Open Access Article
Open Access ArticleMicrofluidic quantification and separation of yeast based on surface adhesion†
Kristina
Reinmets
 a,
Amin
Dehkharghani
b,
Jeffrey S.
Guasto
a,
Amin
Dehkharghani
b,
Jeffrey S.
Guasto
 b and
Stephen M.
Fuchs
b and
Stephen M.
Fuchs
 *a
*a
aDepartment of Biology, Tufts University, Medford, MA 02155, USA. E-mail: Stephen.fuchs@tufts.edu
bDepartment of Mechanical Engineering, Tufts University, MA 02155, USA
First published on 16th September 2019
Abstract
Fungal adhesion is fundamental to processes ranging from infections to food production to bioengineering. Yet, robust, population-scale quantification methods for yeast surface adhesion are lacking. We developed a microfluidic assay to discriminate and separate genetically-related yeast strains based on adhesion strength, and to quantify effects of ionic strength and substrate hydrophobicity on adhesion. This approach will enable the rapid screening and fractionation of yeast based on adhesive properties for genetic protein engineering, anti-fouling surfaces, and a host of other applications.
1. Introduction
Yeast are single-celled eukaryotes that possess the ability to adhere to a range of biotic and abiotic surfaces.1 Adhesion contributes to pathogenicity in clinical yeast such as Candida albicans and Candida glabrata,2 but also imparts desirable qualities to industrial yeast such as Saccharomyces cerevisiae.3 Quantifying the properties of yeast adhesion, as well as environmental factors that mediate interactions between cells and surfaces, is therefore crucial to understanding a host of environmental, medical, and industrial processes. Fungal adhesion is mediated by surface proteins known as adhesins (Fig. 1a), which participate either in the interaction with sugar molecules and proteins on the surface of other yeasts and mammalian cells, or nonspecific binding to abiotic surfaces.1,4–7While yeast adhesin genes exhibit broad genetic diversity, they also show significant structural similarities across related species, including both non-infectious and pathogenic yeast.8,9 Adhesion of yeast cells to surfaces is an important early step in biofilm formation, which provides an ecological advantage to yeast through nutrient availability and protection from environmental hazards, such as fluid flow, changes in pH, or the presence of toxic antifungal agents. For example, surface attached C. albicans, adhered to an abiotic dental prosthetic, were shown to be significantly more resistant to a range of antifungals than the same cells grown planktonically.10 Likewise, S. cerevisiae exhibit the ability to adhere to one another (flocculation), which is important for protection from environmental stresses.11 Adhesion is also an attribute for certain industrial purposes from beer and wine production to chemical synthesis.12,13 The availability of vast molecular tools and the robustness of S. cerevisiae have made budding yeast an ideal model system for studying the mechanisms of fungal adhesion as well as for exploiting their well-known genetics for bioengineering purposes. However, as surface adhesion may either impede or promote various applications, robust and rapid screening methods for adhesion phenotypes are needed in order to improve our understanding of fungal adhesion.
Previous studies of yeast adhesion have primarily focused on either multicellular cell–cell and cell–surface interactions within biofilms or precision measurements of the adhesive properties of single yeast cells and adhesin proteins.14 Fungal biofilms encompass complex cellular structures composed of extracellular matrix (ECM) containing polysaccharides and proteins, including fungal adhesins that are shed from cell surface, as well as yeast cells in various differentiation and life stages.15,16 Standard approaches for studying yeast biofilm adhesion include long term culturing of yeast on agar plates or in liquid media using polystyrene plates, and analysis of biofilm release from the surfaces upon the topical application of a water stream.17 Adhesion or agar invasion is also approximated using colorimetric cell-staining dyes to stain cells adhered to a surface.18 Biofilm investigations have greatly contributed to our understanding of complex interactions among cells within the biofilm as well as environmental and biological signals regulating the formation of fungal mats.19 However, unravelling the fundamental mechanisms of surface attachment of single cells from a complex biofilm community of cells embedded in ECM is challenging. Single-cell studies have helped to illuminate the nanoscale mechanics of yeast adhesion, including the re-localization of adhesin proteins in response to surface contact.20–22 Such studies often rely on atomic force microscopy (AFM), which enables accurate quantification of attachment forces but is limited by extremely low throughput due to individual cell measurements. Fungal adhesive traits are known to be stochastic on an individual basis and can cause uncertainty in precision measurements of adhesion strength when considering a broad population of cells.9,23,24 Hence, a need exists for assays which can provide population scale statistics on individual cell adhesion properties.
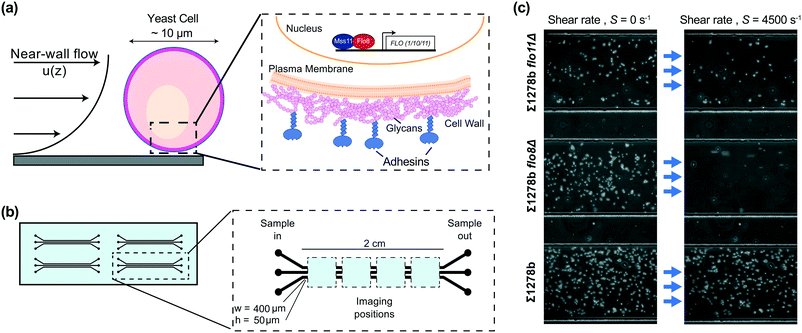
Here, we present a microfluidic assay (Fig. 1) for measuring surface adhesion properties of yeast strains, comprising customized microchannels and image analysis. The assay enables the direct quantification of the adhesive properties of three different yeast strains or replicates simultaneously by monitoring the adhesion state of thousands of cells per channel. Our approach is based on the application of precisely controlled hydrodynamic shear (Fig. 1a), which is proportional to the drag force on the cells (see ESI†) and systematically induces detachment from the surface (Fig. 1c). We demonstrate the efficacy of our assay by quantitatively comparing the differential adhesion of nine common S. cerevisiae laboratory strains and mutants lacking FLO family genes, which contribute to fungal adhesion. We show the distinct effects of ionic strength of the buffer solution and surface hydrophobicity on adhesion of the investigated yeast strains. Furthermore, we demonstrate how strain characteristics obtained from our assay can be applied for the separation and enrichment of cells with different adhesive properties – a feature of great potential for bioengineering applications.
