Open Access Article
Open Access ArticleDual-phone illumination-imaging system for high resolution and large field of view multi-modal microscopy
Sara
Kheireddine
 a,
Ayyappasamy
Sudalaiyadum Perumal
a,
Ayyappasamy
Sudalaiyadum Perumal
 a,
Zachary J.
Smith
a,
Zachary J.
Smith
 b,
Dan V.
Nicolau
b,
Dan V.
Nicolau
 *a and
Sebastian
Wachsmann-Hogiu
*a and
Sebastian
Wachsmann-Hogiu
 *ac
*ac
aDepartment of Bioengineering, McGill University, Montreal, Quebec H3A 0E9, Canada. E-mail: dan.nicolau@mcgill.ca; sebastian.wachsmannhogiu@mcgill.ca
bDepartment of Precision Machinery and Precision Instrumentation, University of Science and Technology of China, Hefei, China
cDepartment of Pathology and Laboratory Medicine, University of California Davis, Davis, CA, USA
First published on 30th January 2019
Abstract
In this paper we present for the first time a system comprised of two mobile phones, one for illumination and the other for microscopy, as a portable, user-friendly, and cost-effective microscopy platform for a wide range of applications. Versatile and adaptive illumination is made with a Retina display of an Apple mobile phone device. The phone screen is used to project various illumination patterns onto the specimen being imaged, each corresponding to a different illumination mode, such as bright-field, dark-field, point illumination, Rheinberg illumination, and fluorescence microscopy. The second phone (a Nokia phone) is modified to record microscopic images about the sample. This imaging platform provides a high spatial resolution of at least 2 μm, a large field-of-view of 3.6 × 2.7 mm, and a working distance of 0.6 mm. We demonstrate the performance of this platform for the visualization of microorganisms within microfluidic devices to gather qualitative and quantitative information regarding microorganism morphology, dimension, count, and velocity/trajectories in the x–y plane.
1. Introduction
There is a compelling need for an imaging system that can provide simultaneously high spatial resolution and large field of view (FOV) at a moderate cost. This need manifests itself in biomedical application areas such as reading well plates for colorimetric assays or cell cultures, where high spatial resolution is required for observing cell morphology and subcellular structures. In addition, a large FOV is required to survey large sample areas for high throughput screening,1 histopathology,2 embryology, where phenotypic screening in transgenic mice is performed by monitoring of embryo organogenesis,3 and developmental biology.4 Moreover, in neuroscience, brain sectioning is used as a means to identify specific brain networks involved in behaviours or functions via reconstruction of neural wiring diagrams,5 which also requires a large FOV for accurate reconstruction and high spatial resolution for precise identification of these paths.Lens-based imaging approaches are faced with a trade-off between resolution and field-of-view (FOV). Generally, the greater the resolvable detail in a sample, the smaller the FOV we can observe. To delineate this, in a totally incoherent imaging system (such as in fluorescence microscopy), a 100× microscope objective lens with numerical aperture (NA) of 1.4 offers a theoretical resolution of 0.25 μm, with a FOV diameter of ≈60 μm, whereas a typical 2× objective with NA of 0.05 provides a theoretical resolution of 6 μm, with a FOV diameter of 1.5 cm. While low magnification, large NA objectives are available, they typically have large costs and may lead to pixel-limited rather than optically limited performance due to their high space-bandwidth products. Furthermore, beyond fluorescence imaging, in a partially coherent system such as in bright-field or phase imaging, both the NA of the objective and the NA of the condenser need to be taken into consideration. The resolving power of the microscope in this case is determined by:
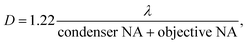 | (1) |
Due to this imposed trade-off between resolution and field of view, at high resolutions, the sample would have to be imaged in sections and then the image mosaic-assembled via stitching techniques. Image stitching or mosaicking includes two steps: image registration and image merging, with the latter adjusting for non-uniform brightness within the images and blending the images in a way that ensures uniform transitions from one tile to the next.7 Merging the individual partitions into a large FOV in this manner, however, takes a long time and often results in a checkboard-like pattern in the final mosaic because of differences in illumination in adjacent images, with the inhomogeneities being most apparent at the edges of the stitched images, as well as image focus errors within parts of the sample having various depths, as is common in biological samples.3 While low-cost three-dimensional microscope systems have been reported, their increased complexity presents a barrier to adoption.8 Flatbed scanners have also been used to achieve ultra-widefield imaging at high resolution,9–15 but often require extensive post-purchase modification, and have slow scanning speeds. This is particularly relevant for applications that require recording image sequences or real-time tracking where partitioning the FOV would lead to loss of valuable data.
On the other hand, lens-free approaches allow for better resolution and larger FOV than lens-based ones since they can make use of the full potential of the image sensor, without being limited by lens specifications. As such, using an image sensor with a large area and a small pixel size is crucial for large FOV and high resolution, respectively. Lens-free microscopy also offers cost-effectiveness, portability, and depth-resolved three-dimensional (3D) imaging.16 This approach is particularly useful for applications that require heavy statistical analysis. Moreover, the lens-free system design can also be adapted for a number of applications, such as fluorescence imaging,17 and imaging S. pombe yeast cells18 and C. elegans19 within microfluidic devices. However, they require extensive data post-processing and complex reconstruction methods that may present a barrier for non-experts.
Cameras of mobile phones, meanwhile, create a lens-based platform that is more than the sum of its parts – a standalone imaging system capable of data transmission, which in addition to being portable, makes it invaluable specifically in resource-limited settings. Consequently, mobile phone based imaging has been applied in areas related to health, the environment, and education.20 In terms of biomedical applications, mobile phone imaging has been used as tool for partial blood counts,21 complete blood counts,22 fluorescence detection of Giardia lamblia water-borne protozoan parasites,23 and soil-transmitted helminths (Ascaris lumbricoides, Trichuris trichiura, and hookworm) in stool samples,24 as well as diagnosis of malaria,25 sickle cell anaemia,1 and dermatological disease.26 As for environmental applications, a mobile phone based system was developed for imaging a plasmonic colorimetric assay that performs mercury(II) detection in water samples.27 Phone based systems have also been used for imaging microfluidic devices, such as for detection of Salmonella,28 and imaging microfluidic-based enzyme-linked immunosorbent assay (ELISA) used for the detection of a food contaminant.29 Thus, mobile-phone microscopy might be seen as an intermediate point between traditional microscopy and lens-free methods. However, mobile phone microscopy has traditionally used simple LED illumination, generally optimized for a single imaging type, such as fluorescence, bright field, or dark field.
