insight into the different nucleic acid extraction methods based on magnetic beads, silicon-based materials, and paper-based materials, in the context of their use with microfluidics. The magnetic beads method is characterized by controllability and high flux along with simple and fast procedures. Silicon material is especially suitable for SPE. Paper-based nucleic acid extraction is characterized by low cost and rapid completion time, offering several advantages over other methodologies. Meanwhile, there are many different ways to achieve nucleic acid extraction, many of which can be integrated with microfluidics for optimal outcome. The choice of nucleic acid extraction method determines the methods of integration with downstream nucleic acid amplification and the speed of detection for the entire process, which are crucial parts of nucleic acid detection.
For nucleic acid amplification methods on microfluidic systems, this study introduced dPCR, LAMP and RPA methods which are currently widely used and have high potential for POC application. dPCR, as the current mainstream nucleic acid amplification method, is suitable for microfluidic systems due to its technical maturity given that 20 years have passed since it was first proposed. The application of this technology to microfluidic systems requires less reaction completion time and results in higher sensitivity. LAMP-based instruments are simpler, cheaper, and consume less energy during the heating steps. However, RPA is the only current nucleic acid amplification technology that can rapidly react at room temperature. In the microfluidic system, the analysis performance, throughput, detection accuracy and dynamic range of the digital nucleic acid amplification should be further explored.
6. Future outlook
The microfluidic chip revolutionized and integrated the process of nucleic acid extraction, amplification, and detection but many challenges are yet to be resolved. For sample preparation, recent technology allows for the extraction of nucleic acids on the microfluidic system from simple samples but cannot readily handle more complex samples such as bone, animal and plant tissue. Therefore, handling complex samples on microfluidic systems is still an urgent problem. Moreover, the complete encapsulation of nucleic acid extraction will be a particularly effective advancement to prevent nucleic acid contamination during processing. In addition, the challenge of optimal integration of the sample preparation with digital nucleic acid amplification to achieve SIDAO needs further exploration. Further development of INEAD systems using the microfluidic chip will help us to realize more accessible “sample-in-answer-out” to improve rapid and accurate test results in POC. However, the velocity, reliability, accuracy and cost of the integrated systems will continue to be key hurdles to progress and broad use of these technologies in POC.Moving forward, for better service in POC and clinical application, any commercialized integrated system should also be miniaturized and affordable. Although there are presently many products, the cost is high and presents a barrier for broad accessibility to some economically disadvantaged and rural areas. To make nucleic acid extraction-microfluidics an integral part of rural or emergency care, POC will require a significant improvement in costs associated with the sample prep and technology. In future, the application of INEAD systems for POC must have the characteristics of automation, low cost, versatility and miniaturization. It would also be important to have a visualized data output module and information transmission module to allow for improved communication of results between POC and potential clinical partners at more metropolitan hubs. Therefore, future POC personnel would not need specialized, operational training beyond careful sample collection and basic instrument handling. In addition, for most rural and remote areas, lack of resources and economical constraints remain limiting factors. In the future, the application of the INEAD system to mobile detection vehicles will enable them to serve more people in the medical and food safety fields, which will promote the application of POC systems. This puts higher requirements on the stability, operability, and energy requirements for any system developed. In many medical fields, accurate test results are required and SIDAO is an ideal testing process leading to improved sample preparation and nucleic acid detection technologies. It is our hope that this review will encourage further dialogue and exploration towards the development of microfluidic systems for NAAT integration. It is anticipated that future research will provide further integrative solutions for improving POC and establishing broader accessibility for microfluidics-based technology across clinical applications.
Conflicts of interest
The authors declare that there are no conflicts of interest associated with the current work.Acknowledgements
The authors are grateful for the financial support from the National Key R&D Program of China (2018YFF01012100), the Open Research Project of the State Key Laboratory of Industrial Control Technology, Zhejiang University, China (No. ICT1900327) and the Fundamental Research Funds for the Central Universities. JXZ and DD are supported by grants from the National Science Foundation, USA (CHE 1709160) and the National Institutes of Health, USA (COBRE-4020GM104360-04, Vaughan, R. as PI), respectively.Notes and references
- G. Mtove, B. Nadjm, I. C. E. Hendriksen, B. Amos, F. Muro, J. Todd and H. Reyburn, Clin. Infect. Dis., 2011, 53, 548–554 CrossRef PubMed.
- E. Lencova, Z. Broukal and J. Spizek, Folia Microbiol., 2010, 55, 559–568 CrossRef CAS PubMed.
- J. D. Tucker, C. H. Bien and R. W. Peeling, Curr. Opin. Infect. Dis., 2013, 26, 73–79 CrossRef PubMed.
