Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Integrated elastomer-based device for measuring the mechanics of adherent cell monolayers†
Francesca
Sorba
 ab,
Alexandre
Poulin
ab,
Alexandre
Poulin
 b,
Réal
Ischer
a,
Herbert
Shea
b,
Réal
Ischer
a,
Herbert
Shea
 b and
Cristina
Martin-Olmos
b and
Cristina
Martin-Olmos
 *a
*a
aSwiss Center for Electronics and Microtechnology, CSEM SA, Neuchâtel, Switzerland. E-mail: cmartin@cellstrates.ch
bSoft Transducers Laboratory, École Polytechnique Fédérale de Lausanne, Neuchâtel, Switzerland
First published on 22nd May 2019
Abstract
Cells in the body collectively sustain mechanical deformations in almost all physiological functions. From the morphogenesis stage, cells' ability to sustain stress is essential for the body's well-being. Several pathologies have been associated with abnormal mechanical properties, thus suggesting the Young's modulus as a biomarker to diagnose diseases and determine their progression. Advancements in the field are quite slow because current techniques for measuring cell and tissue mechanics rely on complex and bulky measurement platforms that have low repeatability rates and limited measurement time-scales. We present the first miniaturized system that allows accurate quantification of the Young's modulus of adherent cell monolayers over a longer time (1–2 days). Our approach is based on tensile testing and optical read-out. Thanks to a thoughtful design and material choice, we are able to miniaturize tensile testing platforms into a 1 cm × 2 cm device. We provide highly repeatable Young's modulus measurements in the relevant range between 3 kPa and 300 kPa, over time and under physiological conditions, thus representing an interesting alternative to existing measurement platforms. Furthermore, the compatibility with standard biological equipment, continuous optical imaging and measurements on all types of adherent cells make this device highly versatile. Measurements on human sarcoma osteogenic (SaOS2) and Madin–Darby canine kidney cells (MDCK) are reported. The demonstrated capability to measure real-time changes in mechanical properties, such as after chemical treatment, opens the door for investigating the effects of drugs on cell mechanics.
Introduction
Within tissues and organs, individual cells actively interact with the surrounding extracellular matrix (ECM) and neighbouring cells through chemical exchange and mechanotransduction mechanisms.1–3 Most of the physiological functions in everyday life such as breathing, muscle contraction, and blood circulation rely on the ability of single cells to collectively organize and sustain mechanical deformation.4–6 Resistance to deformation is typically measured through the Young's modulus, whose specific value is determined by the physiological function7,8 and has been proven to be an effective biomarker in detecting diseases and their progression.9–12Cell monolayers are the simplest tissues in the body and are useful systems to characterize collective cell behaviours,13,14 cell–cell interactions15,16 and collective mechanical response to deformation.17 Within monolayers, cells undergo strong mechanical coupling with each other thus leading to active interactions among cells which are not simply the sum of individual responses.18,19 Cell monolayers play a critical role in the body from the embryogenesis stage as they drive the differentiation into organs. In adults, they contribute to maintaining homeostasis by acting as physical barriers to partition organs.20,21 Because of their specific interface location, cell monolayers stabilize tissues by sustaining external physiological stresses, for example stretching of the skin, peristaltic motion in the gastro-intestinal tract and urothelium stretching due to hydrostatic pressure.22 The current understanding of cell monolayers was mainly obtained using cell stretching devices to study the morphological and biochemical responses of cell populations to cyclic dynamic loading over time.23–26 A main limitation of existing cell stretching devices is the inability to measure the Young's modulus of the cells. This measurement is very challenging because the substrate is much stiffer than the cell; the substrate has a Young's modulus of the order of hundreds of kPa to a few MPa,27–29 compared to the tens of kPa for cells,30–32 and so the substrate dominates the mechanical response of the cell stretcher, masking the cell's contribution.33,34
The few approaches proposed up to now to easily decouple substrate and cell mechanical responses make use of out-of-plane deformations of thin membranes31,35,36 or the use of fully suspended cell monolayers.30,37,38 In both cases, these methods use bulky and complex set-ups that come with limitations: difficulty in observing the sample when out-of-plane deformations are imposed, fragility, low repeatability and, in suspended cell monolayers, strong experimental time limitation.
In this work, we present the first miniaturized device for quantitative Young's modulus measurements of adherent cell monolayers in-plane and over time. We achieve this by careful design of the substrate in terms of its mechanical properties and geometry in order to obtain a clearly distinguishable mechanical response when cells are adhered to the substrate. Thanks to the compatibility with standard cell culture equipment and real-time monitoring of the sample, long time measurements are possible. Therefore, the proposed device opens new possibilities for more relevant investigation of cell mechanics.