2. Materials and methods
2.1 Growth and preparation of yeast suspensions
The Saccharomyces cerevisiae strains used in this work were: Σ1278b, Σ1278b flo8Δ, Σ1278b flo11Δ, W303-1A, SK1, SK1 flo8Δ, and EBY100 (see Table S1†).25 For cell detachment assays, all strains were grown overnight in liquid YPD (yeast extract (1% w/v), peptone (2% w/v), dextrose (2% w/v)) medium at 30 °C with shaking, diluted into fresh media on the following day to OD600 = 0.2, and grown for 3 more hours or until OD600 = 1.0–1.5. A 1 mL volume of cell culture equivalent to OD600 = 3.0 was then washed twice with sterile water, once with 10 mM citric acid phosphate buffer pH 6.0 containing 150 mM, 75 mM, or 10 mM NaCl and resuspended in 3 mL of the same buffer at final concentration around 3 × 107 cells per mL. In yeast strain sorting experiments, cells were grown and washed as described above, resuspended in 10 mM citric acid phosphate buffer supplemented with 150 mM NaCl and mixed in desired proportions based on OD600 readings.2.2 Device fabrication
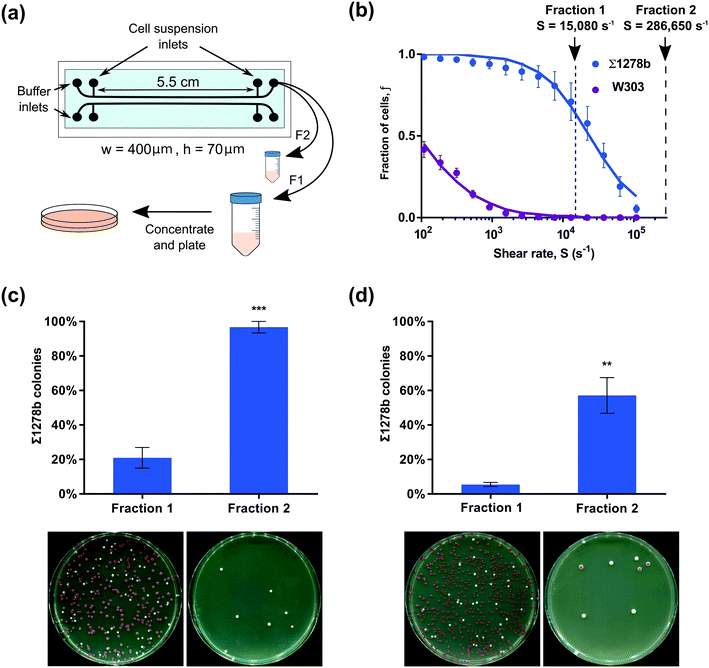
Microfluidic devices used in the cell detachment assay consisted of three parallel rectangular channels (400 μm wide, 50 μm deep, and 2 cm long) for multiplexing measurements, with four devices per chip (Fig. 1b). The devices used in yeast strain sorting experiments had two parallel channels with dimensions of 400 μm in width, 70 μm in height and 5.5 cm in length, and were equipped with two inlet and two outlet openings (Fig. 5a). All devices were fabricated using standard soft-lithography techniques.26 Briefly, a mold was prepared by spin coating photoresist (SU-82050, MicroChem Corp.) on a silicon wafer and patterning the devices through photolithography. The microchannels were then cast in polydimethylsiloxane (PDMS, Dow Corning Corp.) and the devices were plasma-bonded to a standard glass microscope slide using hand-held plasma generator (BD-20AC Laboratory Corona Treater, ETP) for experiments shown on Fig. 1 through 4 or using a plasma cleaner (PE-25, Plasma Etch) for experiments shown on Fig. 5. For modified surface hydrophobicity, microchannels were bonded to a flat PDMS sheet27 (see Fig. S4†).2.3 Experimental setup
PDMS-on-glass channels were filled with 1 M NaOH, incubated for 20 minutes, and rinsed with 8 mL of water to produce a uniform hydrophilic surface.28,29 Non-toxic polyethylene tubing was linked to the channels, which were connected to a syringe pump (Harvard Apparatus Inc.) with glass syringes (Hamilton™ Gastight™ Syringes) containing buffer and to syringes containing yeast cell suspensions through a three-way valve. The microchannels were first wetted by flowing buffer through the system at a flow rate of 100 μL min−1. Next, 200 μL of yeast cell suspension was injected into the channels, and the cells were allowed to settle for 30 minutes, whereby attachment occurs spontaneously. For the cell detachment assay, the flow rate was exponentially increased in 14 steps from 1–1000 μl min−1, corresponding to shear rates 110–108![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 550 s−1 (see ESI†) with each shear rate being held for 4 minutes in duration.
550 s−1 (see ESI†) with each shear rate being held for 4 minutes in duration.