Illumination is often one of the most critical determinants of image quality in microscopy, and may need to be dynamically adapted based on the requirements of the sample. Unstained mammalian cells, for instance, can be more clearly distinguished using dark-field (DF) or phase-contrast (PC) illumination, rather than using bright-field (BF) illumination. Other samples may find certain wavelengths phototoxic, or require narrow-band excitation to provide greater contrast. In general, such illumination modes require additional microscopy hardware such as condenser annuli in multiple sizes to accommodate objectives with different numerical apertures (NAs), and specialized PC objectives, which makes such setups relatively restrictive in how adaptive they can be. To overcome this, programmable light-emitting diode (LED) arrays have been used as adaptive illumination sources that offer more imaging flexibility.30 These arrays have also been adapted to allow for real-time acquisition of image sequences using time-multiplexed LED illumination.31 Moreover, specific modifications of the LED illumination allows for extracting quantitative phase information via differential phase contrast (DPC).32
In this paper, we demonstrate for the first time that two mobile phones can be combined to create a simple microscopy platform with high resolution, large field-of-view, and dynamically adaptive illumination. For our imaging system, we used the Retina display of an Apple device (iPhone 6) as a versatile illumination source. The screen, which is composed of high density RGB LEDs, is used to project various illumination patterns onto the specimen being imaged, each corresponding to a different illumination mode, with a second phone (a Nokia Lumia 1020) capturing the resulting image. The patterns include BF, DF, point illumination (PI), Rheinberg illumination (RI), and fluorescence microscopy. Not only does this illumination setup allow for design and projection of a variety of patterns with ease, but it permits creativity in terms of production and testing of novel illumination patterns or structures, and merging multiple illumination modes together, and is overall simpler to control and use than an LED array.
We use a Nokia Lumia 1020 mobile phone as a portable, user-friendly, and cost-effective imaging platform with high spatial resolution and large FOV. As such, our system presents great potential to cater for a wide range of applications. We show that, in contrast with traditional microscopy, for an objective lens with a comparable FOV, the Nokia phone offers better resolution. Moreover, the system can capture standard (720p) or full (1080p) high-definition (HD) image sequences (videos) at rates of 24, 25, and 30 frames per second (fps), in addition to images at long exposure times of up to 4 seconds.
2. Results and discussion
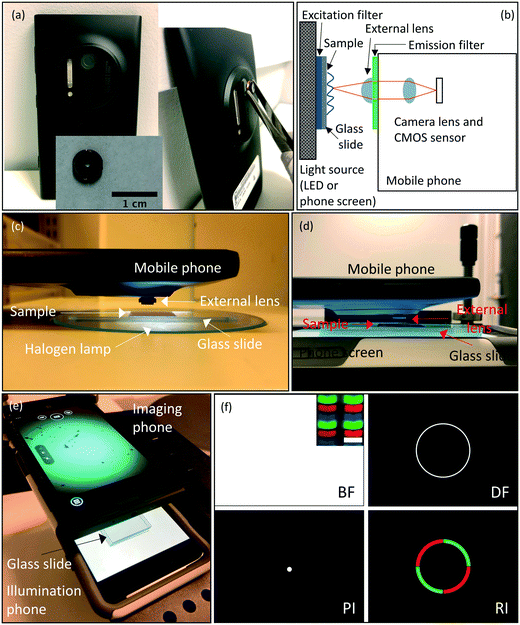
2.1 Description of the imaging platform
To build an imaging system that is capable of high spatial resolution and a large FOV, we used the Nokia Lumia 1020, which has a 41.3-megapixel (MP) image sensor with a pixel size of 1.12 μm, and attached an external lens (iPhone 5 lens: NA = 0.23, f2.2) for an overall 2× magnification and a theoretical spatial resolution of 1.3 μm. We assembled this system to image biological specimens in free space or confined in microfluidic devices. For imaging, we place the sample in between the Nokia-lens system (Fig. 1(a)) and the illumination source, as shown in the sketch of the optical setup (Fig. 1(b)). Fig. 1(c) shows the imaging system in use with halogen lamp illumination, and Fig. 1(d) and (e) show the imaging system in use with Retina display illumination.While the imaging phone-lens system is held in place by a clamp, the sample along with the illumination phone are able to be moved in 3D via a translation stage. More specifically, to image microfluidic devices, we place them onto a glass slide, which in turn is placed directly onto the screen of the illumination phone. For imaging thin samples such as diatoms, E. gracilis, or human epithelial cheek cells (HECC), we placed the specimen on a glass slide underneath a cover slip, we need to add three extra glass slides between the slide carrying the sample and the illumination phone screen to provide the necessary distance such that the screen is not in the focus of the imaging system.
2.2 Description of the illumination system
Microsoft PowerPoint was used to generate various illumination patterns to be displayed on an Apple Retina display as shown in Fig. 1(f). For the BF imaging mode, we project a white screen, at the highest screen brightness, where the sample receives uniform illumination throughout. In addition, we designed patterns for PI, DF, and RI. PI was achieved by creating a very small, white full circle on a black background. For DF illumination, on the other hand, we drew a ring of a size just outside the NA of the imaging lens. We created single (DF-single), double (DF-double), and triple (DF-triple) ring illumination patterns by drawing concentric white rings on a black background.RI is similar to DF, but whereas DF relies on a ring of white light contrasted against a dark specimen background, the RI utilizes a ring of colored light contrasted against a specimen background of a different color. These contrasting colors are specimen-dependent, and are typically chosen to maximally enhance the specimen image without the need for specimen staining. RI also includes rings that have multiple colors (two-sector or four-sector patterns) against a dark specimen background. We designed RI patterns where we used colored rings, and colored the area inside the ring as well. We also sectioned rings into halves or quarters to make two- or four-sector patterns.
Retina display illumination allows for ease of pattern design without any limitations in terms of the shape, size, or color. Since multiple shapes can be created with PowerPoint, virtually endless patterns can be generated and tested with different samples. This makes our illumination platform more versatile than an LED array. Moreover, this platform is just as portable as our imaging system and thus this combined setup can be tested in various settings outside the laboratory, which makes it particularly useful in low-resource environments.