Experimental
Device design and fabrication
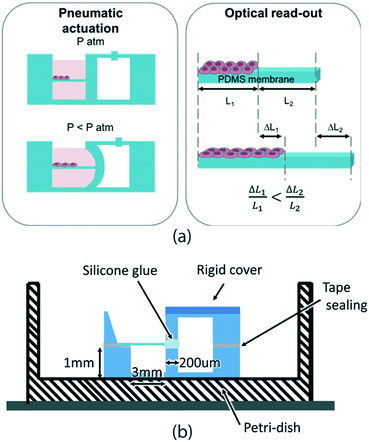
The key requirements that our device needs to meet are: compatibility with cell culture standard protocols (sterilization and cytocompatibility), cell culture environment, and incubator conditions (high humidity and 37 °C) as well as compatibility with real-time imaging of the sample (i.e. transparent materials). Also, the device should allow cell culture over at least 1–2 days and should ideally be compact and miniaturized not only for better manipulation but also for enabling measurement parallelization.Our approach for measuring the mechanical properties of cell monolayers on-chip is shown in Fig. 1a. The device is composed of two parallel chambers separated by a 200 μm thick vertical wall. One chamber is dedicated to cell culture: it can be filled with liquid, it is easily accessible for pipetting and it hosts a horizontally suspended membrane where cells are cultured (Fig. S4†). The adjacent chamber is sealed and is connected to a vacuum pump. When negative pressure is applied in the right chamber, the middle thin wall deforms, thus stretching the membrane. In this configuration, the cells can be exposed to cyclic uniaxial tensile strain. When cells are cultured on the membrane, the overall rigidity of the composite (membrane with cells) is increased and thus affects the membrane deformation. The essential aspect to make cell mechanical properties measurable from the overall substrate mechanical response relies on the accurate choice of the membrane elasticity and thickness by choosing values similar to the ones expected from the cell layers. We use 5 μm thick membranes made of biomedical grade silicone MED4086 (Nusil™) with a Young's modulus of 27.8 ± 9.7 kPa (more detailed characterization in Fig. S3†). By doing this, the mechanical responses of the cell layer and the membrane are in the same order of magnitude and their individual mechanical contribution can be easily decoupled.
Unless otherwise stated, the engineering strain is reported as ε = ΔL/L0, where ΔL is the length change and L0 the original monolayer length. The core of our technology is the use of differential strain measurements between a region covered with cells and a bare region of the membrane. This allows measurement of the mechanical properties of the adherent cell layers by subtracting the effect of the membrane. As the membrane mechanical properties are characterized prior to cell experiments, the differential read-out allows a direct measure of the force exerted during the deformation to be obtained (Fig. 1a).
Furthermore, using a differential approach makes the system independent of many variables (surface covered by the cells and pressure variations) that would otherwise significantly affect its precision. Considering that the force exerted on both regions is the same (eqn (1)) and that the calibrated sample and the membrane have the same width, the cell layer Young's modulus can therefore be measured as (see the ESI† for more details):
| εbareEbaretbare = εcompositeEcompositetcomposite | (1) |
| Ecompositetcomposite = Ecelltcell + Ebaretbare | (2) |
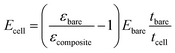 | (3) |
The devices are fabricated using polydimethylsiloxane (PDMS) S186 (Dow Corning®) by a moulding technique, while the membrane is made by casting39 and it is incorporated within two moulded pieces using silicone glue as shown in Fig. 1b.
The final device fits standard cell culture dishes which are engineered with connectors for the negative pressure tube. The device is compatible with inverted microscopy as well as measurement within an incubator environment (Fig. 2a). The sealed chamber of the device is connected to a pressure sensor (general fluid pressure sensor, PSE560 from SMC) and a pressure controller (electronic vacuum regulator, ITV009 from SMC) situated outside the incubator. Images of the samples during the deformation are acquired at a 10 frames per s rate (inverted microscope Etaluma™, LS460) and the strain is measured by a pattern recognition algorithm based on digital image correlation (DIC, developed using National Instruments, Vision Assistant). By tracking the membrane edges as well as the border between the bare and the cell regions, it is possible to quantify the strains of the whole membrane and the individual regions (bare and cell regions) over time.