- F. Tinajeros, D. Grossman, K. Richmond, M. Steele, S. G. Garcia, L. Zegarra and R. Revollo, Sex. Transm. Infect., 2006, 82, V17–V21 CrossRef PubMed.
- N. Ravi, G. Rizzi, S. E. Chang, P. Cheung, P. J. Utz and S. X. Wang, Biosens. Bioelectron., 2019, 130, 338–343 CrossRef CAS PubMed.
- M. Book, L. E. Lehrmann, X. Zhang and F. Stueber, Anasthesiol. Intensivmed. Notfallmed. Schmerzther., 2010, 45, 732–738 CrossRef PubMed.
- N. K. Tran, Z. Godwin and J. Bockhold, Point of Care, 2012, 11, 180–183 Search PubMed.
- N. P. Pai, C. Vadnais, C. Denkinger, N. Engel and M. Pai, PLoS Med., 2012, 9, e1001306 CrossRef PubMed.
- A. Manz, N. Graber and H. M. Widmer, Sens. Actuators, B, 1990, 1, 244–248 CrossRef CAS.
- L. Gervais, R. N. De and E. Delamarche, Adv. Mater., 2011, 23, 151–176 CrossRef PubMed.
- V. Gubala, L. F. Harris, A. J. Ricco, M. X. Tan and D. E. Williams, Anal. Chem., 2012, 84, 487 CrossRef CAS PubMed.
- W. J. Lu, Y. P. Chen, Z. Liu, W. B. Tang, Q. Feng, J. S. Sun and X. Y. Jiang, ACS Nano, 2016, 10, 6685–6692 CrossRef CAS PubMed.
- P. Toren, E. Ozgur and M. Bayindir, Lab Chip, 2016, 16, 2572–2595 RSC.
- L. Ninove, A. Nougairede, C. Gazin, C. Zandotti, M. Drancourt, X. de Lamballerie and R. N. Charrel, J. Clin. Virol., 2010, 49, 304–305 CrossRef PubMed.
- C. K. Y. Ng, G. G. D. Costanzo, L. M. Terracciano and S. Piscuoglio, Front. Med., 2018, 5, 78 CrossRef PubMed.
- K. C. Chan and Y. M. Lo, Histol. Histopathol., 2002, 17, 937–943 CAS.
- J. C. Tsang and Y. M. Lo, Pathology, 2007, 39, 197–207 CrossRef CAS PubMed.
- C. A. Holland and F. L. Kiechle, Curr. Opin. Microbiol., 2005, 8, 504–509 CrossRef PubMed.
- L. Magro, C. Escadafal, P. Garneret, B. Jacquelin, A. Kwasiborski, J. C. Manuguerra, F. Monti, A. Sakuntabhai, J. Vanhomwegen and P. Lafaye, Lab Chip, 2017, 17, 2347–2371 RSC.
- C. E. Jin, T. Y. Lee, B. Koo, K. C. Choi, S. Chang, S. Y. Park, J. Y. Kim, S. H. Kim and Y. Shin, Anal. Chem., 2017, 89, 7502–7510 CrossRef CAS PubMed.
- L. Zhang, B. Ding, Q. Chen, Q. Feng, L. Lin and J. Sun, TrAC, Trends Anal. Chem., 2017, 94, 106–116 CrossRef CAS.
- I. H. K. Basha, E. T. Ho, C. M. Yousuff and N. H. Bin Hamid, Micromachines, 2017, 8, 266 CrossRef PubMed.
- F. Shen, E. K. Davydova, W. Du, J. E. Kreutz, O. Piepenburg and R. F. Ismagilov, Anal. Chem., 2011, 83, 3533–3540 CrossRef CAS PubMed.
- J. Chen, Y. Xu, H. Yan, Y. Zhu, L. Wang, Y. Zhang, Y. Lu and W. Xing, Lab Chip, 2018, 18, 2441–2452 RSC.
- F. Schuler, F. Schwemmer, M. Trotter, S. Wadle, R. Zengerle, F. von Stetten and N. Paust, Lab Chip, 2015, 15, 2759–2766 RSC.
- Z. Li, Y. Liu, Q. Wei, Y. Liu, W. Liu, X. Zhang and Y. Yu, PLoS One, 2016, 11, e0153359 CrossRef PubMed.
- Y. Zhang and N.-T. Nguyen, Lab Chip, 2017, 17, 994–1008 RSC.
- T. Gou, J. Hu, W. Wu, X. Ding, S. Zhou, W. Fang and Y. Mu, Biosens. Bioelectron., 2018, 120, 144–152 CrossRef CAS PubMed.