Whereas the patterns we used in screen illumination were mostly classic illumination patterns, this platform provides the option for simple experimentation with a variety of creative patterns, as well as combinations of multiple patterns. For example, Zheng et al. recently developed a method of obtaining large FOV, high-resolution intensity and phase images from stained and unstained samples, using a computation method dubbed Fourier ptychography microscopy (FPM).33–36 Critically, the illumination must come from sets of LEDs turned on and off by an LED array. The method could be easily adapted to the dynamic illuminator described here, substantially simplifying implementation of FPM for applications in low-resource settings.
Similarly, Tian and Waller recently described the use of an LED array to create a quantitative phase imaging system with rather high image quality.37 Again, by replacing the LED array requiring an external microcontroller with a simple phone screen that can be controlled through an app, the implementation of their method in low-resource settings where trained experts may be lacking could be a substantial benefit.
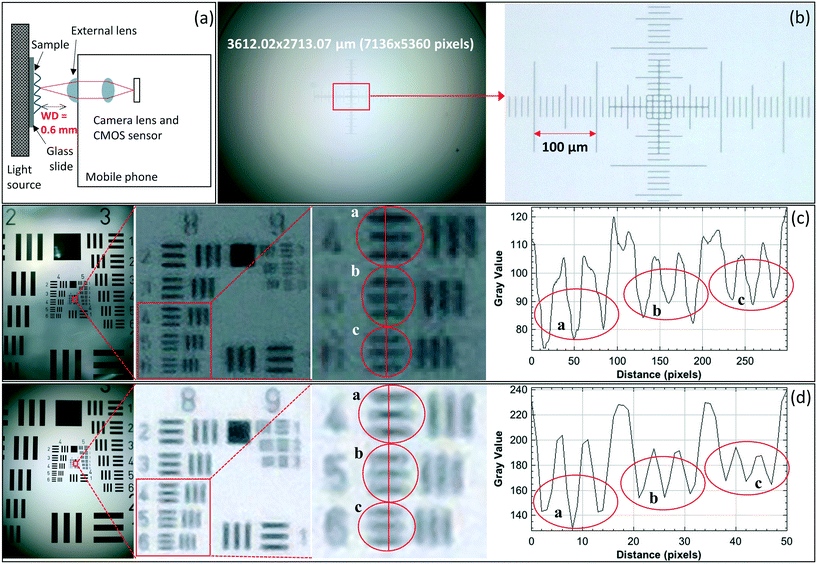
2.3 System characterization for spatial resolution, field-of-view (FOV) and working distance (WD)
We determined the spatial resolution of the Nokia phone-lens system under different illumination conditions by mounting the external lens onto the Nokia phone camera, and imaging a 1951 USAF resolution test chart, once using halogen lamp illumination (Fig. 2(c)), and another time using the Retina display illumination (Fig. 2(d)). The Nokia-lens system was held in place by a clamp, and we placed the resolution chart, along with the illumination phone, if applicable, onto a translation stage. We then moved the stage until the line pairs on the chart were in focus, and took the images.Fig. 2 also shows a plot of intensity profiles of the line pairs in the resulting images generated by using the Fiji (ImageJ) software. We observed that our system is capable of spatially resolving 406.4 line pairs per millimeter (lp per mm) (group 8, element 5) with halogen lamp illumination (Fig. 2(c)), and 362.0 lp per mm (group 8, element 4) with Retina display illumination (Fig. 2(d)); therefore, both illumination approaches provide us with a comparable spatial resolution of below 2 μm. If the spatial resolution were assessed qualitatively, however, up to element 1 of group 9 can be resolved by eye in both cases, which corresponds to a spatial resolution of 512.0 lp per mm. Moreover, we used a stage micrometer and Fiji (ImageJ) software to measure the FOV, as shown in Fig. 2(b), and determined it to be ≈3.6 × 2.7 mm. Furthermore, we characterized the WD to be 0.6 mm in air (Fig. 2(a)).
Given the low magnification in our system (2×), the system requires small pixels in order to Nyquist sample the optical point spread function. Therefore, to achieve high resolution with large FOV, a system with a large number of small pixels is required. In terms of system hardware, the image sensor of the Nokia Lumia 1020 is currently one of the largest on the market (41.3 MP), that in combination with a 1.12 μm pixel size provides both high spatial resolution and large FOV.
2.4 Application to imaging of microorganisms in microfluidic devices
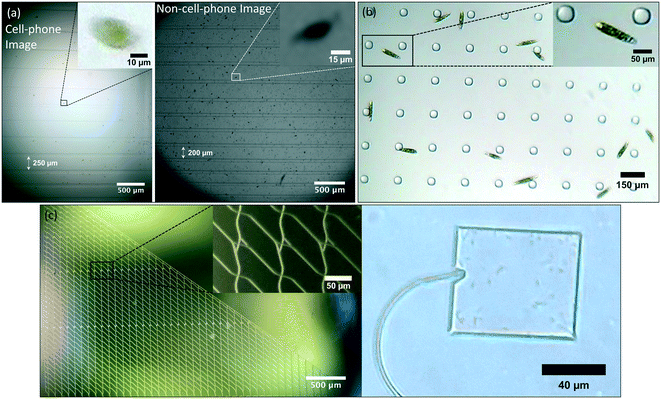
As a potential application, we imaged multiple microorganisms in various microfluidic structures under different illumination conditions. We chose to image two species: E. gracilis and E. coli. Since E. gracilis have relatively large physical dimensions (≈15 μm width), this makes them a good sample for testing the image quality of the microscopy system. Moreover, because these microorganisms are pigmented, and their bodies scatter light well, they serve as an ideal sample for use in testing the DF and RI capabilities of the imaging system. On the other hand, in the case of E. coli, because of their small physical dimensions (≈0.5 μm width), they are a good candidate for demonstrating the high spatial resolution of the system.These organisms are studied in complex environments to learn about their motility and behavior. In this article, we show that our illumination-imaging system can be used to observe them in confined spaces, such as channels or plazas of microfluidic devices, to provide valuable data about their behavior. Depending on the device design, different aspects of the microorganism can be studied. For instance, we used devices with straight channels of various widths ranging from 50 to 450 μm that provide information about the movement trajectory of microorganisms. We also used for our experiments microfluidic devices that have micro-fabricated pillars (50 μm pillar diameter), which are typically used for observing traffic and group motility behavior in microorganisms. Moreover, we imaged a microfluidic device with 2 μm-wide channels that was previously designed for parallel exploration by biological agents.38,39 Finally, wide areas with borders, such as plazas, provide an idea of the affinity of microorganisms for walls or corners, and can also be used to perform particle counts and dimension measurements.