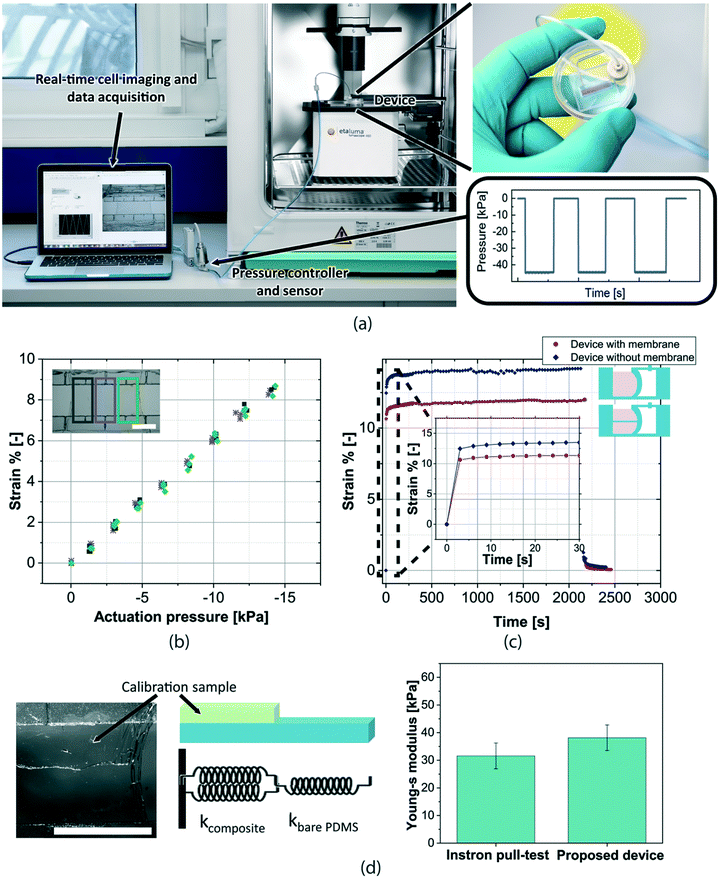 | ||
| Fig. 2 Experimental set-up and device characterization: (a) the device is placed in a physiological environment allowing long time experiments under sterile conditions. Negative pressure cycles are applied through a LabView interface to the device in order to stretch in-plane the membrane. Real-time optical images of the sample deformation are acquired and analysed to measure the strain versus time. (b) Calibration of each device is performed by measuring the strain versus pressure curve. In the reported graph, 2 cycles measured in different regions of the membrane are shown (centre and membrane sides), indicating the uniformity of the strain over the whole membrane. (c) Creep response of the device over 30 minutes shows the stability of the strain response. When the membrane is not present, the strain is higher for the same applied pressure because of the decreased rigidity. A zoom-in of the curve shows the rising time needed before the strain reaches its maximum value. (d) Picture and schematic representing the known-elasticity thin membranes that are used as calibration samples instead of the cells. The sample can be modelled as two springs in series,42 one representing the bare membrane and the other one, the composite (in this case, the measurement membrane plus the known elasticity membrane). The measured values of the Young's modulus at 5% strain taken with our differential strain read-out are well within the experimental uncertainty and therefore validate our approach (measurement on 5 devices each) (s.b. = 2 mm). | ||
Device characterization
The devices are characterized to obtain pressure versus strain curves, which represent the calibration curves specific to every device. This curve is used to determine the pressure to apply to obtain a desired membrane strain. We experimentally confirmed that for the same applied pressure, the strain is uniform in different regions of the membrane (Fig. 2b). Although the device allows strains higher than 10% to be obtained, we used 5% strain for the Young's modulus measurements in order to minimize the possibility of inducing cell morphological modifications because of the applied strain.We also investigated the device creep response. The obtained results show a stable response of the device over at least 30 minutes (Fig. 2c). When the membrane is not present, the device has a very similar response with a higher strain for the same applied pressure, as expected. Because of the intrinsic viscoelasticity of PDMS, it is possible to observe that the strain does not instantaneously reach its maximum level after a change in pressure. A rising time of 3 seconds is needed before the strain reaches its maximum value. We validated our Young's modulus measurement approach by measuring samples of known elasticity in place of the cell layer (Fig. 2d). Previously characterized thin PDMS membranes are attached to the suspended membrane inside the device and their Young's modulus is measured through the differential strain read-out method using eqn (3). The measured Young's modulus has been compared to the one measured using a commercial pull-test device (Single Column Universal Testing System, 3340 from Instron). As shown in Fig. 2d, the results from the two measurement methods agree very well, within the experimental uncertainty, validating our differential approach for elasticity measurements of thin cell layers.