Yeast strain sorting experiments were performed as follows. NaOH-treated and washed microchannels were flushed with buffer, and the yeast cells were introduced through the cell suspension inlets as indicated on Fig. 5(a). The buffer outlet was kept closed during that procedure to avoid premature introduction of yeast cells into the collection tubing. Cells were allowed to settle for 30 minutes prior to flow initiation. First, a shear rate of 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 080 s−1 was applied for 4 minutes and the flow-through was collected into a sterile 50 mL tube prefilled with 10 mL of buffer. The outlet tubing was then removed from the first collection tube, the outside surface of the tubing was cleaned with 70% ethanol, and the tubing was then inserted into a new 50 mL collection tube prefilled with 10 mL buffer. Cells remaining on the channel surface at that point were collected by applying shear rate of 286
080 s−1 was applied for 4 minutes and the flow-through was collected into a sterile 50 mL tube prefilled with 10 mL of buffer. The outlet tubing was then removed from the first collection tube, the outside surface of the tubing was cleaned with 70% ethanol, and the tubing was then inserted into a new 50 mL collection tube prefilled with 10 mL buffer. Cells remaining on the channel surface at that point were collected by applying shear rate of 286![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 650 s−1 for 1 minute. Fractionation shear rates were informed by the relative detachment rates of each yeast strain (Fig. 5b). Cells were captured on the membrane of a sterile 0.2 μm polyether sulfone syringe filter (GE Healthcare Whatman™ Puradisc™ 25 mm filter) and collected by flushing the filter with 1 mL of sterile water to remove cells from the membrane. Eluate of the first fraction was diluted ten-fold and 150 μl of resulting dilution was plated in triplicate on YPD agar plates. The eluate of the second fraction was plated directly on the YPD plates in triplicate by 150 μl for experiments shown on Fig. 5(c) or 1 mL of eluate was divided between 6 plates in experiments shown in Fig. 5(d) to plate all collected cells.
650 s−1 for 1 minute. Fractionation shear rates were informed by the relative detachment rates of each yeast strain (Fig. 5b). Cells were captured on the membrane of a sterile 0.2 μm polyether sulfone syringe filter (GE Healthcare Whatman™ Puradisc™ 25 mm filter) and collected by flushing the filter with 1 mL of sterile water to remove cells from the membrane. Eluate of the first fraction was diluted ten-fold and 150 μl of resulting dilution was plated in triplicate on YPD agar plates. The eluate of the second fraction was plated directly on the YPD plates in triplicate by 150 μl for experiments shown on Fig. 5(c) or 1 mL of eluate was divided between 6 plates in experiments shown in Fig. 5(d) to plate all collected cells.
2.4 Image acquisition and analysis
For each applied shear rate, single images were captured at four different positions prior to flow initiation and three minutes after each increase in flow rate. Four positions were used to monitor a statistically significant number of cells in the microchannels, which provides greater accuracy in the evaluation of phenotypic diversity within cell population and increased statistical power of the collected data. The maximum Reynolds number in our assays was Re = 80, which suggests that the entrance length was L = 0.06 Dh Re = 1.9 mm for the highest shear rate examined, where Dh is taken as the channel width. Therefore, the first position of imaging was chosen at a distance larger than 5 mm to eliminate any entrance effects. The three-minute timepoint was selected based on experiments showing that cell detachment reaches steady state within approximately 2.5 minutes following a step change in shear rate (Fig. S1 and ESI†). The syringe pump and image acquisition were automated (Nikon Elements) and imaging was performed on an inverted microscope (Nikon Ti-E, Nikon Instruments Inc.) using phase contrast (10×, 0.3 NA objective) and a sCMOS camera (Zyla 5.5, Andor Technology). Images of surface attached yeast cells were analysed using custom particle identification algorithms (MATLAB, MathWorks Inc.) incorporating background subtraction, intensity thresholding, and centroid detection.30 Yeast cell detachment events were determined by comparing the number of cells on the microchannel surface following a step change in the shear rate at all four imaged positions. Monitoring cell detachment at multiple positions allows us to evaluate the uniformity of the results produced along the microchannels (see Fig. S2†). The number of cells on the surface at all four positions were then combined for appropriate representation of the examined cell populations and used to calculate fractions of cells on the channel surface. We thus report the fraction of cells remaining on the surface at a given shear rate as f(S) = N(S)/Ntotal, where N(S) is the total number of attached cells at a given shear rate S, and Ntotal was typically in the range of 2–8 × 103 cells.3. Results and discussion
3.1 Establishing microscale adhesion profiles of S. cerevisiae strains
