Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
SIMS imaging in neurobiology and cell biology
Paola
Agüi-Gonzalez
 ab,
Sebastian
Jähne
ab,
Sebastian
Jähne
 ab and
Nhu T. N.
Phan
ab and
Nhu T. N.
Phan
 *ab
*ab
aCenter for Biostructural Imaging of Neurodegeneration, University Medical Center Göttingen, von-Siebold-Straße 3a, 37075 Göttingen, Germany. E-mail: thi.phan@med.uni-goettingen.de
bDepartment of Neuro- and Sensory Physiology, University Medical Center Göttingen, Humboldtallee 23, 37073 Göttingen, Germany
First published on 9th May 2019
Abstract
Secondary ion mass spectrometry (SIMS) has been increasingly recognized as a powerful technique for visualizing molecular architectures in the fields of neurobiology and cell biology. There are two main platforms of SIMS, namely ToF-SIMS and nanoscale SIMS (nanoSIMS), which are capable of imaging different types of biomolecules with resolution at the single cell and organelle level, respectively. In this review, we focus on the fundamental aspects of SIMS, as well as on the current ongoing instrumental developments of this technology. Selective applications of SIMS in neurobiological and cell biological research are provided to demonstrate its strengths, limitations, and future potential in the field. We add several examples of correlative imaging techniques that combine SIMS with other technologies, while highlighting the current trend for comprehensive and specific bio-imaging.
Introduction
Secondary ion mass spectrometry (SIMS), one of the three major platforms of mass spectrometry imaging (MSI), has become a valuable chemical analysis tool in biological research providing information on chemical distribution of the cells and tissues of interest at subcellular spatial resolution. The technique features high chemical specificity, sensitivity, and capability of molecular structure elucidation. SIMS offers multiplexed analysis that allows detection of various biomolecules in single experiments. In addition, it does not require a priori knowledge of the target analytes, unlike optical fluorescence microscopy or electron microcopy approaches, although labeling of target molecules with isotopic compounds is also commonly used, especially in nanoSIMS imaging. SIMS has been continuously developed and expanded in terms of its applications to image different biomolecules for fundamental studies of cell biology and pharmaceuticals, and in medical research.1–3Given the high complexity and heterogeneity of the cells in the brain, spatial information on the molecular organization is essential for the understanding of and insights into cellular processes. A particular cellular mechanism or a disease is not caused by a simple change of a single molecule, but most likely is the result of a complex interaction of various key biomolecules such as lipids, proteins, peptides, and metabolites. SIMS provides advantages in that multiple molecules can be analyzed with high throughput to obtain comprehensive data of the molecular composition and distribution inside cells and organs, often with subcellular or organelle level spatial resolution, and therefore it is an excellent approach for the study of brain and cell biology.
In this review, we provide a fundamental overview of SIMS and updates on recent technological advances in this field. We also introduce the relevant applications of SIMS in neurological and cell biological research, particularly for single cell imaging, three-dimensional imaging, pharmaceuticals, and neurodegenerative diseases and pathology. In addition, correlative imaging approaches combining SIMS and other optical and non-optical imaging modalities will be presented as the current trends in mass spectrometry imaging in an effort to obtain a comprehensive understanding of complex biological questions.
Principles of SIMS
In SIMS, an energetic primary ion beam impacts a solid surface to eject and ionize secondary ions from the sample surface. The sputtering and ionization of the secondary ions have been explained by collision cascade theory.4 The secondary ions are then extracted by an electric field into a mass spectrometer and subsequently separated into different mass per charge (m/z) using a mass analyzer (Fig. 1A). Afterwards, the separated ions reach the detector producing a mass spectrum in which the signal intensity of the detected ions is plotted versus m/z.To obtain an ion image, the focused primary ion beam scans the sample surface spot by spot or pixel by pixel. A mass spectrum is then obtained for each pixel. By recording the positions of the beam while scanning, a color map of the signal intensity for each ion can be constructed across the scanned area, providing a so-called ion image. These ion images show the distribution of the ions on the surface. In addition, a three dimensional (3D) reconstruction from a series of consecutive two dimensional (2D) ion images can be performed allowing the visualization of the molecular distribution underneath the sample surface. Alternatively, depth profiling is the method of choice to observe the molecular compositional change across the sample depth. The depth resolution in SIMS is typically in a range of 1–10 nm.1
What influences SIMS measurements?