Results & discussion
Cell patterning and adhesion to the membrane
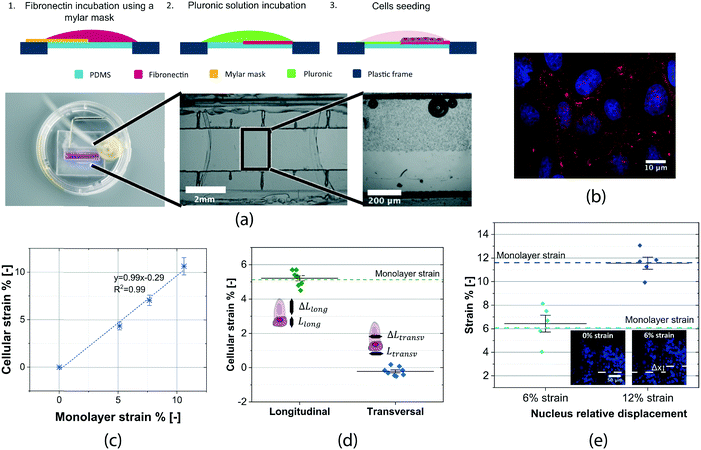
Two different cell lines have been measured using our device: sarcoma osteogenic cells (SaOS2) and kidney epithelial cells (MDCK II) (ATCC). In order to have cells only on a part of the membrane so that differential strains can be measured, we developed a patterning method. This protocol uses thin Mylar masks to achieve dual functionalization on the membrane surface (Fig. 3a). As a result, cell adhesion is confined to half of the membrane. Both functionalizations have been found to have a negligible effect on the membrane mechanical properties. As cell adhesion to the membrane is an essential requirement for our measurement, we performed several validation tests. Confocal images of vinculin protein (Fig. 3b) show that cellular focal adhesions are evenly distributed on the PDMS surface, implying that the whole cell monolayer is tightly anchored to the membrane. No cell detachment due to the applied deformation was observed thanks to the low amount of strain and its limited duration. In general, the cell conformation on our suspended membranes did not exhibit a major difference from standard polystyrene cell culture dishes (Fig. S7†). We investigated the relation between the monolayer and cellular strain. For this, membranes fully covered with cells have been used to relate the strains of the membrane and strain on a single cell within the monolayer. Cellular strain was measured by tracking the cell border using the DIC algorithm. Cell strain was tightly distributed around the value of the imposed engineering strain throughout the membrane. The linear correlation between the monolayer and cellular strain in the direction parallel to actuation confirms that the membrane strain is transferred to the cells (Fig. 3c). Also, we measured the cellular strain in the direction perpendicular to the actuation (Fig. 3d) and found that cell strain was tightly distributed around zero, ε = −0.22% ± 0.24%. These results indicate that the cells deform uniaxially with the membrane and that the perpendicular deformation can be neglected. The out-of-plane motion of the membrane during the actuation was minimal. Cells remained in focus during the entire loading cycle up to 20× magnification, meaning that the out-of-plane displacement was smaller than 5 μm (objective's depth of field).The presented device is compatible with live fluorescence imaging. A live cell fluorescent dye (NucBlue, Thermo Fisher Scientific) was used to stain cell DNA and track the nuclei displacement during the stretching. Fig. 3e shows images of the nuclei on the membrane at 0%, 6% and 12% strain. The images were captured at 20× magnification to have a wide field of view; a higher magnification may be used (long working distance objectives). By representing the nuclei strains at 6% and 12% membrane strains, we observed that they undergo the same deformation as the membrane, giving another validation that the cells and the membrane deform together as a composite.
Cell monolayer mechanical response measurements
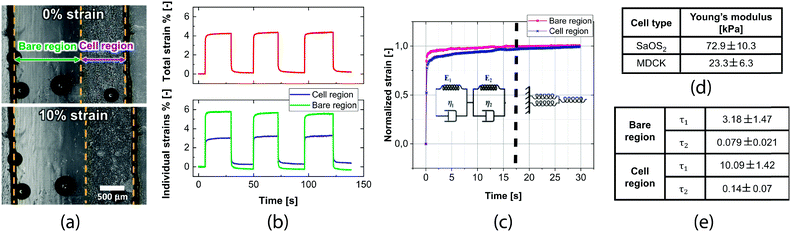
We performed measurements with cells to quantify their mechanical properties. The cells' presence on the membrane resulted in a lower strain of the cell covered region compared to the bare one because of the stiffening effect of cells. Fig. 4b shows that when the whole membrane strain is 4%, the individual strains are 5.5% for the bare region and 2.5% for the cell region. This result confirms that the membrane design allows measurement of the cell contribution to the mechanics of the composite. From the difference in strains, we calculate the Young's modulus of the cell layer using eqn (3). Cell thickness was measured from a z-stack over a large area of the cell monolayer (Fig. S1†).A resulting 8 μm thickness was measured from averaging the different vertical cuts. The measured Young's moduli for the two cell lines are 72.9 ± 10.3 kPa for SaOS2 and 23.3 ± 6.3 kPa for MDCK (Fig. 4d). The Young's modulus of the cell monolayer was 15 and 30 times higher for SaOS2 and MDCK respectively than the elasticity of single cells measured with AFM.40,41
The lower Young's modulus obtained with AFM can be explained by the fact that only a local point on the single cell surface is measured through compressive strain and in the transversal direction while with our method we consider the global response of cells as a population and in the in-plane direction. Our results indicate therefore the big effect of anisotropy on cell mechanics as well as the cruciality of cell–cell contacts in the overall mechanical properties.