We selected common laboratory yeast strains with known qualitative differences in adhesive properties, caused by a point mutation in the FLO8 gene.31 While qualitative differences of investigated yeast strains are readily observable (Fig. 1c), our microfluidic assay enables the precise quantitative comparison of various strains. To provide a comprehensive comparison of yeast surface adhesion strength across the different strains assayed, we calculated the detachment shear rates for all measured strains in several ways. First, by linearly interpolating the measured fraction of surface attached cells to f(S) = 0.5 to determine the median detachment shear rate Sm (Fig. S3 and Table S1†). The median detachment shear rate is a robust metric that enables the determination of the adhesive strength over a broad range of cell adhesion phenotypes. However, the median detachment shear rate is difficult to measure for some weakly adhering cells exhibiting small Sm < 100 s−1, for example W303 (Fig. 2a). We found that the sigmoidal characteristics of the detachment curves are well fit by a Hill equation f(S) =1 /(1 + (S/Sm)n), where Sm is the median detachment shear rate, and n is the slope of the linear decay in log–log scale. Nonlinear regressions show excellent agreement (R2 > 0.99) with the measured detachment curves (see Table S1†) and enable the extrapolation of Sm to small shear rates. Calculation of the mean value of the attachment survival probability from the measured f(S) (Table S1†) as well as exponential fits (analysis not shown) showed qualitative agreement with the observed trends for Sm (see ESI†). Thus, from our microfluidic cell detachment assay, we report results in terms of the measured median detachment shear rate below and provide fitted detachment shear rates in Table S1.†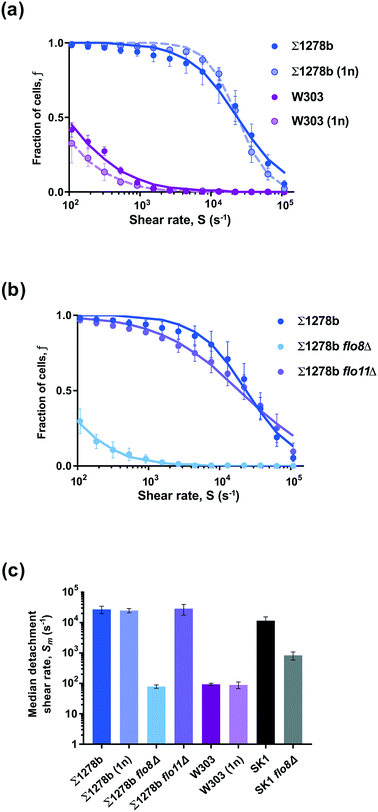 | ||
| Fig. 2 Differential adhesion strength in S. cerevisiae strains is due to variations in genetic background. (a and b) Starting from no shear, the fraction of surface attached cells remaining on the microchannel surface (glass) is measured as a function of the applied shear rate. Data points show detected cells on the surface, error bars are standard error of the mean (SEM, n = 3), and curves are fits of the Hill equation to the data (fitted slopes shown in Table S1†). (a) Strains Σ1278b and W303 (diploid, 2n) show strongly differential adhesion. However, haploid (1n) strains of the same genetic background show comparable detachment rates to their diploid counterparts. (b) Strong adhesion of strain Σ1278b is not affected by the deletion of a single surface protein Flo11p in Σ1278b flo11Δ but is diminished by the deletion of FLO8 gene in Σ1278b flo8Δ mutant. (c) Characteristic adhesive strength for each strain is quantified by the median detachment shear rate Sm for the cell population, which varies over two orders of magnitude. Error bars are SEM (n = 3). | ||
We first measured the detachment from glass surface of wildtype and flo8 mutant diploid strains Σ1278b and W303, respectively (Fig. 2a). The fraction f of Σ1278b cells remaining on the substrate decreased slowly in a sigmoidal fashion with increasing shear rate S, while W303 cells lacking a functional FLO8 gene were rapidly detached from the glass surface. For W303 cells, a fraction f ≈ 50% remained attached to the surface at a shear rate Sm ≈ 100 s−1. For comparison, the median detachment shear rate of W303 was over two orders of magnitude lower than for strain Σ1278b, which required a shear rate of Sm ≈ 27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 s−1 to achieve f ≈ 50% detachment (Fig. 2a).
000 s−1 to achieve f ≈ 50% detachment (Fig. 2a).
The FLO8 gene encodes a transcription factor responsible for activating the expression of yeast surface adhesins such as FLO1, FLO10 and FLO11, and a glucoamylase gene STA1.32,33 Flo1p adhesin has been shown to specifically bind to mannose molecules on the surface of neighbouring yeast cells and participates in flocculation.5 In addition to flocculation, the surface adhesin Flo11p also mediates agar invasion and adhesion to polystyrene.34–36 To determine whether ploidy affects adhesion, we measured the detachment shear rate of haploid Σ1278b (1n) and W303 (1n) cells in comparison to their diploid counterparts (Fig. 2a). The surface detachment profiles of Σ1278b (1n) and W303 (1n) strains were nearly identical to those of the diploid cells, suggesting that adhesion to an abiotic surface like glass is mediated by yeast cell wall components independent of ploidy.
3.2 Quantification of yeast strain-specific differences in adhesion
If the observed differences in adhesion strength between Σ1278b and W303 strains were indeed due to the mutation of FLO8 gene in W303, then deletion of FLO8 in a strongly adhesive strain, such as Σ1278b, should reduce its adhesive properties. To test this hypothesis, we measured the adhesive strength of Σ1278b flo8Δ cells using our microfluidic assay. The detachment shear rate in this strain (Sm < 100 s−1; Fig. 2b) was reduced by over two orders of magnitude relative to the parental strain. As expected, Σ1278b flo8Δ cells performed similarly to FLO8 deficient strain W303 in our adhesion assay and were released from surface at lower applied shear rates compared to wild-type Σ1278b cells. To evaluate if the strong adhesion of Σ1278b is mediated by Flo8-dependent adhesin Flo11p, we measured the detachment rate of Σ1278b flo11Δ cells and observed virtually no difference in the detachment profile compared to wild-type Σ1278b strain (Fig. 2b and c). This indicates that adhesion of Σ1278b cells to a glass surface is not mediated by Flo11p under tested conditions. To confirm the importance of the FLO8 gene, we tested a second strongly adherent strain, SK1,37 and its FLO8 deletion carrying mutant SK1 flo8Δ (Fig. 2c). The flo8 deletion reduced the attachment strength of SK1 from Sm ≈ 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 s−1 to <1000 s−1. While the attachment strength decreased about ten-fold, interestingly, the effect was not as drastic for the SK1 strain as compared to Σ1278b. These results suggest that Flo8 driven expression of yeast surface adhesins is not the sole mechanism of yeast adhesion to abiotic surfaces, and related S. cerevisiae strains may differ significantly in cell wall surface properties.
