Open Access Article
Open Access ArticleMonitoring transient cell-to-cell interactions in a multi-layered and multi-functional allergy-on-a-chip system†
Mario
Rothbauer‡
 ab,
Verena
Charwat‡§
c,
Barbara
Bachmann‡
abd,
Drago
Sticker‡
ab,
Verena
Charwat‡§
c,
Barbara
Bachmann‡
abd,
Drago
Sticker‡
 e,
Richard
Novak
e,
Richard
Novak
 f,
Heinz
Wanzenböck
f,
Heinz
Wanzenböck
 g,
Richard A.
Mathies
f and
Peter
Ertl
g,
Richard A.
Mathies
f and
Peter
Ertl
 *ab
*ab
aFaculty of Technical Chemistry, Vienna University of Technology, Getreidemarkt 9, 1060 Vienna, Austria. E-mail: peter.ertl@tuwien.ac.at
bAustrian Cluster for Tissue Regeneration, 1200 Vienna, Austria
cDepartment of Biotechnology, University of Agricultural Resources and Life Sciences, Muthgasse 18, 1090 Vienna, Austria
dAUVA Research Centre, Ludwig Boltzmann Institute for Experimental and Clinical Traumatology, 1200 Vienna, Austria
eDepartment of Pharmacy, University of Copenhagen, Universitetsparken 2, 2100 Copenhagen, Denmark
fDepartment of Chemistry, University of California at Berkeley, Lewis Hall, Berkeley, California, USA
gFaculty of Electrical Engineering, Vienna University of Technology, Gußhausstr. 25-25a, 1040 Vienna, Austria
First published on 23rd April 2019
Abstract
We have developed a highly integrated lab-on-a-chip containing embedded electrical microsensors, μdegassers and pneumatically-actuated micropumps to monitor allergic hypersensitivity. Rapid antigen-mediated histamine release (e.g. s to min) and resulting muscle contraction (<30 min) is detected by connecting an immune compartment containing sensitized basophile cells to a vascular co-culture model.
1. Introduction
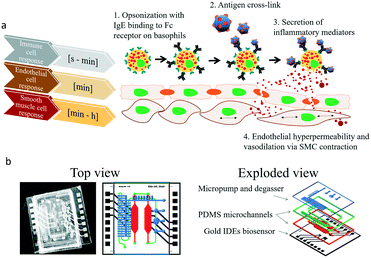
Cell to cell communication and transient cell response to external stimuli are vital in all multicellular organisms to react to changing environments.1 Cross-talk between cells, either by direct cell–cell contact or indirectly via signalling molecules, plays a crucial role in human health2–6 and disease.7–12 These cell–cell interactions can be of permanent nature in the case of tight or gap junction formation, or based on fast, transient signalling between the same and different cell types. Especially short-lived transient cell-to-cell interactions via secretion of signalling molecules are essential, since multicellular assemblies must coordinate complex physiological mechanisms. These mechanisms are known to include neuronal signaling,13 bone formation14 and immune response15,16 but also pathological conditions such as tumour growth,17 cardiovascular disease18 and allergies.19 In particular, during allergic reactions the interaction between immune and different tissue cell types occur immediately following the exposure to an antigen, which leads to release of soluble factors resulting in early phase allergic reactions including vascular dilation and smooth muscle contractions within minutes as illustrated in Fig. 1a.20,21 In the most common type of allergic reaction, type I (e.g., IgE-mediated or anaphylactic-type) hypersensitivity (20% of adults) repeated exposure to an allergen results in an overproduction of IgE-antibodies by B-cells,20 follow on sensitization,22 and binding of these antibodies to the FcεRI23 receptors of tissue mast cells and blood basophils. Upon re-exposure to the antigen and crosslinking to the antibodies triggers the release of histamine24 and other chemical mediators as well as the synthesis of inflammatory molecules, causing the early phase of allergic reactions resulting in contraction of smooth muscle cells and increased vascular permeability.25 This signalling cascade ultimately leads to the typical symptoms related to allergies from rhinitis and mucus secretion in mild to moderate cases to hypotension and shock in anaphylaxis. With allergies becoming increasingly common,26 research into novel treatment options such as vaccines and immunotherapy has gained world-wide momentum.27 Although a deeper understanding of the complex cellular processes that govern immune responses are crucial in basic and applied research, such transient cell-to-cell interactions are exceedingly difficult to monitor using conventional optical methods. To overcome these