Fig. 3 shows microscopic images of these microorganisms in the microfluidic devices described before. In order to demonstrate the utility of the illumination with the Retina display, we show images of E. gracilis and E. coli with Retina display illumination (Fig. 3(a) – left) and halogen lamp illumination (Fig. 3(b) and (c)). We also compared images of E. gracilis in a microfluidic device with multi-sized channels taken with the phone imaging system (Fig. 3(a) – left) and with an Olympus IX83 microscope (4× magnification with NA = 0.16) (Fig. 3(a) – right). The results suggest that for a comparable FOV, the phone system demonstrates superior image quality compared with the traditional microscope (Fig. 3(a) – insets). We also took images of E. gracilis in a microfluidic device with pillars and were able to observe chloroplasts in individual E. gracilis (Fig. 3(b)). Moreover, we imaged E. coli within a microfluidic device. Our system was able to resolve intricate details within the device junctions (Fig. 3(c) – left), as well as individual E. coli in the plaza areas (Fig. 3(c) – right).
2.5 Application to imaging cells with different illumination patterns
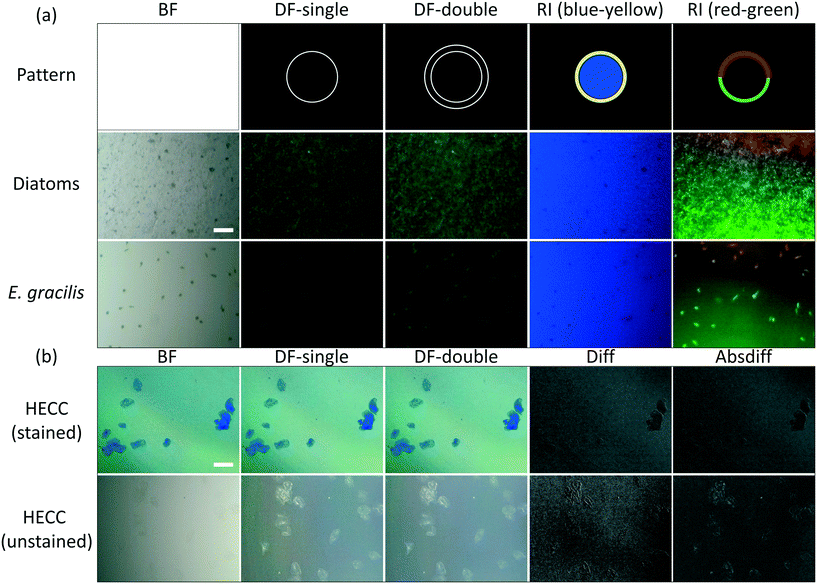
In the previous section we presented images of microorganisms in microfluidic devices with a Retina display illumination in BF mode and a Nokia phone microscope. In order to observe the range of capabilities of Retina display illumination, we used our microscopy-illumination system with different types of cells, and imaged them under various illumination conditions. We imaged thin samples of diatoms, E. gracilis, and human epithelial cheek cells (HECC) under a cover slip. Diatoms were tested in powder form because they form highly variable and heterogeneous samples. HECC are particularly interesting because, as translucent mammalian cells, they are difficult to observe in BF mode. We used illumination patterns such as white screen (BF), single-, double-, and triple-rings (DF), and colored rings encircling differently colored backgrounds (RI), as well as two-sector or four-sector circles with multi-colored sectors (RI). These contrasting colors in RI are chosen depending on the specimen being imaged to enhance the specimen-background contrast. The resulting images are shown in Fig. 4.Fig. 4(a) shows digitally zoomed and cropped images of diatoms and E. gracilis under BF, DF and RI illumination patterns (Fig. 4(a), second and third rows). The illumination patterns as projected on the Retina display are shown in the top row of the figure. We used single dark-field (DF-single), double dark-field (DF-double) ring illumination patterns (second and third columns). Moreover, we prepared different forms of RI: yellow ring with blue background (fourth column), and red-green half-rings pattern against a dark specimen background (fifth column). Fig. 4(b) shows digitally zoomed and cropped images of stained and unstained HECC under BF and DF illumination patterns. Images formed with single dark-field (DF-single) ring illumination pattern (Fig. 4(b), second column) were subtracted from images formed with double dark-field (DF-double) ring illumination pattern (Fig. 4(b), third column) in MATLAB using regular arithmetic difference (Diff) or absolute difference (Absdiff), which resulted in an effect similar to that of image embossing (Fig. 4(b), fourth and fifth columns).
Our choice of imaging samples was intended to test the extent of our system capabilities in terms of spatial resolution and contrast enhancement. In the case of E. gracilis, a qualitative assessment of the images shows that the microorganisms are easier to observe with the green pattern compared to the blue pattern. This can be attributed to the presence of chloroplasts in E. gracilis, which are green in color, and therefore reflect green light well. Moreover, with diatoms, the blue background highlighted the intact diatoms within the diatom powder mixture. Additionally, for HECC, DF images provided much better contrast than BF images, and the embossing-like effect that we achieved with subtraction of double- and single-ring DF images further enhances visual cell detection. These results verify the adaptability of our imaging-illumination system to different types of specimens with varying optical properties.