000 s−1 to <1000 s−1. While the attachment strength decreased about ten-fold, interestingly, the effect was not as drastic for the SK1 strain as compared to Σ1278b. These results suggest that Flo8 driven expression of yeast surface adhesins is not the sole mechanism of yeast adhesion to abiotic surfaces, and related S. cerevisiae strains may differ significantly in cell wall surface properties.
3.3 Electrostatic effects on yeast adhesion
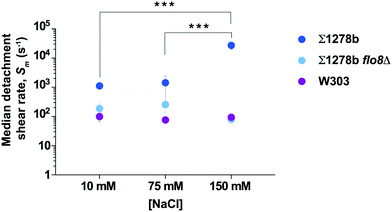
Cell surface properties, such as hydrophobicity and surface charge, have been shown to vary significantly among industrial and laboratory yeast strains.38 Non-specific cell adhesion to a surface depends upon the balance of van der Waals attractive forces and attractive or repulsive electrostatic interactions, which are influenced by the ionic strength of the culture medium through charge screening.39,40 To determine the effect of ionic strength on yeast adhesion, we measured the median detachment shear rate of Σ1278b cells on a glass surface at NaCl concentration of 75 mM, and 10 mM, in addition to the concentration of 150 mM (Fig. 3). At low ionic strength, the median detachment shear rate of Σ1278b cells decreased significantly from Sm ≈ 27![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 s−1 at [NaCl] = 150 mM to Sm ≈ 1000 s−1 at [NaCl] = 10 mM. This effect was not observed for W303 and Σ1278b flo8Δ, which lack a functional FLO8 gene. Moreover, to confirm that differences in adherence seen at different NaCl concentrations are not due to osmosis-driven changes in yeast cell volume, we measured cell size in 10 mM, 75 mM, and 150 mM NaCl over the course of 2 hours and observed no significant deviation from normal size variation (Fig. S5†). The cell surface of yeast is heavily modified with sugars and negatively charged41 at pH 6.0. Thus, this result is consistent with the theory that at low ionic strength, cell adhesion may be inhibited by electrostatic repulsion between the negatively-charged cell surface and the negatively-charged glass.
000 s−1 at [NaCl] = 150 mM to Sm ≈ 1000 s−1 at [NaCl] = 10 mM. This effect was not observed for W303 and Σ1278b flo8Δ, which lack a functional FLO8 gene. Moreover, to confirm that differences in adherence seen at different NaCl concentrations are not due to osmosis-driven changes in yeast cell volume, we measured cell size in 10 mM, 75 mM, and 150 mM NaCl over the course of 2 hours and observed no significant deviation from normal size variation (Fig. S5†). The cell surface of yeast is heavily modified with sugars and negatively charged41 at pH 6.0. Thus, this result is consistent with the theory that at low ionic strength, cell adhesion may be inhibited by electrostatic repulsion between the negatively-charged cell surface and the negatively-charged glass.
3.4 Effects of surface properties on yeast adhesion
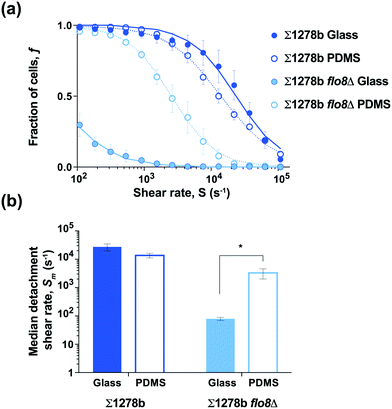
The expression of FLO1 and FLO11 genes in yeast is known to increase cell surface hydrophobicity in industrial S. cerevisiae strains, which is in concordance with observed Flo11-mediated adhesion to plastics.36,42 The expression of both FLO1 and FLO11 genes is controlled by the Flo8 transcription factor, suggesting that FLO8 wild-type and mutant cells may exhibit differential adhesion strength to hydrophobic versus hydrophilic surfaces. In order to test the effect of substrate hydrophobicity on yeast adhesion, microfluidic devices were bonded to a flat sheet of PDMS, which is known to be hydrophobic (contact angle θc > 100°; Fig. S4†),38,43 to compare with the hydrophilic glass substrate used above (contact angle θc ≈ 10°; Fig. 1–3 and S4†). The wild-type strain Σ1278b showed no significant difference in the median detachment shear rate between the glass and PDMS substrates (Fig. 4). Surprisingly, the median detachment shear rate of Σ1278b flo8Δ strain increased dramatically from Sm = 80 s−1 to ≈ 3000 s−1, when assayed on a hydrophobic PDMS surface (Fig. 4 and Table S1†). These results suggest that in the absence of electrostatic repulsive forces, even cells lacking major surface adhesins retain affinity for a more hydrophobic surface.3.5 Separation of yeast cells based on adhesive properties
Emergent yeast surface display techniques show great promise for the development of antibodies and small peptide therapeutics.25 Such approaches exploit sequential rounds of expression of protein libraries on yeast surfaces, selection of cells to recognize soluble or solid-phase conjugated targets, and collection and regrowth of selected clones.44,45 The microfluidic yeast cell screening techniques demonstrated here, have great potential for yeast display and bioengineering applications to perform clone selection on surface-conjugated targets. To demonstrate the potential of our assay to separate yeast strains based on surface attachment strength, we performed a series of sorting experiments to fractionate strongly adherent yeast from weakly adherent cells.Two yeast strains displaying differential median detachment shear rates (W303 and Σ1278b, see Fig. 2a) were combined to form an artificially mixed cell population, and then fractionated using our microfluidic assay. The W303 strain carries a mutation in ADE2, resulting in the accumulation of a coloured intermediate. Consequently, W303 colonies grown on agar plates in aerobic conditions gradually turn red allowing them to be easily distinguished from white-coloured Σ1278b colonies and to determine the efficacy of the yeast cell separation. W303 and Σ1278b cells were combined in a 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 ratio, respectively, and allowed to attach to the channel surface (Fig. 5a). A shear rate of 15