drawbacks a number of microfluidic cell cultures systems containing embedded biosensors28 have been developed over the last decade to allow efficient cell manipulation, stimulation and analysis of cell-to-cell interactions in vitro at the micro-scale under physiological29 and highly controllable microenvironment conditions.30,31 For instance, microfluidic devices have been employed to analyse B- and T-cell repertoires within populations of single cells,32 to conduct antibody screening,32 aid in allergy diagnosis33,34 and accelerate cancer immunology.35 Among these, microfluidic devices containing integrated electrical biosensors have demonstrated the ability to e.g. study dynamic interactions between tumor cells, immune cells and resident tissue36 as well as a variety of hetero- and homogenous cell-to-cell interactions.37–39To monitor transient immunological reactions that occur in type I allergy (see Fig. 1), we present a multi-layered, membrane-based and sensor-integrated lab-on-a-chip microdevice that performs automated fluid handling routines, cell-loading protocols and continuous, non-invasive bioimpedance spectroscopy. While the integration of micropumps and micro-degassers enable rapid control over the cellular microenvironment with nano-liter dead volume, the embedded electrical microsensors enable the detection of cellular responses with millisecond resolution (see Fig. S1† for detailed set-up). This highly integrated microfluidic device contains two spatially separated compartments to (1) maintain a vascular co-culture model comprising of endothelial cell monolayers and smooth muscle cells and (2) an immune cell cultivation chamber located upstream, where the allergic reaction takes place. Both tissue chambers contain embedded impedance sensors to (a) monitor rapid basophil degranulation during type I and time-dependent regeneration events as well as (b) to detect dynamic cell-to-cell interactions between endothelial and smooth muscle as a result to antigen-mediated histamine release of basophile cells. Using the highly integrated biochip design shown in Fig. 1b, we now attempt to demonstrate the technological ability to evaluate cell signalling cascades and to show that secondary cellular responses such as muscle contraction can be used to amplify otherwise difficult to detect cellular signals (e.g. release of histamines).
2. Results and discussion
2.1. Characterization of on-chip micro-degassers, micropumps and thin-film gold impedance sensors
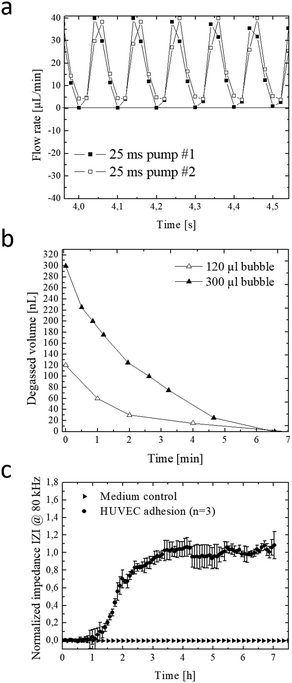
The integrated micropump provides programmable perfusion of medium and reagents at nL dead volume and is based on a four-step actuation procedure of three serially-connected microvalves.40,41 Fluid displacement is defined by the geometry of the ellipsoid central microvalve featuring a diameter of 2 mm length and 1 mm width and 100 μm height, which corresponds to a volume of 0.6 μl per pump stroke. Actuation pressures and step times between valves can be adjusted to control net flow velocities, peak shear forces and peristaltic profiles (see Fig. S-2† for detailed characterizations). We characterized micropump operation over a range of actuation times and pressures in order to optimize cell culture perfusion conditions. Fig. 2a shows five actuation cycles and actuation step time of 25 ms resulting in positive pump strokes with peak values around 38 ± 2 μl and actuation duration for individual strokes of around 100 ms. Increase of actuation step time to 75 ms as shown in Fig. S-2a† decreases the peak values to 31 ± 1 μl with an average actuation time around 300 ms. Incomplete execution of the actuation protocol due to inertia of the flexible membrane resulted in negative backflow of around 20 ± 1 μl with an average backflow duration of 50 ms. In a next set of experiments, the influence of actuation pressure on micropump peak values was evaluated in the presence of linear increase in actuation pressure between 10–40 kPa. Fig. S-2c–f† shows that not only step time but also actuation pressures can effectively influence micropump performance resulting in varying flow profiles and flow rates. As an example, highest peak values were observed for 40 kPa actuation