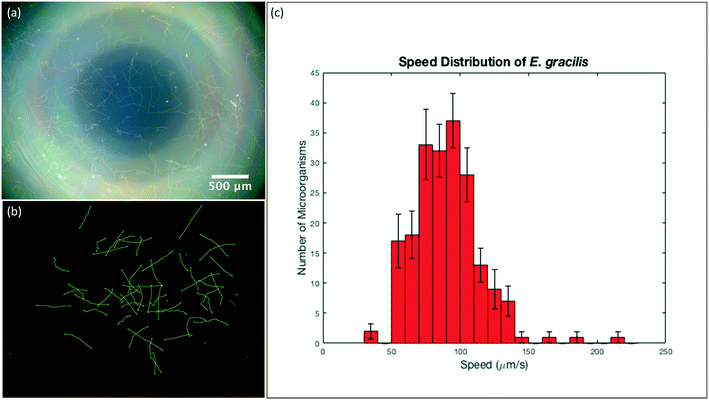
2.6 Application to quantitative analysis of microorganism motility
The still images we showed previously can provide us with qualitative information regarding cell/microorganism morphology, and quantitative information such as cell/microorganism dimensions and count. However, when it comes to calculating velocities of microorganisms, image sequences (videos) would have to be taken instead, and particle tracking would have to be done between successive video frames. Particle tracking is typically time consuming and can be particularly difficult in areas where there is a large number of microorganisms with overlapping trajectories. In order to overcome this, we took still images of motile specimens with long exposure times to help us determine their velocities and trajectories in the x–y plane. In a long-exposure image, the trajectory that a microorganism follows is highlighted over a predetermined period of time.First, we took a DF image with the maximum exposure time of 4 seconds of the motile E. gracilis sample, and then converted it to 8-bit in Fiji (ImageJ) and performed background correction to better highlight the trajectories. Using the Simple Neurite Tracer plug-in40 in the Fiji (ImageJ) software, we traced the microorganism trajectories semi-automatically, where we specified various beginning and end points for each trajectory, and the plug-in would then predict the overall trajectory from these points. After tracing several hundred trajectories of E. gracilis, we exported the trace length values we got in pixels, converted these values to micrometers using the predetermined scale of our imaging system, and calculated the respective speeds by taking the exposure time used into account. We then plotted a histogram with the speed distributions of the microorganisms, with the error bars indicating the standard deviation from the mean trajectory length of all the analyzed images per bin.
Fig. 5(a) shows the DF image we took of E. gracilis with 4 s exposure time, where the microorganism trajectories can be observed. Fig. 5(b) shows the processed image after background correction and tracing of the E. gracilis paths, where the individual trajectories are highlighted in green. After using data from 10 different images and exporting 20 trajectories per image, we constructed a histogram of the speed distribution of E. gracilis using these 200 data points (Fig. 5(c)). The error bars in the histogram indicate the standard deviation from the mean of all the images per bin. As such, we determined the average speed of E. gracilis to be ≈90 μm s−1, with a standard deviation of ≈24 μm s−1, which is consistent with values previously reported in the literature.41
2.7 Application to fluorescence microscopy
Fluorescence is a valuable imaging technique for biological samples that can be used to determine cell viability and activity, to distinguish a specific cell type or multiple cell types within a mixture of cells, and to track cells. The addition of fluorescence capability to this simple and versatile phone illumination-imaging system will make this technique suitable for a range of applications in the field where fluorescence needs to be detected.Here we demonstrate fluorescence imaging of both microfluidic structures and cells using the dual phone illumination-imaging system. The illumination is performed with a solid, blue-colored 15 mm circle generated with the Retina display screen. The luminance of a typical screen is on the order of hundreds of cd m−2, and since we only use the blue emitters, the luminous intensity of the blue illumination circle is in the order of 10−2 cd. This allows us to use the Nokia phone microscope to record fluorescence images of a fluorescein dye solution inside microfluidic devices with varying channel shapes and sizes (Fig. 6(a) and (b)). In addition, we used the same system to record fluorescence images of CMFDA-stained HEK 293 cells and compared those with images obtained with an Olympus IX83 microscope (10× magnification with NA = 0.4) (Fig. 6(c)).
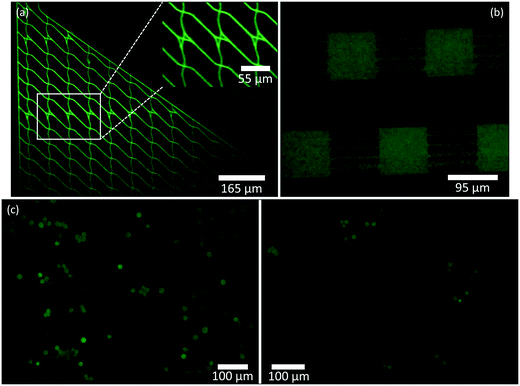 | ||
| Fig. 6 Application to fluorescence microscopy. (a) Zoomed-in and cropped fluorescence image of a microfluidic device taken with the Nokia phone-lens system (Retina display illumination) with fluorescein dye solution in the plaza regions and channels. The inset shows a zoomed-in image of the device junctions, similar to Fig. 3(c) – left. (b) Zoomed-in and cropped fluorescence image of a microfluidic device with fluorescein dye solution in plaza regions and multi-shaped channels (Retina display illumination). (c) Comparison of zoomed-in and cropped fluorescence images of CMFDA-stained HEK 293 cells taken with the Nokia phone-lens system (Retina display illumination) (left) and with the Olympus IX83 microscope (10× magnification with NA = 0.4) (right). | ||
3. Experimental
3.1 Imaging
For imaging, we used a Nokia Lumia 1020, which has a 41.3-megapixel (MP) image sensor with a pixel size of 1.12 μm, and attached an external lens (iPhone 5 lens: NA = 0.23, f2.2, 2× magnification) to the camera to improve focus, which limits the theoretical spatial resolution of our system to 1.3 μm. For illumination, we used a 120 mm Retina display of an Apple device (iPhone 6), with a pixel density of 326 pixels per inch (PPI) which translates to an RGB pixel size of ≈78 μm. The imaging phone-lens system is held in place by a clamp, and the sample along with the illumination phone is placed onto an XYZ-translation stage (Thorlabs Inc.) for simple directional manipulation. The pictures were taken using the Lumia Camera application. Characterization of the phone system for spatial resolution was done using 1951 USAF resolution test chart (Edmund Optics). Characterization of FOV and WD was done using a stage micrometer (Motic) and cover slips (Fisherbrand, 150 μm thickness), respectively.3.2 Illumination
Patterns to be projected onto the Retina display are designed using the Microsoft PowerPoint application for iOS. Different patterns correspond to different imaging illumination modes. BF mode is attained by projecting a white screen, at the highest brightness, where the sample has uniform illumination throughout. DF illumination is typically achieved with ring illumination, where the ring lies just outside the NA of the objective lens. We created single (DF-single), double (DF-double), and triple (DF-triple) ring illumination patterns by drawing concentric white rings on a black background.RI is similar to DF, but whereas DF relies on a ring of white light contrasted against a dark specimen background, RI utilizes a ring of colored light contrasted against a specimen background of a different color. These contrasting colors are specimen-dependent, and are typically chosen to maximally enhance the specimen image without the need for specimen staining. RI also includes rings that have multiple colors (two-sector or four-sector patterns) against a dark specimen background. Designs can be expanded to include more colored sectors per circle. PI can be accomplished by drawing and filling a small white circle on a black background.