For the applications of SIMS in neurobiology and cell biology, several important elements that influence the measurement need to be considered. First, the properties of the primary ion beam determine the mass range of the detected molecules.5 A higher energy of the primary ions produces higher secondary ion yield which increases sensitivity; however it causes higher fragmentation, meaning that the molecular information on the detected ions can be lost. A highly energetic beam is also more destructive to samples. In addition, the size of the primary ions influences the spatial resolution of the imaging. Small ions provide a superior spatial resolution to large cluster ions although the latter possesses a much lower energy per atom in each cluster and therefore is more advantageous in terms of molecular structure preservation and sample damage. Various types of primary ion sources have been developed and are available in the SIMS community, from monatomic (Au+, Cs+, O−) and polyatomic sources (C60+),6 and liquid metal ion guns (LMIG) Bi3+ and Au3+,7 to gas cluster ion beams (GCIB) (Ar4000+,8 (CO2)2000+,9 and (H2O)1000+).10 The choice of the primary ion source depends on the purposes of the applications and the molecules of interest.Second, the sensitivity for the molecules of interest depends on several factors, particularly the abundance of the molecules in the analyzed volume and their ionization probability (typically lower than ∼10−4).11 This is a critical element especially for biological imaging at subcellular resolution, where the amount of analytes can become too low for detection. In this case, gaining better spatial resolution is a compromise with limit of detection. In addition, the properties of the target analyte and its chemical and morphological environment also heavily affect the sensitivity. The limit of detection obtained by SIMS is in the range of parts per million (ppm) to parts per billion (ppb).12
Lastly, the success of SIMS analysis is also heavily dependent on the sample preparation. It is essential to preserve the native states of the biological samples in order to obtain reliable information about the spatial distribution of the biomolecules. The common sample preparation methods for SIMS imaging are typically called frozen-hydrated, freeze-dried, and chemical fixation.13,14 Freeze-drying is the most widely used method due to its simplicity; however, there is a risk of molecular rearrangement of the subcellular structure. The samples are plunged into liquid nitrogen (77 K) or liquid propane (85 K) quickly to avoid the formation of ice crystals which may cause ultrastructural damage. The samples are then inserted into a high vacuum (∼10−5 Torr) where water is sublimed slowly to prevent molecular rearrangement. Frozen-hydrated sample preparation provides the safest method for the preservation of the morphology and localization of the cellular components. However, a more laborious and complicated instrumental setup at near liquid nitrogen temperature is needed and there is a high risk of ice condensation on the sample surface which can restrict its applications in biology. On the other hand, chemical fixation and embedding in resin is routinely used, especially for nanoSIMS. Glutaraldehyde is the gold standard for protein fixation while osmium tetroxide is common for lipids.15 It should be noted that the addition of fixative chemicals might introduce artifacts in the analysis due to the fact that the fixation involves chemical modification of the sample constituents. The selection of suitable fixatives should therefore be considered carefully. Other less common methods, namely freeze-fracture, etching, and trehalose vitrification have also been proposed in the literature.2,13,14
Different configurations of SIMS and recent developments
There are several configurations developed and available for SIMS imaging; each has its own capacities and limitations, and hence is suitable for different applications. The most common is the time-of-flight SIMS (ToF-SIMS) approach which utilizes a time-of-flight mass analyzer to sort the secondary ions of different m/z that travel to reach the detector at different time points in a flight cycle. To produce separate cycles for ToF-SIMS, the primary ion beam needs to be pulsed as in the configuration of most commercial instruments (Fig. 1B).11 Alternatively, the secondary ions need to be bunched into small pulses before going to the ToF mass analyzer as in the configuration of the Ionoptika J105.16 For a pulsed primary ion beam, the mass resolution and spatial resolution are inversely related; however, a recently developed method using delayed extraction has been used