2 ratio, respectively, and allowed to attach to the channel surface (Fig. 5a). A shear rate of 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 080 s−1 was applied for four minutes to remove the bulk of the weakly attached W303 cells. Based on detachment profiles measured in our microfluidic adhesion assay and confirmed in single shear rate experiments (see ESI† and Fig. S6), up to 70% of strain Σ1278b cells from the mixed population were expected to remain surface attached up to this shear rate (Fig. 5b). The detached fraction of cells was captured in a sterile collection tube. Subsequently, the remaining fraction of surface attached cells – comprising mostly Σ1278b – was then collected by applying a shear rate of 286
080 s−1 was applied for four minutes to remove the bulk of the weakly attached W303 cells. Based on detachment profiles measured in our microfluidic adhesion assay and confirmed in single shear rate experiments (see ESI† and Fig. S6), up to 70% of strain Σ1278b cells from the mixed population were expected to remain surface attached up to this shear rate (Fig. 5b). The detached fraction of cells was captured in a sterile collection tube. Subsequently, the remaining fraction of surface attached cells – comprising mostly Σ1278b – was then collected by applying a shear rate of 286![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 650 s−1, which is well beyond the Σ1278b median detachment rate for 1 minute (Fig. 5a and b). Notably, we have confirmed that such a high shear rate (286
650 s−1, which is well beyond the Σ1278b median detachment rate for 1 minute (Fig. 5a and b). Notably, we have confirmed that such a high shear rate (286![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 650 s−1) used to collect the second fraction has no deleterious effects on yeast cells (Fig. S7†). To evaluate strain composition of the two collected fractions, both cell suspensions were concentrated and plated on YPD agar plates. As predicted based on the measurements in our adhesion assay, 96% of cells in the strongly-attached fraction were Σ1278b, while only 4% were W303, showing that characteristics obtained above can be used to sort cells based on adherence (Fig. 5c). We speculate that the presence of W303 cells in the strongly-adherent fraction could be explained in two possible ways – first, W303 and Σ1278b could be attaching to each other, causing the less adherent W303 cells to withstand high shear rates relying on Σ1278b strain's adherence to glass. Second, the adhesion assay is performed by gradually increasing shear rates over time while strain separation was done by applying a single shear rate and ultimately reducing the period of time over which the cells were sheared. Although we did not observe cell detachment after four minutes of constant shear in preliminary experiments (Fig. S1†), it is possible that increasing shear rate over longer periods of time may facilitate cell detachment.
650 s−1) used to collect the second fraction has no deleterious effects on yeast cells (Fig. S7†). To evaluate strain composition of the two collected fractions, both cell suspensions were concentrated and plated on YPD agar plates. As predicted based on the measurements in our adhesion assay, 96% of cells in the strongly-attached fraction were Σ1278b, while only 4% were W303, showing that characteristics obtained above can be used to sort cells based on adherence (Fig. 5c). We speculate that the presence of W303 cells in the strongly-adherent fraction could be explained in two possible ways – first, W303 and Σ1278b could be attaching to each other, causing the less adherent W303 cells to withstand high shear rates relying on Σ1278b strain's adherence to glass. Second, the adhesion assay is performed by gradually increasing shear rates over time while strain separation was done by applying a single shear rate and ultimately reducing the period of time over which the cells were sheared. Although we did not observe cell detachment after four minutes of constant shear in preliminary experiments (Fig. S1†), it is possible that increasing shear rate over longer periods of time may facilitate cell detachment.
To evaluate the robustness of this method, we tested our approach by reducing the number of Σ1278b cells in the sorting experiment and changed the W303 to Σ1278b ratio to 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, respectively. Only 5% of observed colonies from fraction 1 were Σ1278b, while 57% of the second fraction were Σ1278b (Fig. 5d). Taking into account the small concentration of Σ1278b cells in the initial suspension, the observed ratio of Σ1278b in the second fraction further emphasises the utility of the strain characteristics obtained earlier for screening and selection processes. Based on these observations, the selected fraction could be further enriched through additional rounds of fractionation to isolate Σ1278b cells from the total population. Furthermore, we have been able to successfully collect fractions of weakly adherent W303 cells from a mixture of W303 and Σ1278b, albeit with slightly lower efficiency after the first round of fractionation (see ESI† and Fig. S8), using the present approach. Taken together, we demonstrated that surface adhesion traits quantified through our assay can be used to select the optimal shear rates for cell separation based on adhesive properties.