- T. Tangchaikeeree, D. Polpanich, A. Elaissari and K. Jangpatarapongsa, Colloids Surf., B, 2017, 158, 1–8 CrossRef CAS PubMed.
- V. Kamat, S. Pandey, K. Paknikar and D. Bodas, Biosens. Bioelectron., 2018, 99, 62–69 CrossRef CAS PubMed.
- L. Zhang, R. N. Deraney and A. Tripathi, Biomicrofluidics, 2015, 9, 064118 CrossRef PubMed.
- R.-Q. Zhang, S.-L. Hong, C.-Y. Wen, D.-W. Pang and Z.-L. Zhang, Biosens. Bioelectron., 2018, 100, 348–354 CrossRef CAS PubMed.
- J.-C. Yu, C.-C. Hu, W.-H. Chang, P.-C. Chen, M. S. Lee, K.-T. Peng and G.-B. Lee, Microfluid. Nanofluid., 2018, 22, 13 CrossRef.
- X. Shi, C.-H. Chen, W. Gao, S.-h. Chao and D. R. Meldrum, Lab Chip, 2015, 15, 1059–1065 RSC.
- U. Lehmann, C. Vandevyver, V. K. Parashar and M. A. Gijs, Angew. Chem., Int. Ed., 2006, 45, 3062–3067 CrossRef CAS PubMed.
- S. M. Berry, E. T. Alarid and D. J. Beebe, Lab Chip, 2011, 11, 1747–1753 RSC.
- K.-Y. Lien, W.-C. Lee, H.-Y. Lei and G.-B. Lee, Biosens. Bioelectron., 2007, 22, 1739–1748 CrossRef CAS PubMed.
- O. Mosley, L. Melling, M. D. Tarn, C. Kemp, M. M. Esfahani, N. Pamme and K. J. Shaw, Lab Chip, 2016, 16, 2108–2115 RSC.
- B. Shu, Z. Li, X. Yang, F. Xiao, D. Lin, X. Lei, B. Xu and D. Liu, Chem. Commun., 2018, 54, 2232–2235 RSC.
- N. C. Cady, S. Stelick and C. A. Batt, Biosens. Bioelectron., 2004, 19, 59–66 CrossRef.
- T.-H. Kim, J. Park, C.-J. Kim and Y.-K. Cho, Anal. Chem., 2014, 86, 3841–3848 CrossRef CAS PubMed.
- B. Bruijns, A. van Asten, R. Tiggelaar and H. Gardeniers, Biosensors, 2016, 6, 41 CrossRef PubMed.
- C. W. Price, D. C. Leslie and J. P. Landers, Lab Chip, 2009, 9, 2484–2494 RSC.
- T. Gou, J. Hu, W. Wu, X. Ding, S. Zhou, W. Fang and Y. Mu, Biosens. Bioelectron., 2018, 120, 144–152 CrossRef CAS PubMed.
- N. C. Cady, S. Stelick and C. A. Batt, Biosens. Bioelectron., 2003, 19, 59–66 CrossRef CAS PubMed.
- L. A. Christel, K. Petersen, W. McMillan and M. A. Northrup, J. Biomech. Eng., 1999, 121, 22–27 CrossRef CAS PubMed.
- Y. C. Chung, M. S. Jan, Y. C. Lin, J. H. Lin, W. C. Cheng and C. Y. Fan, Lab Chip, 2004, 4, 141–147 RSC.
- B. H. Park, S. J. Oh, J. H. Jung, G. Choi, J. H. Seo, D. H. Kim, E. Y. Lee and T. S. Seo, Biosens. Bioelectron., 2017, 91, 334–340 CrossRef CAS PubMed.
- L. Christel, K. Petersen, W. McMillan and M. Northrup, J. Biomech. Eng., 1999, 121, 22–27 CrossRef CAS PubMed.
- J. Khandurina, S. C. Jacobson, L. C. Waters, R. S. Foote and J. M. Ramsey, Anal. Chem., 1999, 71, 1815–1819 CrossRef CAS PubMed.
- S. Petralia, E. L. Sciuto and S. Conoci, Analyst, 2017, 142, 140–146 RSC.
- G. Gunal, C. Kip, S. E. Ogut, D. D. Usta, E. Senlik, G. Kibar and A. Tuncel, Mater. Sci. Eng., C, 2017, 74, 10–20 CrossRef CAS PubMed.
- J. T. Connelly, J. P. Rolland and G. M. Whitesides, Anal. Chem., 2015, 87, 7595–7601 CrossRef CAS PubMed.
- A. K. Yetisen, M. S. Akram and C. R. Lowe, Lab Chip, 2013, 13, 2210–2251 RSC.