to obtain both good spatial and mass resolution.17 In addition, the mass resolution of the ION-TOF and similar instruments is also affected by the topography of the sample. These issues are overcome by pulsing the secondary ions as in the J105. Nevertheless, higher spatial resolution for imaging can be routinely obtained by the ION-TOF by the use of a liquid metal ion gun (LMIG) (∼250 nm).18 ToF-SIMS allows parallel detection of the entire mass range; therefore, it is an ideal choice for non-targeted imaging.Another SIMS platform, nanoSIMS, employs a magnetic sector mass analyzer and a high energy and highly focussed ion beam. In the magnetic sector, the secondary ions travel in a circular path under the influence of a magnetic field. The radius of the path is different based on the m/z of the ions (Fig. 1C). By varying the magnetic field, the detectable mass range, although narrow, can be adjusted. High mass resolution of about 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 can be obtained.19 NanoSIMS typically allows parallel detection of up to 7 different secondary ions, although more secondary ions can be detected using magnetic switching. The most significant feature of nanoSIMS is the coaxial configuration between the primary and secondary ion beams (Fig. 1C), which improves the focus of the primary ion beam, decreases aberrations, and increases the transmission efficiency of secondary ions (∼80%).20 In addition, the instrument can be equipped with both O− and Cs+ primary ion beams which are highly focused and energetic, so both positive and negative ions can be imaged. A spatial resolution of ∼50 nm can be achieved;21 however, to obtain sufficient signals of the secondary ions from biological samples (e.g. lipids), a lateral resolution of ∼100 nm is more commonly used.15 On the other hand, owing to the intense fragmentation that occurs, only elements and small ions are observed in nanoSIMS from the larger parent molecules. The technique is suitable for the analysis of the chemical organization of the entire sample (such as 31P, 32S, 12C14N, etc.). To study specific target biomolecules, labeling the biomolecules of interest with less abundant isotopic compounds is needed. A technique, known as multi-isotope imaging mass spectrometry (MIMS), was first applied by the Lechene group to track the protein turnover in cells and tissues.12
000 can be obtained.19 NanoSIMS typically allows parallel detection of up to 7 different secondary ions, although more secondary ions can be detected using magnetic switching. The most significant feature of nanoSIMS is the coaxial configuration between the primary and secondary ion beams (Fig. 1C), which improves the focus of the primary ion beam, decreases aberrations, and increases the transmission efficiency of secondary ions (∼80%).20 In addition, the instrument can be equipped with both O− and Cs+ primary ion beams which are highly focused and energetic, so both positive and negative ions can be imaged. A spatial resolution of ∼50 nm can be achieved;21 however, to obtain sufficient signals of the secondary ions from biological samples (e.g. lipids), a lateral resolution of ∼100 nm is more commonly used.15 On the other hand, owing to the intense fragmentation that occurs, only elements and small ions are observed in nanoSIMS from the larger parent molecules. The technique is suitable for the analysis of the chemical organization of the entire sample (such as 31P, 32S, 12C14N, etc.). To study specific target biomolecules, labeling the biomolecules of interest with less abundant isotopic compounds is needed. A technique, known as multi-isotope imaging mass spectrometry (MIMS), was first applied by the Lechene group to track the protein turnover in cells and tissues.12
Other noticeable configurations have been developed and applied in biological SIMS such as Fourier transform ion cyclotron resonance mass spectrometry (FT-ICR)-SIMS which offers an outstanding mass resolution (m/Δm 67![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 at m/z 750),22 and hybrid quadrupole orthogonal ToF for matrix-assisted laser desorption ionization (MALDI) and electrospray ionization (ESI) combined with SIMS (QSTAR XL-QqTOF) which allows the detection of very large molecules up to m/z 40
500 at m/z 750),22 and hybrid quadrupole orthogonal ToF for matrix-assisted laser desorption ionization (MALDI) and electrospray ionization (ESI) combined with SIMS (QSTAR XL-QqTOF) which allows the detection of very large molecules up to m/z 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000.23 However, the spatial resolution is sufficient for only tissue imaging, perhaps not for cellular imaging.