In addition, we investigated the creep response of the cell monolayer. We observed an initial rising time followed by stabilization. By observing the normalized response of the cell and bare regions over the first 30 seconds after imposing the deformation, it is possible to clearly distinguish between the time constants of the cell region and the bare region (Fig. 4c). These curves can be fitted with a Kelvin–Voigt model, as typically done for viscoelastic material creep response modeling.32,43,44 We chose to use this model with two time-constants. The results, summarized in Fig. 4e, show a clear difference between the bare region and the cell region in terms of viscoelasticity. The cell region has both time constants higher than the bare region, in particular, τ1 is around three times higher and τ2 two times higher. The cell mechanics is therefore also measurable on our device in terms of their higher viscoelastic properties with respect to the substrate.
Measuring cell monolayer elasticity changes over time
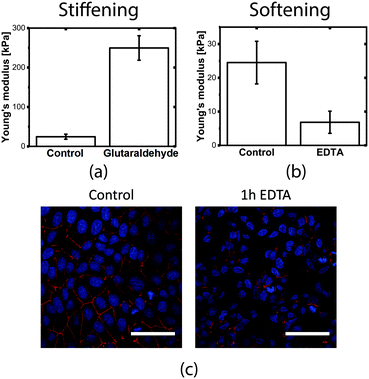
Cell elasticity is known to change at different stages of several diseases.32 Measuring these changes in elasticity is therefore of great interest because of the applications in diagnosis and drug screening.In order to measure the Young's modulus variation of the MDCK cell population, we used chemical treatments to induce known modifications in the cell layer structure. The experimental procedure consists of comparing the Young's modulus before and after treatment on the same device (control experiments under the same conditions but without cells show that the chemicals themselves do not affect the membrane mechanical properties, as shown in Fig. S6†). To simulate an increase in cell stiffness, cells were incubated with 4% glutaraldehyde, a chemical that is normally used to fix cells and is known for its stiffening effect without altering the cell shape morphology and internal structure.45 Cells treated with glutaraldehyde showed an increase in the Young's modulus of more than 10-fold, with a final value of 249.6 ± 31.0 kPa, which was easily measured by our device (Fig. 5a).
We then investigated the contribution of cellular junctions by treating the cell monolayer with ethylenediaminetetraacetic acid (EDTA), a chemical that disrupts cell–cell tight junctions. The choice of the EDTA treatment incubation time was made by comparing confocal images of the tight junctions at 0 minutes, 30 minutes and 1 hour (Fig. S5†).
After 1 hour of incubation, the junctions are almost completely disrupted in comparison to the control sample (Fig. 5c), however, the cells were still adhered to the substrate. The modification of the tight junctions led to a 4-fold decrease of the measured Young's modulus to 6.86 ± 3.27 kPa (Fig. 5b), indicating the importance of tight junctions in the mechanical stability of the monolayer.
Our device allows measurement of both an increase and decrease in cell elasticity due to chemical modification by targeting a known protein in the cell layer. The results obtained with EDTA and softening of the monolayer because of cell junction rupture prove the importance of cell–cell proteins to the mechanical stability of cell layers. This further highlights the relevance of measuring cells within a higher structural level than single cells.
Materials and methods
Device fabrication and assembly
Sylgard 186 (Dow Corning®) was mixed in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 ratio and cured inside custom Teflon moulds to reproduce the bottom and top parts of the device. Suspended membranes of 2 mm × 6 mm, made of biomedical grade silicone MED4086 (Nusil™), were fabricated using a previously published casting method.24 The membranes were pre-stretched (λ = 1.5) to avoid the loss of mechanical tension during the actuation; their thickness and Young's modulus were measured through a custom-made interferometer and pull-test set-up respectively. Double-sided tape and silicone glue (RTV, room-temperature-vulcanizing silicone, E43 from Elastosil) were used to assemble and seal the device. Glass slides were glued to both the bottom and top parts of the device for stiffening purposes and thus avoid their deformation.