pressure corresponding to 38 ± 3 μl pump strokes. When reducing the actuation pressure displaced volume marginally decreased to 22 ± 1, 14 ± 1 and 9 ± 1 μl for 30 to 10 kPa, respectively.In a more detailed study (see Fig. S-2c†) average micropump flow rates were evaluated for 10–45 kPa actuation pressure and 1–100 Hz actuation frequency or 0.3–8.5 ms step time. Results of the study revealed linear increase of average net flow when using actuation frequencies below 20 Hz. Above this 20 Hz threshold, flow rates gradually decreased, despite the increasing actuation pressures indicating that valves did not fully close in the presence of higher actuation frequencies. This means that at 10 Hz actuation frequencies similar flow rates of approximately 6 μl min−1 (0.25 dyn cm−2) are obtained for 45 kPa and 30 kPa actuation pressures. The ability to tune flow rates adjust peak shear forces and regulate fluid flow directions using integrated micropumps provides a close controlled and, reproducible cell culture conditions that can be optimized for any cell type.
The formation of microbubbles as shown in Fig. S-3† constitutes a prevalent obstacle to microfluidic cell cultures systems in general, since bubbles can form on nucleation sites and obstruct flow and increase shear stress to cell layers. Systems that rely on pneumatically-driven micropumps where pressurized air actuation is used face an additional challenge since, PDMS membranes valves are gas-permeable and facilitate evaporation of medium while the pressurized gas for actuation can dissolve into and potentially saturate the medium. To address these challenges, we have developed an integrated low dead volume degasser design. The on-chip integrated micro degassing system was evaluated for its capacity to remove microbubbles from the cell cultivation chamber. Fig. S-4ab† shows that nucleated bubbles tend to form, accumulate and increase at 37 °C within the microfluidic cultivation chamber with an average growth rate of 15 nl min−1 leading to several kOhms increase in impedance signal. A negative pressure of −60 kPa was applied to the semipermeable μdegassing layer see Fig. 2b resulting in an average degassing time of around 6.5 min corresponding to a degassed bubble volume of 32 ± 16 μl min−1. Interestingly, when increasing the microchannel height from 60 μm to 100 μm at a constant channel area the time for bubble removal was similar even though the bubble volume increased from 120 nl to 300 nl. Results of a long-term study demonstrated that bubble-free operation of the microfluidic cell cultures system can be accomplished over several weeks when using a serpentine-shaped degasser. The elimination of microbubble formation in the cell culture chamber as shown in Fig. S-4c–e† is particularly crucial when employing bioimpedance sensors that readily detect dielectric changes in the sensing volume to investigate most transient soluble cell interactions under constant conditions.
Next, the embedded bioimpedance sensors were characterized to determine optimum AC frequencies for detecting substrate adhesion and monolayer formation of human umbilical vein endothelial target cells. Sensitivity was calculated as a change in impedance of confluent endothelial cell monolayers in comparison to blank electrodes. Fig. S-4f† shows that the silicon-nitride coated, interdigitated electrode structures showed highest sensitivity to endothelial cell adhesion at a frequency range between 80 kHz and 100 kHz. Normalized impedance-time traces recorded at 80 kHz of endothelial cells adhesion and spreading is shown in Fig. 2c. A rapid increase of 290 ± 60 Ohms per h (100%) was observed during the first 3.5 hours indicating efficient cell adhesion at the sensor surface, followed by cell spreading events that resulted in a plateau phase indicating full sensor coverage and establishment of a stable cell barrier after 8 hours. Results of the sensor evaluation demonstrate the ability of the passivated and throughout silicon nitride-coated (50 nm) impedance sensors to continuously monitor the endothelial cell barrier integrity during allergic reactions resulting in endothelial hyperpermeability (see also Fig. 1a for cascade overview).