3.3 Fluorescence imaging
To adapt the system for fluorescence, we used a fluorescence excitation filter (Chroma, 470/40 nm) inserted between the illumination phone screen and the sample, and a fluorescence emission filter (Chroma, 525/50 nm) between the imaging phone camera and the external lens.For illumination of fluorescent samples, we prepared and used a blue-colored filled circle with a diameter of approximately 15 mm on the illumination phone screen. To observe fluorescence within microfluidic devices, we added a 2.7 mM solution of fluorescein dye (Sigma-Aldrich) into the channels. As for cells, we used CellTracker™ Green CMFDA dye (Thermo Fisher Scientific) and followed the staining protocol for cells in suspension specified by the manufacturer to stain HEK 293 cells. The final working concentration of the dye was 25 μM. All fluorescence images were recorded with an exposure time of 4 s with the mobile phone and 10 ms with the Olympus IX83 microscope.
3.4 Microfluidics
Our microfluidic devices are fabricated using soft lithography. Polydimethylsiloxane (PDMS) (Sylgard® 184) is mixed in a 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 polymer to crosslinker ratio, poured onto silica masters (designed and written at the Kavli Institute of Nanoscience Delft, The Netherlands), and cured overnight in an oven at 60 °C. The cured PDMS is then cut and peeled from the masters, and bonded to glass cover slips (Fisherbrand, 150 μm thickness) using plasma treatment for 30 to 45 seconds. Next, the finished device is pre-wet with a medium suitable for the microorganism to be used, by adding 50 to 100 μL of medium onto the cover slip of the device, and placing it in a vacuum chamber for 30 minutes. The device is then ready for use in experiments.
1 polymer to crosslinker ratio, poured onto silica masters (designed and written at the Kavli Institute of Nanoscience Delft, The Netherlands), and cured overnight in an oven at 60 °C. The cured PDMS is then cut and peeled from the masters, and bonded to glass cover slips (Fisherbrand, 150 μm thickness) using plasma treatment for 30 to 45 seconds. Next, the finished device is pre-wet with a medium suitable for the microorganism to be used, by adding 50 to 100 μL of medium onto the cover slip of the device, and placing it in a vacuum chamber for 30 minutes. The device is then ready for use in experiments.
3.5 Microorganisms
For the microorganisms that we used in testing our imaging system, namely Escherichia coli and Euglena gracilis, we acquired the E. coli 437 strain from Prof. Howard C. Berg's laboratory at Harvard University, and we purchased E. gracilis from Carolina Biological Supply Company (USA). We grow E. coli colonies on Luria-Bertani (LB)-agar Petri dishes at 37 °C and E. gracilis in liquid Provasoli's enriched seawater (PES) medium at room temperature. For using E. coli in experiments, we inoculate a single colony in 4–6 mL of LB and incubate it at 37 °C overnight. On the day of the experiment, the liquid culture can either be directly pipetted onto the microfluidic device, or if its optical density is too high, then it can be diluted in LB to a more suitable concentration. The diluted mixture should then be further incubated at 37 °C for anywhere between half an hour to three hours, depending on the concentration, until the bacteria regain their growth and motility. On the other hand, E. gracilis can be directly used in experiments by pipetting a few microliters of the microorganism from its liquid culture onto the microfluidic device. LB and PES media were prepared according to the protocols mentioned in the Handbook of Culture Media for Food and Water Microbiology.423.6 Cell lines
To prepare unstained human epithelial cheek cell (HECC) samples, we pipette 50 μL of Milli-Q water onto a glass slide (Globe Scientific Inc., 1.1 mm thickness), and place the cells directly onto the water drop, then add a cover slip on top. The HECC were harvested by swabbing the inside of the cheek using a toothpick. To prepare stained HECC samples, we follow the same procedure, but we use 50 μL of methylene blue (Bio Basic Inc.) solution (0.1% W/V) instead of Milli-Q water. For diatoms and E. gracilis, we pipette 50 μL from a diatom suspension in Milli-Q water (1% W/V) or from E. gracilis suspension in PES medium, respectively, directly onto a glass slide, and then add a cover slip on top. The HEK 293 cells we used for fluorescence imaging were obtained from Prof. Amine Kamen's laboratory at McGill University.4. Conclusion
In this manuscript we report for the first time the combination of two mobile phones to provide a simple, versatile, and portable microscopy system that uses the screen of one phone for illumination and the camera of the other phone for imaging. While the phone screen illumination allows for facile generation of illumination patterns that can be used in different microscopy modalities, the imaging phone provides high spatial resolution and large FOV. With this system we demonstrate imaging of cells, microorganisms in microfluidic structures, as well as accurately determine their velocities. Overall, the design and construction of this system allows for substantial integration and miniaturization of the illumination-imaging components into a portable and user-friendly device.The imaging system is based on a Nokia Lumia 1020 with an attached external lens (iPhone 5 lens: NA = 0.23, f2.2) yielding an overall 2× magnification and a theoretical spatial resolution limit of 1.3 μm. This platform has an image sensor with 41.3 MP and 1.12 μm pixel size, with a spatial resolution of at least 2 μm, a field-of-view of 3.6 × 2.7 mm, and a working distance of 0.6 mm. We used this device in combination with a Retina display (iPhone 6) illumination system to project multiple illumination patterns such as bright-field, dark-field, Rheinberg, and point illumination. To demonstrate the capabilities of this system, we imaged E. gracilis and E. coli within microfluidic devices, diatoms, and human epithelial cheek cells. We also measured the velocities of E. gracilis using images recorded with long exposure times, and determined that they agree with previously reported data. Finally, we demonstrated that with this simple screen illumination, fluorescence microscopy can be performed as well.