10 ratio and cured inside custom Teflon moulds to reproduce the bottom and top parts of the device. Suspended membranes of 2 mm × 6 mm, made of biomedical grade silicone MED4086 (Nusil™), were fabricated using a previously published casting method.24 The membranes were pre-stretched (λ = 1.5) to avoid the loss of mechanical tension during the actuation; their thickness and Young's modulus were measured through a custom-made interferometer and pull-test set-up respectively. Double-sided tape and silicone glue (RTV, room-temperature-vulcanizing silicone, E43 from Elastosil) were used to assemble and seal the device. Glass slides were glued to both the bottom and top parts of the device for stiffening purposes and thus avoid their deformation.
Device measurement procedure
All measurements are performed in liquid and inside the incubator after a 30 minute conditioning time to let the device and liquid adapt to the temperature and humidity. A first conditioning loading cycle is performed to let the sample adapt to the mechanical deformation. The cells are then measured by deforming them through three consecutive pressure cycles. The applied pressure is chosen after a sample calibration to reach a membrane strain of around 5%. Each cycle consists of a pressure step followed by 30 seconds of constant pressure and then by another step to return to the initial position. The Young's moduli obtained at constant strain over the 3 cycles are then averaged. Pictures are continuously taken during the cycles at 10 frames per s.Cell culture on device
A fibronectin solution (50 mg ml−1 in HEPES 10 mM) was incubated on the membrane using a Mylar mask (12 μm thick, from DuPont) for 1 hour. The mask was carefully placed on the membrane and peeled off after the fibronectin incubation without causing any damage to the suspended PDMS. The surface was blocked with a 4% Pluronic solution in DI water for 1 hour. Cells were seeded on the membrane in a concentration of 250![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per ml and left to adhere for 2 hours. The membrane surface was then washed to remove non-adherent cells in the area functionalized with Pluronic. The cell culture chamber of the device was filled with medium and left to rest for at least 5 hours in a biological incubator.
000 cells per ml and left to adhere for 2 hours. The membrane surface was then washed to remove non-adherent cells in the area functionalized with Pluronic. The cell culture chamber of the device was filled with medium and left to rest for at least 5 hours in a biological incubator.
Cell elasticity modification protocols
Cell stiffening was achieved by 4 minutes of cell incubation with 4% glutaraldehyde followed by three washes in PBS. The removal of tight junction proteins was accomplished by incubating with 3 mM EDTA in serum-free cell medium for 1 hour.Conclusion
We presented a miniaturized cell stretcher device able to determine Young's modulus changes on adherent cell monolayers. By designing the cell substrate with similar mechanical properties to the ones expected from the cell layer, we can decouple the mechanical responses of the two. This allows measurements on cells that are not only mechanically coupled among each other but also with a substrate, thus recreating similar conditions to those inside the body. By performing a differential strain read-out between the bare and cell-covered substrate regions, we can have a direct measure of both the force exerted to the substrate and the consequent deformation without the need of a bulky set-up. This measurement method is implemented within a compact fluidic device using pneumatic actuation to deform the substrate and cells. Our transparent device can be mounted on an inverted microscope for real-time monitoring of the sample. Also, the device can fit standard Petri-dishes thus allowing measurements inside the incubator in a sterile and physiological environment. We validate the device performance on two confluent cell monolayers, SaOS2 and MDCK, by stretching them between 0% and 5% strain at 0.025 Hz while acquiring pictures at 10 frames per s. The deformations of the two regions were measured by digital image correlation analysis of the pictures. Results showed a 50% higher strain in the bare region compared to the region covered with cells when subjected to the same force, thus demonstrating the capability to decouple the mechanical properties of cells from the substrate ones. We demonstrate accurate Young's modulus measurements within 3 kPa and 300 kPa; this range is mainly limited by the optical resolution when the sample is very soft while it has no limitation for stiffer samples.The resulting cell monolayer Young's modulus is around one order of magnitude bigger compared to single cell measurements, which can be explained by the difference in cytoskeletal organization between single cells and cells within a tissue as well as their anisotropy between the in-plane and out-of-plane responses. The possibility to measure the in-plane cell mechanics contribution when they are attached to a substrate is a great advantage of this technology as it makes this measurement principle highly versatile and adaptable to all type of adherent cells. Also, as cells are kept under physiological conditions, our device opens the door for long time scale monitoring of cell mechanics which was not feasible before. We envision this technology to be easily scalable to multi-well plates for higher throughput experiments relying on coupled pneumatic actuation and optical read-out through automatized scanning microscopes. Alternatively, electrical detection could be implemented for a high throughput acquisition embedding strain sensors46 in the membrane. Thanks to its miniaturization, versatility and applicability to most of adherent cell types, our approach paves the way for quantitative measurements of cell population elasticity and its changes over time.