2.2. On-chip degranulation of of RBL-2H3 basophil cells
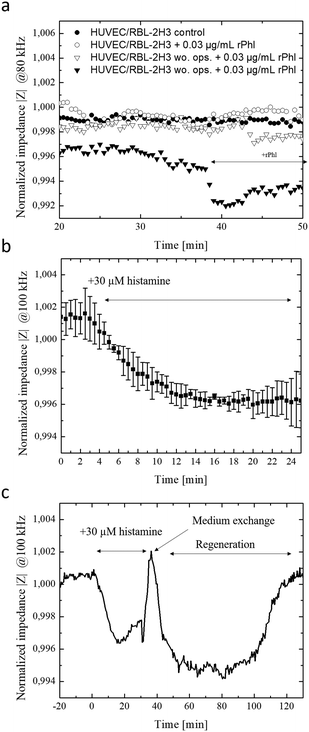
Since the crucial step in our chip-based allergic signaling cascade is the release of inflammatory mediators such as histamine by basophil cells, speed and efficiency of antigen-mediated degranulation of IgE opsonized RBL-2H3 basophils was evaluated using a methylene blue assay and impedance measurements. Degranulation of IgE opsonized basophil cells was initiated with the addition of 0.03 μg ml−1 of rPhI antigen into DPBS and supplemented RPMI 1640 medium. Fig. S-5a† shows that independent from the media composition opsonized basophil cells degranulated within 7 min of exposure to the rPhI antigen. However, while complete degranulated was observed in DPBS, 40% of the basophil cells remained granulated in the presence of in RPMI 1640 medium. These results indicate that fully supplemented culture medium is a more competitive environment for antigen binding due to the abundance of serum. To confirm these results basophil degranulation was also monitored using the embedded bioimpedance sensors. Fig. S-5b† shows time-impedance traces of degranulation events using our silicon nitrate-coated integrated impedance biosensors. The observed increase in impedance signal by 0.175 ± 0.03% following the addition of the rPhI antigen points at rapid basophil degranulation over a few minutes and the ability of the embedded impedance biosensors to detect transient cell responses in the range of seconds. Interestingly, impedance values rapidly normalized following media change to standard cell culture medium indicating efficient removal of rPhI antigen-containing media using the integrated micropumps, thus regranulation of basophil cells over the next period of 45 minutes. Another hallmark of type I allergy signaling cascade is concerned with endothelial cell responses to the release of histamine by basophil, which triggers the contraction of endothelial actomyosin fibers and results in vascular hyperpermeability.25 To evaluate endothelial cell responses in our lab-on-a-chip system, human endothelial cell monolayers are established over a 10 hours cultivation period and exposed to two subsequent additions of 30 μM histamine. Impedance-time traces shown in Fig. S-5c† reveal a rapid signal drop by 1.1% within the first 10 minutes of histamine exposure followed by a slow recovery upon histamine removal over a period of 60 min pointing at a slow regeneration of a tight endothelial barrier. A sequential second histamine exposure resulted in a similar decrease of resistance thus increases in permeability by around 0.9% within another 10 min.2.3. On-chip monitoring of transient inflammatory cell-to-cell interactions
In a final set of experiments, the allergy-on-a-chip system containing integrated liquid handling and embedded impedance biosensors was used to non-invasively monitor the cascade of transient cell-to-cell interactions that take place during IgE mediated type I allergic responses between basophil, endothelial and smooth muscle cells. Fig. 3a shows that within seconds of antigen exposure and cross-linking, opsonized RBL-2H3 basophils release inflammatory mediators to the underlying HUVEC endothelial cells. In turn, these endothelial cells react within 2 minutes with a decrease in impedance by 0.2% because of endothelial permeability. In the absence of rPhI antigen, no transient cellular responses were observable between basophils and endothelial cells. Additionally, impedance-time traces demonstrate that no alterations on the permeability of endothelial barriers are observed when using unprimed basophil cells (no IgE opsonization took place) even in the presence of the antigen. Impedance signals dropped by 0.7% within 5 minutes following the addition of 1 μg ml−1 rPhI antigen to a co-culture of IgE opsonized basophils and endothelial cells as shown in Fig. S-5e.