These results verify the aforementioned capabilities of our Nokia-lens imaging system with Retina display illumination and demonstrate a range of applications that this system can be used for. While our choice of phones was based on their specifications, other phones and combinations of phones can also be used. In particular, various patterns of illumination can be easily obtained with other smartphone displays as well, adding to the versatility of the microscopy platform. Obvious imaging applications for this platform include microfluidics, motility studies of microorganisms, and cell analysis. Based on the results we showed, potential applications for our systems could include blood counts, downstream cell analysis, and reading colorimetric assays. This last application may especially benefit from a well-controlled illuminator, as quantifying colorimetric assays depends critically on an illumination source with stable, well-calibrated color characteristics.43
This new imaging-illumination microscopy system, by virtue of its portability and versatility, can complement or even replace heavy and complex microscopes, in addition to being relatively inexpensive and adaptive. Our system can serve as a platform for further development, where it can be adapted for other applications as well, and would be especially advantageous in low-resource settings.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was financially supported by the Defense Advanced Research Projects Agency (DARPA) under Grant Agreement No. HR0011-16-2-0028 (to D. V. N.) and by the National Sciences and Engineering Research Council of Canada (NSERC), Discovery Grant RGPIN-2018-05675 (to S. W.-H.). S. K. acknowledges the McGill Engineering Doctoral Award (MEDA).References
- D. N. Breslauer, R. N. Maamari, N. A. Switz, W. A. Lam and D. A. Fletcher, Mobile Phone Based Clinical Microscopy for Global Health Applications, PLoS One, 2009, 4(7), e6320 CrossRef PubMed.
- S. A. Sekula-Gibbs and M. A. Shearer, Sentinel node biopsy should be offered in thin melanoma with mitotic rate greater than one, Dermatol. Surg., 2011, 37(8), 1080–1088 CrossRef CAS PubMed.
- G. McConnell, J. Tragardh, R. Amor, J. Dempster, E. Reid and W. B. Amos, A novel optical microscope for imaging large embryos and tissue volumes with sub-cellular resolution throughout, eLife, 2016, 5, e18659 CrossRef PubMed.
- Z. Liu and P. J. Keller, Emerging Imaging and Genomic Tools for Developmental Systems Biology, Dev. Cell, 2016, 36(6), 597–610 CrossRef CAS PubMed.
- S. Pacheco, C. Wang, M. K. Chawla, M. Nguyen, B. K. Baggett and U. Utzinger, et al., High resolution, high speed, long working distance, large field of view confocal fluorescence microscope, Sci. Rep., 2017, 7(1), 13349 CrossRef PubMed.
- D. B. Murphy and M. W. Davidson, Fundamentals of light microscopy and electronic imaging, Wiley-Blackwell, Hoboken, N.J., 2nd edn, 2013, vol. xiii, p. 538 Search PubMed.
- F. Legesse, O. Chernavskaia, S. Heuke, T. Bocklitz, T. Meyer and J. Popp, et al., Seamless stitching of tile scan microscope images, J. Microsc., 2015, 258(3), 223–232 CrossRef CAS PubMed.
- Q. Lu, G. Liu, C. Xiao, C. Hu, S. Zhang and R. X. Xu, et al., A modular, open-source, slide-scanning microscope for diagnostic applications in resource-constrained settings, PLoS One, 2018, 13(3), e0194063 CrossRef PubMed.
- Z. Gorocs, Y. Ling, M. D. Yu, D. Karahalios, K. Mogharabi and K. Lu, et al., Giga-pixel fluorescent imaging over an ultra-large field-of-view using a flatbed scanner, Lab Chip, 2013, 13(22), 4460–4466 RSC.
- Z. Gorocs and A. Ozcan, Biomedical imaging and sensing using flatbed scanners, Lab Chip, 2014, 14(17), 3248–3257 RSC.
- X. Ou, G. Zheng and C. Yang, Erratum: 0.5 gigapixel microscopy using a flatbed scanner: erratum, Biomed. Opt. Express, 2016, 7(2), 646–647 CrossRef PubMed.
- T. Shimobaba, H. Yamanashi, T. Kakue, M. Oikawa, N. Okada and Y. Endo, et al., In-line digital holographic microscopy using a consumer scanner, Sci. Rep., 2013, 3, 2664 CrossRef PubMed.
- C. Zhang and K. S. Suslick, A colorimetric sensor array for organics in water, J. Am. Chem. Soc., 2005, 127(33), 11548–11549 CrossRef CAS PubMed.
- G. Zheng, X. Ou and C. Yang, 0.5 gigapixel microscopy using a flatbed scanner, Biomed. Opt. Express, 2013, 5(1), 1–8 Search PubMed.
- K. Sullivan, J. Kloess, C. Qian, D. Bell, A. Hay and Y. P. Lin, et al., High throughput virus plaque quantitation using a flatbed scanner, J. Virol. Methods, 2012, 179(1), 81–89 CrossRef CAS PubMed.
- A. Ozcan and E. McLeod, Lensless Imaging and Sensing, Annu. Rev. Biomed. Eng., 2016, 18, 77–102 CrossRef CAS PubMed.
- A. F. Coskun, T. W. Su and A. Ozcan, Wide field-of-view lens-free fluorescent imaging on a chip, Lab Chip, 2010, 10(7), 824–827 RSC.
- S. Seo, T. W. Su, D. K. Tseng, A. Erlinger and A. Ozcan, Lensfree holographic imaging for on-chip cytometry and diagnostics, Lab Chip, 2009, 9(6), 777–787 RSC.
- X. Cui, L. M. Lee, X. Heng, W. Zhong, P. W. Sternberg and D. Psaltis, et al., Lensless high-resolution on-chip optofluidic microscopes for Caenorhabditis elegans and cell imaging, Proc. Natl. Acad. Sci. U. S. A., 2008, 105(31), 10670–10675 CrossRef CAS PubMed.
- J. C. Contreras-Naranjo, Q. Wei and A. Ozcan, Mobile Phone-Based Microscopy, Sensing, and Diagnostics, IEEE J. Sel. Top. Quantum Electron., 2016, 22(3), 1–14 Search PubMed.