1, respectively. Only 5% of observed colonies from fraction 1 were Σ1278b, while 57% of the second fraction were Σ1278b (Fig. 5d). Taking into account the small concentration of Σ1278b cells in the initial suspension, the observed ratio of Σ1278b in the second fraction further emphasises the utility of the strain characteristics obtained earlier for screening and selection processes. Based on these observations, the selected fraction could be further enriched through additional rounds of fractionation to isolate Σ1278b cells from the total population. Furthermore, we have been able to successfully collect fractions of weakly adherent W303 cells from a mixture of W303 and Σ1278b, albeit with slightly lower efficiency after the first round of fractionation (see ESI† and Fig. S8), using the present approach. Taken together, we demonstrated that surface adhesion traits quantified through our assay can be used to select the optimal shear rates for cell separation based on adhesive properties.
Conclusions
In this work, we developed and validated the efficacy of a microfluidic assay for quantifying yeast surface adhesion strength via the precise control of hydrodynamic shear and image analysis. The resolution of the step-wise increase of shear rate in this assay can be arbitrarily adjusted to capture a wide range of adhesive strengths across a variety of yeast strains. We demonstrated that our assay enables the detection of differences in adhesion based on genetic background; the FLO8 gene deletion decreases adhesive strength to a varying degree in Σ1278b and SK1 strains. This methodology was applied to determine effects of environmental conditions and surface composition on yeast adhesive strength. Furthermore, by capturing the cells detaching at specific shear rates, we demonstrated the ability of our assay to separate cells with differential surface adhesion strength from a mixed cell population. Specifically, combining our assay with an outgrowth phase or multi-stage microfluidic separation would lead to separation of cells from mixtures of more than two strains if differences in adherence exist. Such approaches can be applied even when the detachment profiles of cells under investigation are not known, as fractionation can be performed based on adhesive properties prior to exact identification of strains in the studied mixture of cells.The median detachment shear rate Sm provides a concrete metric for discerning adhesive strength of cells and can be calibrated for direct force measurements to complement techniques such as AFM. Fungal adhesion is of great concern to both medical and industrial communities, and thus, this approach will have utility in future applications for determining genetic and environmental factors that contribute to surface adhesion in many fungal species. For example, the quantification of pathogenic yeast interactions with hydrophilic and hydrophobic surfaces will aid in testing of anti-fungal compounds and the development of materials used in medical implants.46 Other potential applications include quantitative evaluation of yeast adhesion on a population level for fitness heterogeneity and biofilm studies as well as establishing baseline detachment shear rate of strains used for protein engineering. Display of protein libraries on yeast surface has been widely adapted for antibody and biologics development and screening. Most often, magnetic beads are used as solid carriers of antigens, however, silicon surfaces are amenable to functionalization and subsequent conjugation of peptides and nucleic acids. Our microfluidic assay coupled with antigen-coated channels could be used to determine yeast background adhesion, counterselection, and selection of yeast clones with differential affinity to the target.
Author contributions
K. R. and A. D. performed the experiments and analysed the data. K. R. was responsible for all cell manipulations and imaging and A. D. designed the microfluidic devices, developed microscope automation protocols, and image analysis software. All authors conceived and designed the experiments and wrote and edited the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was funded in part by Army Research Office Grant W911NF-16-1-0175 (to S. M. F.) and by National Science Foundation Grants CAREER-1554095 and CBET-1511340 (to J. S. G.). This work was also supported by at Tufts Collaborates grant (to S. M. F. and J. S. G.).References
- K. J. Verstrepen and F. M. Klis, Mol. Microbiol., 2006, 60, 5–15 CrossRef CAS PubMed.
- J. R. Blankenship and A. P. Mitchell, Curr. Opin. Microbiol., 2006, 9, 588–594 CrossRef CAS PubMed.
- S. E. Van Mulders, M. Ghequire, L. Daenen, P. J. Verbelen, K. J. Verstrepen and F. R. Delvaux, Appl. Microbiol. Biotechnol., 2010, 88, 1321–1331 CrossRef CAS PubMed.
- O. Kobayashi, N. Hayashi, R. Kuroki and H. Sone, J. Bacteriol., 1998, 180, 6503–6510 CAS.
- K. V. Goossens, C. Stassen, I. Stals, D. S. Donohue, B. Devreese, H. De Greve and R. G. Willaert, Eukaryotic Cell, 2011, 10, 110–117 CrossRef CAS PubMed.
- M. L. Zupancic, M. Frieman, D. Smith, R. A. Alvarez, R. D. Cummings and B. P. Cormack, Mol. Microbiol., 2008, 68, 547–559 CrossRef CAS PubMed.
- D. J. Rigden, L. V. Mello and M. Y. Galperin, Trends Biochem. Sci., 2004, 29, 335–339 CrossRef CAS PubMed.
- X. Xie and P. N. Lipke, Yeast, 2010, 27, 479–488 CrossRef CAS PubMed.
- M. Fidalgo, R. R. Barrales and J. Jimenez, Yeast, 2008, 25, 879–889 CrossRef CAS PubMed.
- J. Chandra, P. K. Mukherjee, S. D. Leidich, F. F. Faddoul, L. L. Hoyer, L. J. Douglas and M. A. Ghannoum, J. Dent. Res., 2001, 80, 903–908 CrossRef CAS PubMed.
- S. Smukalla, M. Caldara, N. Pochet, A. Beauvais, S. Guadagnini, C. Yan, M. D. Vinces, A. Jansen, M. C. Prevost, J. P. Latge, G. R. Fink, K. R. Foster and K. J. Verstrepen, Cell, 2008, 135, 726–737 CrossRef CAS PubMed.
- V. Nedovic, B. Gibson, T. F. Mantzouridou, B. Bugarski, V. Djordjevic, A. Kalusevic, A. Paraskevopoulou, M. Sandell, D. Smogrovicova and M. Yilmaztekin, Yeast, 2015, 32, 173–216 CAS.