000.23 However, the spatial resolution is sufficient for only tissue imaging, perhaps not for cellular imaging.
Recent developments in SIMS
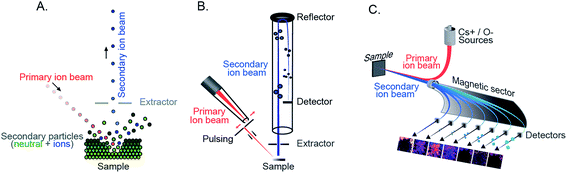
With the rapid technological development of SIMS, it has been increasingly embraced by the biological and medical community. Recently, improving the capability of SIMS for bioimaging, especially for molecular identification, improving the mass resolving power and sensitivity for biomolecules, and targeted imaging of specific molecules have been the focus of several developments. Given the high complexity of typical biological samples where various interferences can affect analysis and data interpretation, it is sometimes important to elucidate the molecular structures of the biomolecules in order to obtain reliable biological information. Tandem MS (MS/MS or MSn) enables the identification of specific molecules of interest based on their typical fragmented species and has become more prevalent in bioimaging with SIMS.24,25 Tandem MS has been a special feature in several SIMS instruments such as the FT-ICR-SIMS, the hybrid QSTAR XL QqTOF, and the ToF-SIMS J105. For example, MS and MS/MS imaging were successfully used to study the lipid distribution in Drosophila brain, particularly diacylglycerols (DAGs) and glycerolphospholipids.24 MS/MS could be used both for profiling to identify the lipid structure and for imaging to observe the distribution of the fragmented ions. The latter would be useful for the study of intact biomolecules and their possible precursors or metabolic products. Another new platform focusing on tandem MS, the so-called TRIFT nanoTOF, has been demonstrated (Fig. 2A).25 The instrument is equipped with a triple ion focusing time-of-flight (TRIFT) analyzer which allows conventional ToF-SIMS imaging and tandem MS analysis to be performed simultaneously with a mass precision of 1 Da (at m/z 500) and a mass resolving power of about m/Δm 3000. TRIFT was used to carry out molecular identification of lipids and fatty acids in discrete nuclei and neuronal connections of zebra finch brain, in the cross-section of the whole body of zebrafish (Fig. 2B), and neuropeptides up to m/z 2000 Da in pituitary gland tissue from rats.26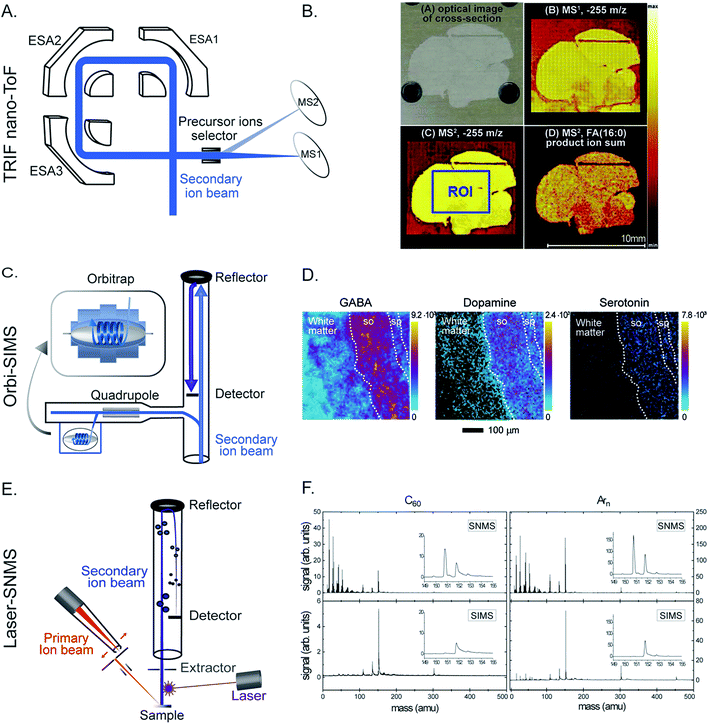 | ||
| Fig. 2 Examples of recent developments in SIMS. (A and B) Parallel imaging tandem MS (MS1, MS2) on a TRIFT nanoTOF. (A) Schematic of a TRIFT nanoTOF. (B) An example of MS1 and MS2 imaging of a fatty acid (16:0) at m/z 255 in a zebra finch brain section using a TRIFT nanoTOF. Reproduced from ref. 25 with permission. (C and D). 3D Orbi-SIMS for MSn imaging with high speed and high mass resolution. (C) Schematic of 3D OrbiSIMS. (D) Molecular maps of neurotransmitters GABA, dopamine, and serotonin in the cornu ammonis region of mouse hippocampus obtained by 3D OrbiSIMS. Reproduced from ref. 27 with permission. (E and F) Laser-SNMS postionization for improving the secondary ion yield. (E) Schematic of a laser-SNMS. (F) Mass spectra of a guanine surface analyzed with a 20 keV C60+ (left) and Arn+ GCIB (right), with and without laser-SNMS postionization (first and second row, respectively). Laser-SNMS clearly increases the intensity of the detected ions. Reproduced from ref. 29 with permission. | ||
Another new development in this field is 3D Orbi-SIMS which couples a ToF and Orbitrap mass analyzer (Fig. 2C). This configuration combines the best features of both analyzers, particularly the high speed of analysis, high spatial resolution of a ToF analyzer (∼2 μm for biomolecules), and high mass-resolving power of an Orbitrap (>240![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 at m/z 200).27 Orbi-SIMS was employed to map the distribution of the neurotransmitters GABA, dopamine, and serotonin (Fig. 2D) in the cornu ammonis region of mouse hippocampus. Additionally, 29 sulfoglycosphingolipids and 45 glycerolphospholipids in different layers of the hippocampus were imaged and identified by tandem SIMS. In addition, the metabolic profile of single rat alveolar macrophages treated with the drug amiodarone was examined. The exponential increase in the abundance of cholesterol and several phospholipids was found to be related to the accumulation of the drug.27