- J. R. Choi, K. W. Yong, R. Tang, G. Yan, T. Wen, L. Fei, B. Pingguan-Murphy, B. Dan and X. Feng, TrAC, Trends Anal. Chem., 2017, 93, 37–50 CrossRef CAS.
- S. B. Boppana, S. A. Ross, Z. Novak, M. Shimamura, R. W. Tolan, A. L. Palmer, A. Ahmed, M. G. Michaels, P. J. Sánchez and D. I. Bernstein, JAMA, 2010, 303, 1375–1382 CrossRef CAS PubMed.
- D. Zhang, D. Broyles, E. A. Hunt, E. Dikici, S. Daunert and S. K. Deo, Analyst, 2017, 142, 815–823 RSC.
- L. Zhang, Y. Zhang, C. Wang, Q. Feng, F. Fan, G. Zhang, X. Kang, X. Qin, J. Sun, Y. Li and X. Jiang, Anal. Chem., 2014, 86, 10461–10466 CrossRef CAS PubMed.
- W. Gan, B. Zhuang, P. Zhang, J. Han, C.-X. Li and P. Liu, Lab Chip, 2014, 14, 3719–3728 RSC.
- R. Tang, H. Yang, Y. Gong, M. You, Z. Liu, J. R. Choi, T. Wen, Z. Qu, Q. Mei and F. Xu, Lab Chip, 2017, 17, 1270–1279 RSC.
- X. Ye, J. Xu, L. Lu, X. Li, X. Fang and J. Kong, Anal. Chim. Acta, 2018, 1018, 78–85 CrossRef CAS PubMed.
- K. A. Hagan, C. R. Reedy, M. L. Uchimoto, D. Basu, D. A. Engel and J. P. Landers, Lab Chip, 2011, 11, 957–961 RSC.
- W. Gan, Y. Gu, J. Han, C. X. Li, J. Sun and P. Liu, Anal. Chem., 2017, 89, 3568–3575 CrossRef CAS PubMed.
- S. A. Byrnes, J. D. Bishop, L. Lafleur, J. R. Buser, B. Lutz and P. Yager, Lab Chip, 2015, 15, 2647–2659 RSC.
- N. M. Rodriguez, J. C. Linnes, A. Fan, C. K. Ellenson, N. R. Pollock and C. M. Klapperich, Anal. Chem., 2015, 87, 7872–7879 CrossRef CAS PubMed.
- Y. Zou, M. G. Mason, Y. Wang, E. Wee, C. Turni, P. J. Blackall, M. Trau and J. R. Botella, PLoS Biol., 2017, 15, 96–104 CrossRef PubMed.
- A. V. Govindarajan, S. Ramachandran, G. D. Vigil, P. Yager and K. F. Böhringer, Lab Chip, 2011, 12, 174–181 RSC.
- J. C. Linnes, A. Fan, N. M. Rodriguez, B. Lemieux, H. Kong and C. M. Klapperich, RSC Adv., 2014, 4, 42245 RSC.
- C. F. Fronczek, S. P. Tu, D. K. Harshman, A. M. Nicolini and J. Y. Yoon, RSC Adv., 2014, 4, 11103–11110 RSC.
- M. S. Cordray and R. R. Richards-Kortum, Malar. J., 2015, 14, 472 CrossRef PubMed.
- M. M. Ali, S. D. Aguirre, Y. Xu, C. D. Filipe, R. Pelton and Y. Li, Chem. Commun., 2009, 6640–6642 RSC.
- B. A. Rohrman and R. R. Richards-Kortum, Lab Chip, 2012, 12, 3082 RSC.
- N. Kaur and B. J. Toley, Analyst, 2018, 143, 2213–2234 RSC.
- Y. Fu, X. Zhou and D. Xing, Lab Chip, 2017, 17, 4334–4341 RSC.
- M. C. Morales and J. D. Zahn, Microfluid. Nanofluid., 2010, 9, 1041–1049 CrossRef CAS.
- R. Zhang, H.-Q. Gong, X. Zeng, C. Lou and C. Sze, Anal. Chem., 2013, 85, 1484–1491 CrossRef CAS PubMed.
- L. A. Marshall, L. L. Wu, S. Babikian, M. Bachman and J. G. Santiago, Anal. Chem., 2012, 84, 9640–9645 CrossRef CAS PubMed.
- K. Han, Y. J. Yoon, S. Yong and K. P. Mi, Lab Chip, 2015, 16, 132–141 RSC.
- J. Yoon, Y.-J. Yoon, T. Y. Lee, M. K. Park, J. Chung and Y. Shin, Sens. Actuators, B, 2018, 255, 1491–1499 CrossRef CAS.