† These results point at efficient yet highly transient cell-to-cell communication between basophil and endothelial cell monolayers.Finally, the direct influence of endothelial hyperpermeability on vascular smooth muscle cells responses was investigated to determine the time-dependence of cellular reactions within the signal transduction cascades. To create a multi-layered cell construct that resembles vasculature, vascular smooth muscle cells and endothelial cells were sequentially layered on top of each other using the integrated liquid handling system. Here, smooth muscle cells were first seeded at confluency and after 24 h, a monolayer of endothelial cells was seeded on top, and cells were allowed form a monolayer over the smooth muscle cell layers (see Fig. S-6†). As a simplified model for allergic response cascade, 30 μM histamine was added to the vascular construct and morphology changes of the underlying smooth muscle layer detected using the embedded electrical microsensors. Results of the endothelial cell-to-smooth muscle cell interaction study are shown in Fig. 3b and c, revealing an impedance signal drop by 0.4% within 15 minutes of histamine exposure indicating vascular hyperpermeability, loss of barrier integrity and stimulation of smooth muscle cells. Overall, a delay in cellular histamine response time from a few minutes to 28 min is obtained when using our smooth muscle co-culture model compared to endothelial monolayers. These results highlight that the proposed integrated lab-on-a-chip approach is capable to investigate transient cell-co-cell interactions via biomolecules more rapidly within minutes in comparison to state-of-the-art electrical biosensing approaches that need several hours.42
Conclusions
In conclusion, the developed highly integrated and multi-functional lab-on-a-chip technology containing embedded liquid handling and biosensing is able to monitor transient cell-to-cell responses in the range of seconds to hours. We show here that the combination of micropumps and-degassers enables robust automated cell culture handling, rapid cell stimulation using nanoliter dosing as well as elimination of microbubbles, while automated seeding of multiple cell layers enables spatial organization of vascular co-culture models. We demonstrated that our highly integrated lab-on-a-chip enables the translation of rapid and small cell responses such as release of histamine by basophile cells into readily measurable muscle contractions by microfluidic connection of a vascular co-culture model to an immune cell compartment. This scalable and automated technology has the potential to accelerate development of more effective and fast-acting allergy therapeutics.Author contributions
All authors conceived and discussed the manuscript. P. E., R. A. M., R. N., V. C., H. W. and D. S. designed and performed the experiments. V. C., D. S. and M. R. analyzed data. M. R., B. B. and P. E. wrote the manuscript and all authors edited the manuscript. P. E. and R. A. M. supervised the project.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors thank BioSensor Technologies unit of AIT Austrian Institute of Technology GmbH (Vienna) and the Medical University Vienna for supporting the project. Rudolf Valenta kindly provided basophil cells, antibodies and antigens.Notes and references
- G. M. Cooper, The Cell - A Molecular Approach, Sinauer Associates, Sunderland (MA), 2nd Edition, 2000 Search PubMed.
- E. Taverna, M. Götz and W. B. Huttner, Annu. Rev. Cell Dev. Biol., 2014, 30, 465–502 CrossRef CAS PubMed.