000 at m/z 200).27 Orbi-SIMS was employed to map the distribution of the neurotransmitters GABA, dopamine, and serotonin (Fig. 2D) in the cornu ammonis region of mouse hippocampus. Additionally, 29 sulfoglycosphingolipids and 45 glycerolphospholipids in different layers of the hippocampus were imaged and identified by tandem SIMS. In addition, the metabolic profile of single rat alveolar macrophages treated with the drug amiodarone was examined. The exponential increase in the abundance of cholesterol and several phospholipids was found to be related to the accumulation of the drug.27
As discussed above, the use of a GCIB in SIMS offers an advantage of reducing fragmentation during the ionization; however, this is accompanied by a decrease of the secondary ion yield. Laser secondary neutral mass spectrometry (Laser-SNMS) has been developed as an effective postionization approach to increase the secondary ion production (Fig. 2E).28 The technique applies a laser beam just before the extraction lens to ionize the neutral species which are co-sputtered with the secondary ions from the sample surface. To adjust the degree of fragmentation and the energy of the sputtered neutral species, the laser power density and the laser pulse delay time can be adjusted. A guanine surface was analyzed with both a 20 keV C60+ and Arn+ GCIB with and without laser-SNMS postionization (Fig. 2F) and the postionization efficiency was even higher when accompanied by a GCIB primary ion source.29,30
Recently, the correlation of the molecular distribution obtained by SIMS imaging and the cellular structures or the localization of specific proteins has been investigated. A main drawback of SIMS is the difficulty in the identification of cellular organelles and protein localization. Several labeling tags for specific proteins utilizing heavy metals such as lanthanides31 and colloidal gold conjugated to antibodies32 have been developed with the goal of imaging specific proteins with SIMS. However, these tags exhibit several disadvantages, especially that their large size causes low labeling precision. Targeted imaging of specific cellular proteins by nanoSIMS has been demonstrated using newly developed labeling probes containing fluorine33 and boron.34 For example, boron probes were used to label proteins of interest which then could be visualized by both nanoSIMS and fluorescence microscopy. Successful imaging of the synaptic protein syntaxin 1 and mitochondrial marker TOM-70 in mammalian cells was carried out. The probes were shown to be well suited for imaging of target proteins with subcellular resolution due to their high selectivity, high sensitivity of detection, and high spatial precision of labeling. In addition, they could be applied for different proteins using different labeling methods. The probes could also be applied for cluster SIMS, allowing the study of lipid–protein domains and their functional relations.
Applications of SIMS in neurobiology and cell biology
Single cell imaging
Single cells are a widely used biological model owing to characteristics such as that they are easy to maintain in culture, quick to propagate, and simple compared to other animal and tissue models. To understand thoroughly the complexity of a biological system, it is essential to understand its molecular mechanism at the cellular and organelle level. Single cells, however, are highly heterogeneous and molecular organization plays an essential role in maintaining and regulating molecular functions. Alteration of the molecular organization can result in dramatic impacts and functional disorders in the cell. In addition, single cells are small, typically in a range of several micrometers to several hundred micrometers in size. This requires the use of imaging techniques with high sensitivity and spatial resolution in order to capture detailed images of cellular substructures with high accuracy. Regarding the characteristics of SIMS and its different configurations as introduced in this review, SIMS imaging provides many advantages for single cell imaging.One of the first significant applications of SIMS in single cell imaging was the imaging of mating Tetrahymena thermophila to visualize the distribution of membrane lipids.35 During mating, the membranes of two complementary Tetrahymena cells form a fusion pore allowing the transportation of micronuclei between the cells. It was found that low curvature lipids, such as phosphatidylcholine (PC), decreased significantly, whereas high curvature lipids, such as 2-amino ethylphosphonolipids, were dominant at the fusion site. Based on the study, it is implied that the heterogeneous distribution of membrane lipids could play a key role in regulating membrane fusion for cellular processes.