- C. Liu, S.-C. Liao, J. Song, M. G. Mauk, X. Li, G. Wu, D. Ge, R. M. Greenberg, S. Yang and H. H. Bau, Lab Chip, 2016, 16, 553–560 RSC.
- C. Liu, E. Geva, M. Mauk, X. Qiu, W. R. Abrams, D. Malamud, K. Curtis, S. M. Owen and H. H. Bau, Analyst, 2011, 136, 2069–2076 RSC.
- S.-C. Liao, J. Peng, M. G. Mauk, S. Awasthi, J. Song, H. Friedman, H. H. Bau and C. Liu, Sens. Actuators, B, 2016, 229, 232–238 CrossRef CAS PubMed.
- J. Song, M. G. Mauk, B. A. Hackett, S. Cherry, H. H. Bau and C. Liu, Anal. Chem., 2016, 88, 7289–7294 CrossRef CAS PubMed.
- J. Song, C. Liu, M. G. Mauk, J. Peng, T. Schoenfeld and H. H. Bau, Anal. Chem., 2017, 90, 1209–1216 CrossRef PubMed.
- D. C. Duffy, S. Oja, G. M. Whitesides and J. C. Mcdonald, Anal. Chem., 1998, 70, 4974–4984 CrossRef CAS PubMed.
- W. Du, L. Li, K. P. Nichols and R. F. Ismagilov, Lab Chip, 2009, 9, 2286–2292 RSC.
- M. Ogura, Y. Agata, K. Watanabe, R. M. McCormick, Y. Hamaguchi, Y. Aso and M. Mitsuhashi, Clin. Chem., 1998, 44, 2249–2255 CAS.
- M. Geissler, J. A. Beauregard, I. Charlebois, S. Isabel, F. Normandin, B. Voisin, M. Boissinot, M. G. Bergeron and T. Veres, Eng. Life Sci., 2011, 11, 174–181 CrossRef CAS.
- K. Liu and Z. H. Fan, Analyst, 2011, 136, 1288–1297 RSC.
- R. K. Jena, C. Yue, Y. Lam and Z. Wang, Sens. Actuators, B, 2010, 150, 692–699 CrossRef CAS.
- T. M. Valentin, E. M. DuBois, C. E. Machnicki, D. Bhaskar, F. R. Cui and I. Y. Wong, Polym. Chem., 2019, 10, 2015–2028 RSC.
- Z. Gan, L. Zhang and G. Chen, Electrophoresis, 2011, 32, 3319–3323 CrossRef CAS PubMed.
- A. E. Vasdekis, M. J. Wilkins, J. W. Grate, R. T. Kelly, A. Konopka, S. S. Xantheas and T.-M. Chang, Lab Chip, 2014, 14, 2072–2080 RSC.
- G. Choi, J. H. Jung, B. H. Park, S. J. Oh, J. H. Seo, J. S. Choi and T. S. Seo, Lab Chip, 2016, 16, 2309–2316 RSC.
- P. L. Quan, M. Sauzade and E. Brouzes, Sensors, 2018, 18, 1271 CrossRef PubMed.
- T. G. Aw and J. B. Rose, Curr. Opin. Biotechnol., 2012, 23, 422–430 CrossRef CAS PubMed.
- J.-H. Wang, L. Cheng, C.-H. Wang, W.-S. Ling, S.-W. Wang and G.-B. Lee, Biosens. Bioelectron., 2013, 41, 484–491 CrossRef CAS PubMed.
- S. A. Bustin, V. Benes, J. Garson, J. Hellemans, J. Huggett, M. Kubista, R. Mueller, T. Nolan, M. W. Pfaffl and G. Shipley, Nat. Methods, 2013, 10, 1063–1067 CrossRef CAS PubMed.
- R. T. Hayden, K. M. Hokanson, S. B. Pounds, M. J. Bankowski, S. W. Belzer, J. Carr, D. Diorio, M. S. Forman, Y. Joshi and D. Hillyard, J. Clin. Microbiol., 2008, 46, 157–163 CrossRef CAS PubMed.
- P. J. Sykes, S. H. Neoh, M. J. Brisco, E. Hughes, J. Condon and A. A. Morley, BioTechniques, 1992, 13, 444–449 CAS.
- M. Baker, Nat. Methods, 2012, 9, 541–544 CrossRef CAS.
- B. Vogelstein and K. W. Kinzler, Proc. Natl. Acad. Sci. U. S. A., 1999, 96, 9236–9241 CrossRef CAS PubMed.