- J. Wu, A. Elliston, G. Le Gall, I. J. Colquhoun, S. R. A. Collins, J. Dicks, I. N. Roberts and K. W. Waldron, Sci. Rep., 2017, 7, 14259 CrossRef PubMed.
- G. A. O'Toole, L. A. Pratt, P. I. Watnick, D. K. Newman, V. B. Weaver and R. Kolter, Methods Enzymol., 1999, 310, 91–109 Search PubMed.
- H. C. Flemming and J. Wingender, Nat. Rev. Microbiol., 2010, 8, 623–633 CrossRef CAS PubMed.
- M. H. Meem and P. J. Cullen, FEMS Yeast Res., 2012, 12, 809–818 CrossRef CAS PubMed.
- B. Guo, C. A. Styles, Q. Feng and G. R. Fink, Proc. Natl. Acad. Sci. U. S. A., 2000, 97, 12158–12163 CrossRef CAS PubMed.
- W. Kueng, E. Silber and U. Eppenberger, Anal. Biochem., 1989, 182, 16–19 CrossRef CAS PubMed.
- S. Bruckner and H. U. Mosch, FEMS Microbiol. Rev., 2012, 36, 25–58 CrossRef PubMed.
- C. X. Chan, S. El-Kirat-Chatel, I. G. Joseph, D. N. Jackson, C. B. Ramsook, Y. F. Dufrene and P. N. Lipke, mSphere, 2016, 1, e00128-16 CrossRef PubMed.
- D. Alsteens, M. C. Garcia, P. N. Lipke and Y. F. Dufrene, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 20744–20749 CrossRef CAS PubMed.
- P. N. Lipke, S. A. Klotz, Y. F. Dufrene, D. N. Jackson and M. C. Garcia-Sherman, Microbiol. Mol. Biol. Rev., 2018, 82, e00035-17 CrossRef PubMed.
- B. L. Chin, O. Ryan, F. Lewitter, C. Boone and G. R. Fink, Genetics, 2012, 192, 1523–1532 CrossRef CAS PubMed.
- K. J. Verstrepen, T. B. Reynolds and G. R. Fink, Nat. Rev. Microbiol., 2004, 2, 533–540 CrossRef CAS PubMed.
- E. T. Boder and K. D. Wittrup, Nat. Biotechnol., 1997, 15, 553–557 CrossRef CAS PubMed.
- Y. Xia and G. M. Whitesides, Annu. Rev. Mater. Sci., 1998, 28, 153–184 CrossRef CAS.
- K. Haubert, T. Drier and D. Beebe, Lab Chip, 2006, 6, 1548–1549 RSC.
- I. Hoek, F. Tho and W. M. Arnold, Lab Chip, 2010, 10, 2283–2285 RSC.
- J. J. Cras, C. A. Rowe-Taitt, D. A. Nivens and F. S. Ligler, Biosens. Bioelectron., 1999, 14, 683–688 CrossRef CAS.
- J. C. Crocker and D. G. Grier, J. Colloid Interface Sci., 1996, 179, 298–310 CrossRef CAS.
- H. Liu, C. A. Styles and G. R. Fink, Genetics, 1996, 144, 967–978 CAS.
- O. Kobayashi, H. Suda, T. Ohtani and H. Sone, Mol. Gen. Genet., 1996, 251, 707–715 CAS.
- O. Kobayashi, H. Yoshimoto and H. Sone, Curr. Genet., 1999, 36, 256–261 CrossRef CAS PubMed.
- W. S. Lo and A. M. Dranginis, Mol. Biol. Cell, 1998, 9, 161–171 CrossRef CAS PubMed.
- K. V. Goossens and R. G. Willaert, FEMS Yeast Res., 2012, 12, 78–87 CrossRef CAS PubMed.
- F. Li and S. P. Palecek, Eukaryotic Cell, 2003, 2, 1266–1273 CrossRef CAS PubMed.
- K. Nowlin, A. Boseman, A. Covell and D. LaJeunesse, J. R. Soc., Interface, 2015, 12, 20140999 CrossRef PubMed.
- M. Schiavone, S. Dejean, N. Sieczkowski, M. Castex, E. Dague and J. M. Francois, Front. Microbiol., 2017, 8, 1806 CrossRef PubMed.
- M. Hermanssonm, Colloids Surf., B, 1999, 14, 105–119 CrossRef.
- D. Kregiel, J. Berlowska and W. Ambroziak, World J. Microbiol. Biotechnol., 2012, 28, 3399–3408 CrossRef CAS PubMed.
- P. Thonart, M. Custinne and M. Paquot, Enzyme Microb. Technol., 1982, 4, 191–194 CrossRef CAS.
- P. Govender, J. L. Domingo, M. C. Bester, I. S. Pretorius and F. F. Bauer, Appl. Environ. Microbiol., 2008, 74, 6041–6052 CrossRef CAS PubMed.
- A. Mata, A. J. Fleischman and S. Roy, Biomed. Microdevices, 2005, 7, 281–293 CrossRef CAS PubMed.
- H. R. Hoogenboom, Nat. Biotechnol., 2005, 23, 1105–1116 CrossRef CAS PubMed.
- J. A. Van Deventer and K. D. Wittrup, Methods Mol. Biol., 2014, 1131, 151–181 CrossRef CAS PubMed.
- M. Ramasamy and J. Lee, BioMed Res. Int., 2016, 2016, 1851242 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9lc00275h |
| This journal is © The Royal Society of Chemistry 2019 |