Author contributions
F. S., H. S. and C. M. conceptualized the study and designed the experiments. F. S. designed and fabricated the devices. A. P. participated in the device design and helped with the pattern recognition algorithm. R. I. contributed to the experimental set-up development. F. S. cultured the cells, performed the cell measurements and analysed the data. F. S., A. P., H. S, and C. M. wrote and edited the manuscript. All the authors have approved the final version of the manuscript.Conflicts of interest
The authors have no conflicts to declare.Acknowledgements
The authors thank Jasmine Kernen (CSEM) for her generous help in setting the protocols for cell fluorescence imaging and her knowledge in cell culture techniques. We acknowledge the help of Michel Despont (CSEM) in the device design and fabrication. We acknowledge the support from the BIOP staff in EPFL for confocal imaging and image analysis. This work was supported by the Swiss National Science Foundation (SNF) grant number 205321_153365, and partially by the CSEM and EPFL.References
- J. M. Barnes, L. Przybyla and V. M. Weaver, J. Cell Sci., 2017, 130, 71–82 CrossRef CAS PubMed.
- C. Guillot and T. Lecuit, Science, 2013, 340(6137), 1185–1189 CrossRef CAS PubMed.
- D. A. Fletcher and R. D. Mullins, Nature, 2010, 463, 485–492 CrossRef CAS PubMed.
- A. Llucia-Valldeperas, R. Bragos, C. Soler-Botija, S. Roura, C. Galvez-Monton, C. Prat-Vidal, I. Perea-Gil and A. Bayes-Genis, Sci. Rep., 2018, 8, 499 CrossRef PubMed.
- A. R. Carvalho and W. A. Zin, Biophys. Rev., 2011, 3, 71 CrossRef PubMed.
- T. J. Roberts, J. Exp. Biol., 2016, 219, 266–275 CrossRef PubMed.
- V. M. Weaver, Mol. Biol. Cell, 2017, 28, 1815–1818 CrossRef CAS PubMed.
- J. A. Park, L. Atia, J. A. Mitchel, J. J. Fredberg and J. P. Butler, J. Cell Sci., 2016, 129, 3375–3383 CrossRef CAS PubMed.
- D. E. Jaalouk and J. Lammerding, Nat. Rev. Mol. Cell Biol., 2009, 10, 63–73 CrossRef CAS PubMed.
- J. Guck and E. R. Chilvers, Sci. Transl. Med., 2013, 5(212), 212fs41 CrossRef PubMed.
- G. Weder, M. C. Hendriks-Balk, R. Smajda, D. Rimoldi, M. Liley, H. Heinzelmann, A. Meister and A. Mariotti, Nanomedicine, 2014, 10, 141–148 CrossRef CAS PubMed.
- M. A. Lizarralde Iragorri, S. El Hoss, V. Brousse, S. D. Lefevre, M. Dussiot, T. Xu, A. R. Ferreira, Y. Lamarre, A. C. Silva Pinto, S. Kashima, C. Lapoumeroulie, D. T. Covas, C. Le Van Kim, Y. Colin, J. Elion, O. Francais, B. Le Pioufle and W. El Nemer, Lab Chip, 2018, 18, 2975–2984 RSC.
- K. R. M, S. Bhattacharya, S. DasGupta and S. Chakraborty, Lab Chip, 2018, 18, 3939–3948 RSC.
- L. Wang, J. Zhu, C. Deng, W. L. Xing and J. Cheng, Lab Chip, 2008, 8, 872–878 RSC.
- M. Rothbauer, H. Zirath and P. Ertl, Lab Chip, 2018, 18, 249–270 RSC.
- F. Guo, J. B. French, P. Li, H. Zhao, C. Y. Chan, J. R. Fick, S. J. Benkovic and T. J. Huang, Lab Chip, 2013, 13, 3152–3162 RSC.
- M. Merkel and M. L. Manning, Semin. Cell Dev. Biol., 2017, 67, 161–169 CrossRef PubMed.
- B. Ladoux and R. M. Mege, Nat. Rev. Mol. Cell Biol., 2017, 18, 743–757 CrossRef CAS PubMed.
- T. E. Angelini, E. Hannezo, X. Trepat, M. Marquez, J. J. Fredberg and D. A. Weitz, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 4714–4719 CrossRef CAS PubMed.