- D. Simant, Q. Jian and R. Ramesh, PLoS One, 2008, 3, e2876 CrossRef PubMed.
- A. S. Whale, S. Cowen, C. A. Foy and J. F. Huggett, PLoS One, 2013, 8, e58177 CrossRef CAS PubMed.
- A. C. Hatch, J. S. Fisher, A. R. Tovar, A. T. Hsieh, R. Lin, S. L. Pentoney, D. L. Yang and A. P. Lee, Lab Chip, 2011, 11, 3838–3845 RSC.
- K. A. Heyries, C. Tropini, M. Vaninsberghe, C. Doolin, O. I. Petriv, A. Singhal, K. Leung, C. B. Hughesman and C. L. Hansen, Nat. Methods, 2011, 8, 649–651 CrossRef CAS PubMed.
- C. A. Milbury, Q. Zhong, J. Lin, M. Williams, J. Olson, D. R. Link and B. Hutchison, Biomol. Detect. Quantif., 2014, 1, 8–22 CrossRef PubMed.
- J. de Jong, R. G. H. Lammertink and M. Wessling, Lab Chip, 2006, 6, 1125–1139 RSC.
- J. C. Linnes, N. M. Rodriguez, L. Liu and C. M. Klapperich, Biomed. Microdevices, 2016, 18, 1–12 CrossRef CAS PubMed.
- F. Shen, W. B. Du, J. E. Kreutz, A. Fok and R. F. Ismagilov, Lab Chip, 2010, 10, 2666–2672 RSC.
- Q. Tian, B. Yu, Y. Mu, Y. Xu, C. Ma, T. Zhang, W. Jin and Q. Jin, RSC Adv., 2015, 5, 81889–81896 RSC.
- Q. Zhu, L. Qiu, B. Yu, Y. Xu, Y. Gao, T. Pan, Q. Tian, Q. Song, W. Jin, Q. Jin and Y. Mu, Lab Chip, 2014, 14, 1176–1185 RSC.
- Q. Zhu, Y. Xu, L. Qiu, C. Ma, B. Yu, Q. Song, W. Jin, Q. Jin, J. Liu and Y. Mu, Lab Chip, 2017, 17, 1655–1665 RSC.
- Y. Ning, X. Cui, C. Yang, F. Jing, X. Bian, L. Yi and G. Li, Anal. Chim. Acta, 2019, 1055, 65–73 CrossRef CAS PubMed.
- C. M. Hindson, J. R. Chevillet, H. A. Briggs, E. N. Gallichotte, I. K. Ruf, B. J. Hindson, R. L. Vessella and M. Tewari, Nat. Methods, 2013, 10, 1003 CrossRef CAS PubMed.
- J. Chen, Z. Luo, L. Li, J. He, L. Li, J. Zhu, P. Wu and L. He, Lab Chip, 2018, 18, 412–421 RSC.
- Z. T. Chen, P. Y. Liao, F. L. Zhang, M. C. Jiang, Y. S. Zhu and Y. Y. Huang, Lab Chip, 2017, 17, 235–240 RSC.
- K. A. Heyries, C. Tropini, M. VanInsberghe, C. Doolin, I. Petriv, A. Singhal, K. Leung, C. B. Hughesman and C. L. Hansen, Nat. Methods, 2011, 8, 649 CrossRef CAS PubMed.
- Z. Cheng, K. Wang, Z. Wu, L. Zhou, Z. Wang, Y. Bai, J. Zhao and H. Mao, Sens. Actuators, B, 2018, 258, 1302–1308 CrossRef CAS.
- J. Yong, Y. Fang, F. Chen, J. Huo, Q. Yang, H. Bian, G. Du and X. Hou, Appl. Surf. Sci., 2016, 389, 1148–1155 CrossRef CAS.
- A. Didelot, S. K. Kotsopoulos, A. Lupo, D. Pekin, X. Y. Li, I. Atochin, P. Srinivasan, Q. Zhong, J. Olson, D. R. Link, P. Laurent-Puig, H. Blons, J. B. Hutchison and V. Taly, Clin. Chem., 2013, 59, 815–823 CrossRef CAS PubMed.
- Q. Zhong, S. Bhattacharya, S. Kotsopoulos, J. Olson, V. Taly, A. D. Griffiths, D. R. Link and J. W. Larson, Lab Chip, 2011, 11, 2167–2174 RSC.
- J. F. Huggett, C. A. Foy, V. Benes, K. Emslie, J. A. Garson, R. Haynes, J. Hellemans, M. Kubista, R. D. Mueller and T. Nolan, Clin. Chem., 2013, 59, 892–902 CrossRef CAS PubMed.