- O. Tornavaca, M. Chia, N. Dufton, L. O. Almagro, D. E. Conway, A. M. Randi, M. A. Schwartz, K. Matter and M. S. Balda, J. Cell Biol., 2015, 208(6), 821–838 CrossRef CAS PubMed.
- V. J. Thannickal and B. L. Fanburg, Am. J. Physiol., 2000, 279, L1005–L1028 CAS.
- Q. Shen, Y. Wang, E. Kokovay, G. Lin, S.-M. Chuang, S. K. Goderie, B. Roysam and S. Temple, Cell Stem Cell, 2008, 3, 289–300 CrossRef CAS PubMed.
- G. Folco and R. C. Murphy, Pharmacol. Rev., 2006, 58, 375–388 CrossRef CAS PubMed.
- M. E. Gleave, J. T. Hsieh, A. C. Von Eschenbach and L. W. K. Chung, J. Urol., 1992, 147, 1151–1159 CrossRef CAS PubMed.
- K. Masaki, Neuropathology, 2015, 35, 469–480 CrossRef CAS PubMed.
- K. Kamińska, C. Szczylik, Z. F. Bielecka, E. Bartnik, C. Porta, F. Lian and A. M. Czarnecka, J. Cell. Mol. Med., 2015, 19, 283–296 CrossRef PubMed.
- A. G. Clark and D. M. Vignjevic, Curr. Opin. Cell Biol., 2015, 36, 13–22 CrossRef CAS PubMed.
- J. Panés and D. N. Granger, Gastroenterology, 1998, 114, 1066–1090 CrossRef.
- N. V. Valeyev, C. Hundhausen, Y. Umezawa, N. V. Kotov, G. Williams, A. Clop, C. Ainali, C. Ouzounis, S. Tsoka and F. O. Nestle, PLoS Comput. Biol., 2010, 6, e1001024 CrossRef PubMed.
- D. L. Chao, L. Ma and K. Shen, Nat. Rev. Neurosci., 2009, 10, 262 CrossRef CAS PubMed.
- Y. Choi, R. Faccio, S. L. Teitelbaum and H. Takayanagi, in Osteoimmunology, ed. J. Lorenzo, M. C. Horowitz, Y. Choi, H. Takayanagi and G. Schett, Academic Press, San Diego, 2nd edn, 2016, pp. 41–70, DOI:10.1016/B978-0-12-800571-2.00004-9.
- M. I. Cuartero, I. Ballesteros, I. Lizasoain and M. A. Moro, Brain Res., 2015, 1623, 53–62 CrossRef CAS PubMed.
- B. Rossboth, A. M. Arnold, H. Ta, R. Platzer, F. Kellner, J. B. Huppa, M. Brameshuber, F. Baumgart and G. J. Schutz, Nat. Immunol., 2018, 19, 821–827 CrossRef CAS PubMed.
- L. Zhong, J. Roybal, R. Chaerkady, W. Zhang, K. Choi, C. A. Alvarez, H. Tran, C. J. Creighton, S. Yan, R. M. Strieter, A. Pandey and J. M. Kurie, Cancer Res., 2008, 68, 7237–7245 CrossRef CAS PubMed.
- C. M. Howard and T. A. Baudino, J. Mol. Cell. Cardiol., 2014, 70, 19–26 CrossRef CAS PubMed.
- G. A. W. Rook and J. L. Stanford, Immunol. Today, 1998, 19, 113–116 CrossRef CAS PubMed.
- P. T. C. A. Janeway Jr., M. Walport and M. J. Shlomchik, Immunobiology: The Immune System in Health and Disease, Garland Science, 5th edn, 2001 Search PubMed.
- D. G. Waller and A. P. Sampson, in Medical Pharmacology and Therapeutics, ed. D. G. Waller and A. P. Sampson, Elsevier, 5th edn, 2018, pp. 439–449, DOI:10.1016/B978-0-7020-7167-6.00038-5.
- J. Wedemeyer, M. Tsai and S. J. Galli, Curr. Opin. Immunol., 2000, 12, 624–631 CrossRef CAS PubMed.