- D. Eder, C. Aegerter and K. Basler, Mech. Dev., 2017, 144, 53–61 CrossRef CAS PubMed.
- M. I. Gary Wilk, P. E. Fuller, K. Kandere-Grzybowska and B. A. Grzybowski, Phys. Rev. Lett., 2014, 112(13), 138104 CrossRef PubMed.
- O. T. Guenat and F. Berthiaume, Biomicrofluidics, 2018, 12(4), 042207 CrossRef PubMed.
- A. Poulin, C. Saygili Demir, S. Rosset, T. V. Petrova and H. Shea, Lab Chip, 2016, 16, 3788–3794 RSC.
- H. Kamble, M. J. Barton, M. Jun, S. Park and N. T. Nguyen, Lab Chip, 2016, 16, 3193–3203 RSC.
- D. Tremblay, S. Chagnon-Lessard, M. Mirzaei, A. E. Pelling and M. Godin, Biotechnol. Lett., 2014, 36, 657–665 CrossRef CAS PubMed.
- C. S. Simmons, J. Y. Sim, P. Baechtold, A. Gonzalez, C. Chung, N. Borghi and B. L. Pruitt, J. Micromech. Microeng., 2011, 21, 54016–54025 CrossRef CAS PubMed.
- H. Kamble, R. Vadivelu, M. Barton, M. J. A. Shiddiky and N.-T. Nguyen, Lab Chip, 2018, 18(5), 765–774 RSC.
- J. Wang, B. Fan, Y. Wei, X. Suo and Y. Ding, Lab Chip, 2016, 16, 360–367 RSC.
- H. Parsa, B. Z. Wang and G. Vunjak-Novakovic, Lab Chip, 2017, 17, 3264–3271 RSC.
- A. R. Harris, L. Peter, J. Bellis, B. Baum, A. J. Kabla and G. T. Charras, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, 16449–16454 CrossRef CAS PubMed.
- E. Kang, J. Ryoo, G. S. Jeong, Y. Y. Choi, S. M. Jeong, J. Ju, S. Chung, S. Takayama and S. H. Lee, Adv. Mater., 2013, 25, 2167–2173 CrossRef CAS PubMed.
- N. D. Evans, R. O. Oreffo, E. Healy, P. J. Thurner and Y. H. Man, J. Mech. Behav. Biomed. Mater., 2013, 28, 397–409 CrossRef PubMed.
- N. R. Chevalier, E. Gazguez, S. Dufour and V. Fleury, Methods, 2016, 94, 120–128 CrossRef CAS PubMed.
- G. Bartalena, Y. Loosli, T. Zambelli and J. G. Snedeker, Soft Matter, 2012, 8, 673–681 RSC.
- J. C. Selby and M. A. Shannon, Rev. Sci. Instrum., 2007, 78, 094301 CrossRef PubMed.
- D. Lee, A. Erickson, T. You, A. T. Dudley and S. Ryu, Lab Chip, 2018, 18(14), 2077–2086 RSC.
- C. Gullekson, M. Walker, J. L. Harden and A. E. Pelling, Mol. Biol. Cell, 2017, 28, 111–119 CrossRef CAS PubMed.
- D. E. Backman, B. L. LeSavage and J. Y. Wong, J. Biomech., 2017, 51, 118–122 CrossRef PubMed.
- S. Rosset, O. A. Araromi, S. Schlatter and H. R. Shea, J. Visualized Exp., 2016, e53423, DOI:10.3791/53423.
- H. Zoellner, N. Paknejad, K. Manova and M. A. Moore, Histochem. Cell Biol., 2015, 144, 533–542 CrossRef CAS PubMed.
- B. R. Bruckner, H. Noding and A. Janshoff, Biophys. J., 2017, 112, 724–735 CrossRef PubMed.
- C. T. Lim, E. H. Zhou and S. T. Quek, J. Biomech., 2006, 39, 195–216 CrossRef CAS PubMed.
- A. Raj and A. K. Sen, RSC Adv., 2018, 8, 20884–20893 RSC.
- G. Tomaiuolo, M. Barra, V. Preziosi, A. Cassinese, B. Rotoli and S. Guido, Lab Chip, 2011, 11, 449–454 RSC.
- D. A. Schreier, O. Forouzan, T. A. Hacker, J. Sheehan and N. Chesler, J. Biomech. Eng., 2016, 138, 021012 CrossRef PubMed.
- F. Sorba and C. Martin-Olmos, Microelectron. Eng., 2018, 191, 38–41 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9lc00075e |
| This journal is © The Royal Society of Chemistry 2019 |