- S. A. Bustin, V. Benes, J. A. Garson, J. Hellemans, J. Huggett, M. Kubista, R. Mueller, T. Nolan, M. W. Pfaffl and G. L. Shipley, Clin. Chem., 2009, 55, 611–622 CrossRef CAS PubMed.
- B. A. Rohrman and R. R. Richards-Kortum, Lab Chip, 2012, 12, 3082–3088 RSC.
- R. K. Daher, G. Stewart, M. Boissinot and M. G. Bergeron, Clin. Chem., 2016, 62, 947–958 CrossRef CAS.
- Z. A. Crannell, B. Rohrman and R. Richards-Kortum, PLoS One, 2014, 9, e112146 CrossRef PubMed.
- J. M. Aliotta, J. J. Pelletier, J. L. Ware, L. S. Moran, J. S. Benner and H. Kong, Genet. Anal.: Biomol. Eng., 1996, 12, 185 CrossRef CAS.
- S. Alamer, S. Eissa, R. Chinnappan, P. Herron and M. Zourob, Talanta, 2018, 185, 275–280 CrossRef CAS PubMed.
- D. Yuan, J. Kong, X. Li, X. Fang and Q. Chen, Sci. Rep., 2018, 8, 8682 CrossRef PubMed.
- Y.-D. Ma, W.-H. Chang, K. Luo, C.-H. Wang, S.-Y. Liu, W.-H. Yen and G.-B. Lee, Biosens. Bioelectron., 2018, 99, 547–554 CrossRef CAS PubMed.
- K. Zhang, D.-K. Kang, M. M. Ali, L. Liu, L. Labanieh, M. Lu, H. Riazifar, T. N. Nguyen, J. A. Zell, M. A. Digman, E. Gratton, J. Li and W. Zhao, Lab Chip, 2015, 15, 4217–4226 RSC.
- A. Gansen, A. M. Herrick, I. K. Dimov, L. P. Lee and D. T. Chiu, Lab Chip, 2012, 12, 2247–2254 RSC.
- J. E. Kreutz, J. Wang, A. M. Sheen, A. M. Thompson, J. P. Staheli, M. R. Dyen, Q. Feng and D. T. Chiu, Lab Chip, 2019, 19, 1035–1040 RSC.
- S. J. Oh, B. H. Park, G. Choi, J. H. Seo, J. H. Jung, J. S. Choi, D. H. Kim and T. S. Seo, Lab Chip, 2016, 16, 1917–1926 RSC.
- N. Shao, J. Chen, J. Hu, R. Li and S. C. Tao, Lab Chip, 2017, 17, 521 RSC.
- T. N. D. Trinh and N. Y. Lee, Lab Chip, 2018, 18, 2369–2377 RSC.
- B. Pang, K. Fu, Y. Liu, X. Ding, J. Hu, W. Wu, K. Xu, X. Song, J. Wang and Y. Mu, Anal. Chim. Acta, 2018, 1040, 81–89 CrossRef CAS PubMed.
- X. Fang, Y. Liu, J. Kong and X. Jiang, Anal. Chem., 2010, 82, 3002–3006 CrossRef CAS PubMed.
- X. Zhi, M. Deng, H. Yang, G. Gao, K. Wang, H. Fu, Y. Zhang, D. Chen and D. Cui, Biosens. Bioelectron., 2014, 54, 372–377 CrossRef CAS PubMed.
- K. Hsieh, A. S. Patterson, B. S. Ferguson, K. W. Plaxco and H. T. Soh, Angew. Chem., Int. Ed., 2012, 51, 4896–4900 CrossRef CAS PubMed.
- N. Tomita, Y. Mori, H. Kanda and T. Notomi, Nat. Protoc., 2008, 3, 877 CrossRef CAS PubMed.
- I. Hongwarittorrn, N. Chaichanawongsaroj and W. Laiwattanapaisal, Talanta, 2017, 175, 135–142 CrossRef CAS PubMed.
- E.-C. Yeh, C.-C. Fu, L. Hu, R. Thakur, J. Feng and L. P. Lee, Sci. Adv., 2017, 3, e1501645 CrossRef PubMed.
- S. Lutz, P. Weber, M. Focke, B. Faltin, J. Hoffmann, C. Müller, D. Mark, G. Roth, P. Munday and N. Armes, Lab Chip, 2010, 10, 887–893 RSC.
- J. Song, C. Liu, M. G. Mauk, S. C. Rankin, J. B. Lok, R. M. Greenberg and H. H. Bau, Clin. Chem., 2017, 63, 714–722 CrossRef CAS PubMed.