- Z. J. Smith, K. Chu, A. R. Espenson, M. Rahimzadeh, A. Gryshuk and M. Molinaro, et al., Cell-phone-based platform for biomedical device development and education applications, PLoS One, 2011, 6(3), e17150 CrossRef CAS PubMed.
- T. Gao, Z. J. Smith, T. Y. Lin, D. Carrade Holt, S. M. Lane and D. L. Matthews, et al., Smart and Fast Blood Counting of Trace Volumes of Body Fluids from Various Mammalian Species Using a Compact, Custom-Built Microscope Cytometer, Anal. Chem., 2015, 87(23), 11854–11862 CrossRef CAS PubMed.
- H. Zhu, O. Yaglidere, T. W. Su, D. Tseng and A. Ozcan, Wide-field fluorescent microscopy on a cell-phone, Conf. Proc. IEEE Eng. Med. Biol. Soc., 2011, 2011, 6801–6804 Search PubMed.
- I. I. Bogoch, J. R. Andrews, B. Speich, J. Utzinger, S. M. Ame and S. M. Ali, et al., Mobile phone microscopy for the diagnosis of soil-transmitted helminth infections: a proof-of-concept study, Am. J. Trop. Med. Hyg., 2013, 88(4), 626–629 CrossRef PubMed.
- C. W. Pirnstill and G. L. Cote, Malaria Diagnosis Using a Mobile Phone Polarized Microscope, Sci. Rep., 2015, 5, 13368 CrossRef CAS PubMed.
- R. R. Jahan-Tigh, G. M. Chinn and R. P. Rapini, A Comparative Study Between Smartphone-Based Microscopy and Conventional Light Microscopy in 1021 Dermatopathology Specimens, Arch. Pathol. Lab. Med., 2016, 140(1), 86–90 CrossRef PubMed.
- Q. Wei, R. Nagi, K. Sadeghi, S. Feng, E. Yan and S. J. Ki, et al., Detection and spatial mapping of mercury contamination in water samples using a smart-phone, ACS Nano, 2014, 8(2), 1121–1129 CrossRef CAS PubMed.
- T. S. Park, W. Li, K. E. McCracken and J. Y. Yoon, Smartphone quantifies Salmonella from paper microfluidics, Lab Chip, 2013, 13(24), 4832–4840 RSC.
- A. Chen, R. Wang, C. R. Bever, S. Xing, B. D. Hammock and T. Pan, Smartphone-interfaced lab-on-a-chip devices for field-deployable enzyme-linked immunosorbent assay, Biomicrofluidics, 2014, 8(6), 064101 CrossRef CAS PubMed.
- G. A. Zheng, C. Kolner and C. H. Yang, Microscopy refocusing and dark-field imaging by using a simple LED array, Opt. Lett., 2011, 36(20), 3987–3989 CrossRef PubMed.
- Z. J. Liu, L. Tian, S. J. Liu and L. Waller, Real-time brightfield, darkfield, and phase contrast imaging in a light-emitting diode array microscope, J. Biomed. Opt., 2014, 19(10), 106002 Search PubMed.
- N. Lue, W. Choi, G. Popescu, T. Ikeda, R. R. Dasari and K. Badizadegan, et al., Quantitative phase imaging of live cells using fast Fourier phase microscopy, Appl. Opt., 2007, 46(10), 1836–1842 CrossRef PubMed.
- S. Dong, Z. Bian, R. Shiradkar and G. Zheng, Sparsely sampled Fourier ptychography, Opt. Express, 2014, 22(5), 5455–5464 CrossRef PubMed.
- S. Dong, R. Shiradkar, P. Nanda and G. Zheng, Spectral multiplexing and coherent-state decomposition in Fourier ptychographic imaging, Biomed. Opt. Express, 2014, 5(6), 1757–1767 CrossRef PubMed.
- L. Tian, X. Li, K. Ramchandran and L. Waller, Multiplexed coded illumination for Fourier Ptychography with an LED array microscope, Biomed. Opt. Express, 2014, 5(7), 2376–2389 CrossRef PubMed.
- G. Zheng, R. Horstmeyer and C. Yang, Wide-field, high-resolution Fourier ptychographic microscopy, Nat. Photonics, 2013, 7(9), 739–745 CrossRef CAS PubMed.
- L. Tian and L. Waller, Quantitative differential phase contrast imaging in an LED array microscope, Opt. Express, 2015, 23(9), 11394–11403 CrossRef CAS PubMed.
- D. V. Nicolau Jr., M. Lard, T. Korten, F. C. van Delft, M. Persson and E. Bengtsson, et al., Parallel computation with molecular-motor-propelled agents in nanofabricated networks, Proc. Natl. Acad. Sci. U. S. A., 2016, 113(10), 2591–2596 CrossRef PubMed.
- F. C. M. J. M. van Delft, G. Ipolitti, D. V. Nicolau, A. S. Perumal, O. Kasper and S. Kheireddine, et al., Something has to give: scaling combinatorial computing by biological agents exploring physical networks encoding NP-complete problems, Interface Focus, 2018, 8(6), 20180034 CrossRef PubMed.
- M. H. Longair, D. A. Baker and J. D. Armstrong, Simple Neurite Tracer: open source software for reconstruction, visualization and analysis of neuronal processes, Bioinformatics, 2011, 27(17), 2453–2454 CrossRef CAS PubMed.
- T. Ogawa, E. Shoji, N. J. Suematsu, H. Nishimori, S. Izumi and A. Awazu, et al., The Flux of Euglena gracilis Cells Depends on the Gradient of Light Intensity, PLoS One, 2016, 11(12), e0168114 CrossRef PubMed.
- J. E. L. Corry, G. D. W. Curtis and R. M. Baird, Handbook of Culture Media for Food and Water Microbiology 3rd Edition Introduction to the Third Edition, Handbook of Culture Media for Food and Water Microbiology, 3rd edn, 2012, pp. Xxiii–Xxiv Search PubMed.
- L. Shen, J. A. Hagen and I. Papautsky, Point-of-care colorimetric detection with a smartphone, Lab Chip, 2012, 12(21), 4240–4243 RSC.
| This journal is © The Royal Society of Chemistry 2019 |