- K. D. Stone, C. Prussin and D. D. Metcalfe, J. Allergy Clin. Immunol., 2010, 125, S73–S80 CrossRef PubMed.
- J. T. Schroeder, Immunol. Rev., 2011, 242, 144–160 CrossRef CAS PubMed.
- M. V. White, J. Allergy Clin. Immunol., 1990, 86, 599–605 CrossRef CAS PubMed.
- T. A. Platts-Mills, J. Allergy Clin. Immunol., 2015, 136, 3–13 CrossRef PubMed.
- R. Valenta, R. Campana, M. Focke-Tejkl and V. Niederberger, J. Allergy Clin. Immunol., 2016, 137, 351–357 CrossRef CAS PubMed.
- H. Zirath, M. Rothbauer, S. Spitz, B. Bachmann, C. Jordan, B. Müller, J. Ehgartner, E. Priglinger, S. Mühleder, H. Redl, W. Holnthoner, M. Harasek, T. Mayr and P. Ertl, Front. Physiol., 2018, 9, 815 CrossRef PubMed.
- M. Rothbauer, J. M. Rosser, H. Zirath and P. Ertl, Curr. Opin. Biotechnol., 2018, 55, 81–86 CrossRef PubMed.
- M. Rothbauer, H. Zirath and P. Ertl, Lab Chip, 2018, 18, 249–270 RSC.
- B. Bachmann, S. Spitz, M. Rothbauer, C. Jordan, M. Purtscher, H. Zirath, P. Schuller, C. Eilenberger, S. F. Ali, S. Mühleder, E. Priglinger, M. Harasek, H. Redl, W. Holnthoner and P. Ertl, Biomicrofluidics, 2018, 12, 042216 CrossRef PubMed.
- Y. F. S. Seah, H. Hu and C. A. Merten, Mol. Aspects Med., 2018, 59, 47–61 CrossRef CAS PubMed.
- Y. Yanase, K. Sakamoto, K. Kobayashi and M. Hide, Opt. Mater. Express, 2016, 6, 1339–1348 CrossRef CAS.
- J. Sinurat, I. Rengganis, C. M. Rumende and K. Harimurti, Asia Pac. Allergy, 2018, 8(1), e10 CrossRef PubMed.
- Q. Ramadan and M. A. Gijs, Lab Chip, 2015, 15, 614–636 RSC.
- V. Charwat, M. Rothbauer, S. F. Tedde, O. Hayden, J. J. Bosch, P. Muellner, R. Hainberger and P. Ertl, Anal. Chem., 2013, 85, 11471–11478 CrossRef CAS PubMed.
- V. Charwat, M. Joksch, D. Sticker, M. Purtscher, M. Rothbauer and P. Ertl, Analyst, 2014, 139, 5271–5282 RSC.
- O. Y. F. Henry, R. Villenave, M. J. Cronce, W. D. Leineweber, M. A. Benz and D. E. Ingber, Lab Chip, 2017, 17, 2264–2271 RSC.
- V. Charwat, M. Rothbauer, S. F. Tedde, O. Hayden, J. J. Bosch, P. Muellner, R. Hainberger and P. Ertl, Anal. Chem., 2013, 85, 11471–11478 CrossRef CAS PubMed.
- J. Kim, M. Kang, E. C. Jensen and R. A. Mathies, Anal. Chem., 2012, 84, 2067–2071 CrossRef CAS PubMed.
- J. Kim, A. M. Stockton, E. C. Jensen and R. A. Mathies, Lab Chip, 2016, 16, 812–819 RSC.
- I. Bischof, M. C. Hornburger, B. A. Mayer, A. Beyerle, J. Wegener and R. Fürst, Sci. Rep., 2016, 6, 23671 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9lc00108e |
| ‡ These authors contributed equally. |
| § Current address: University of California at Berkeley, 370 Hearst Memorial Mining Building #1760, Berkeley, CA 94720. |
| This journal is © The Royal Society of Chemistry 2019 |