- S. Kersting, V. Rausch, F. F. Bier and M. von Nickisch-Rosenegk, Microchim. Acta, 2014, 181, 1715–1723 CrossRef CAS PubMed.
- Y. Shin, A. P. Perera, K. W. Kim and M. K. Park, Lab Chip, 2013, 13, 2106–2114 RSC.
- F. Shen, E. K. Davydova, W. Du, J. E. Kreutz, O. Piepenburg and R. F. Ismagilov, Anal. Chem., 2011, 83, 3533–3540 CrossRef CAS PubMed.
- F. Yin, J. Liu, A. Liu, Y. Li, J. Luo, G. Guan and H. Yin, Vet. Parasitol., 2017, 237, 125–129 CrossRef CAS PubMed.
- K. Tae-Hyeong, P. Juhee, K. Chi-Ju and C. Yoon-Kyoung, Anal. Chem., 2014, 86, 3841–3848 CrossRef PubMed.
- J. F. C. Loo, H. C. Kwok, C. C. H. Leung, S. Y. Wu, I. L. G. Law, Y. K. Cheung, Y. Y. Cheung, M. L. Chin, P. Kwan, M. Hui, S. K. Kong and H. P. Ho, Biosens. Bioelectron., 2017, 93, 212–219 CrossRef CAS PubMed.
- G. Huang, Q. Huang, L. Xie, G. Xiang, L. Wang, H. Xu, L. Ma, X. Luo, J. Xin, X. Zhou, X. Jin and L. Zhang, Sci. Rep., 2017, 7, 6441 CrossRef PubMed.
- J. H. Jung, B. H. Park, S. J. Oh, G. Choi and T. S. Seo, Biosens. Bioelectron., 2015, 68, 218–224 CrossRef CAS PubMed.
- L. Zhang, F. Tian, C. Liu, Q. Feng, T. Ma, Z. Zhao, T. Li, X. Jiang and J. Sun, Lab Chip, 2018, 18, 610–619 RSC.
- X. Ye, J. Xu, L. Lu, X. Li, X. Fang and J. Kong, Anal. Chim. Acta, 2018, 1018, 78–85 CrossRef CAS PubMed.
- G. Choi, T. Prince, J. Miao, L. Cui and W. Guan, Biosens. Bioelectron., 2018, 115, 83–90 CrossRef CAS PubMed.
- S. Santiago-Felipe, L. A. Tortajada-Genaro, R. Puchades and Á. Maquieira, Microchim. Acta, 2016, 183, 1195–1202 CrossRef CAS.
- D. Liu, G. Liang, Q. Zhang and B. Chen, Anal. Chem., 2013, 85, 4698–4704 CrossRef CAS PubMed.
- H. Yang, Z. Chen, X. Cao, Z. Li, S. Stavrakis, J. Choo, P. D. Howes and N. He, Anal. Bioanal. Chem., 2018, 410, 7019–7030 CrossRef CAS PubMed.
- R. Blakemore, E. Story, D. Helb, J. Kop, P. Banada, M. R. Owens, S. Chakravorty, M. Jones and D. Alland, J. Clin. Microbiol., 2010, 48, 2495–2501 CrossRef CAS PubMed.
- D. Helb, M. Jones, E. Story, C. Boehme, E. Wallace, K. Ho, J. Kop, M. R. Owens, R. Rodgers and P. Banada, J. Clin. Microbiol., 2010, 48, 229–237 CrossRef CAS PubMed.
- S. Tanriverdi, L. Chen and S. Chen, J. Infect. Dis., 2010, 201, S52–S58 CrossRef CAS PubMed.
- N. Brenwald, N. Baker and B. Oppenheim, J. Hosp. Infect., 2010, 74, 245–249 CrossRef CAS PubMed.
- E. M. Harding-Esch, E. C. Cousins, S.-L. Chow, L. T. Phillips, C. L. Hall, N. Cooper, S. S. Fuller, A. V. Nori, R. Patel and S. Thomas-William, EBioMedicine, 2018, 28, 120–127 CrossRef CAS PubMed.
- A. Niemz, T. M. Ferguson and D. S. Boyle, Trends Biotechnol., 2011, 29, 240–250 CrossRef CAS PubMed.
- H. Wang, Z. Ma, J. Qin, Z. Shen, Q. Liu, X. Chen, H. Wang, Z. An, W. Liu and M. Li, Biosens. Bioelectron., 2019, 126, 373–380 CrossRef CAS PubMed.
- J. R. Choi, Z. Liu, J. Hu, R. Tang, Y. Gong, S. Feng, H. Ren, T. Wen, H. Yang and Z. Qu, Anal. Chem., 2016, 88, 6254–6264 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2019 |