A more recent example was the imaging and characterization of membrane lipids of single neurons in the sea slug Aplysia californica using TOF-SIMS and a 20 keV C60+ primary ion gun.36 The major lipid species of the neuronal membrane including PC (16:0e/18:1), vitamin E, and cholesterol were detected and their molecular structures were identified by tandem MS directly on the sample surface. The localization of vitamin E was found to be highly correlated with the pigment carotenoids. Vitamin E was also previously detected by SIMS on Aplysia neurons.37 This could imply the biological function of vitamin E in neuronal activity.
Silver nanoparticles (Ag NPs) have been widely used for therapeutics and biomedical purposes; however, their cytotoxicity is not fully known.38 The effects of Ag NPs on the lipid composition of macrophage cells were investigated by ToF-SIMS.39 To increase the sensitivity towards biomolecules, matrix enhanced SIMS is a method in which a matrix 2,5-dihydroxybenzoic acid was deposited on top of the sample surface before freeze drying. It was found that cells exposed to Ag NPs exhibit a significant change in the lipid composition at the cell surface; specifically there was a significant increase in the abundance of monoacylglycerols (MAGs), diacylglycerols (DAGs), and cholesterol. The change was more dramatic as the concentration of Ag NPs was elevated (Fig. 3A). In addition, cholesterol and DAGs were shown to migrate to the surroundings of the cells which indicated cell death.
 | ||
| Fig. 3 Examples of applications of SIMS in neurobiology and cell biology. (A) ToF-SIMS imaging of macrophage cells without (top) and with Ag NP treatment (bottom). From left to right: SIMS images of total ions, cholesterol (C27H45) at m/z 369.38, a fragment of DAG (C39H71O4) at m/z 603.52, the overlay of cholesterol and the DAG fragment, and a score plot of principle component analysis showing good separation between the cell group exposed to a high level of Ag NPs (500–2000 ng mL−1) and those exposed to no or low amount of Ag NPs (>50 ng mL−1). Cholesterol and DAGs were shown to migrate to the surroundings of the cells when exposed to Ag NPs. Reproduced from ref. 39 with permission. (B) SIM and nanoSIMS imaging of tumor tissue of mice pretreated with a Pt drug containing nanocarriers for cancer treatment. Left: SIM and nanoSIMS images showing the localization of the drug in tumor tissue; the nanoSIMS image of 31P and 195Pt (top row), overlay of the SIM fluorescent image of actin filaments (green), DAPI (blue), and the nanocarrier via Cy5.5 dye (red) and the nanoSIMS ratio image of 15N/14N in hue saturation intensity (HSI) showing the enriched area of the nanocarrier in tissue (bottom row). Right: Schematic of the nanocarrier assembly which contains a polymer structure of oxaliplatin drug, 15N label and Cy5.5 fluorescent label of the nanocarrier, and MMP substrate peptide. Reproduced from ref. 50 with permission. (C) ToF-SIMS imaging of drosophila brain treated with cocaine and methylphenidate to study the lipid structural effects of the drug on brain chemistry. Left: SIMS images of PC (34:1) at m/z 760.58 in control and cocaine treated brain. Right: Relative quantification of lipid change in the fly brains under cocaine and methylphenidate treatments. It is shown that both the abundance and localization of the lipids in the brain significantly changed by drug exposure; however, the change is opposite in cocaine and methylphenidate treatments. Reproduced from ref. 59 with permission. | ||
Three dimensional imaging
Three dimensional (3D) imaging has been an important development in the biological field. The method provides a map of the 3D distribution of the biomolecules inside an organism. Dual beam or single beam configurations can be used to perform 3D imaging. A conventional ToF-SIMS instrument utilizes a dual beam configuration in which a highly focused analysis beam, such as a 25 keV LMIG Bi3+, is used to statically image the sample with high spatial resolution, and subsequently a non-focused etching beam, for example a 10 keV Arn+ GCIB, is used to remove the damaged subsurface caused by the LMIG and prepare a fresh surface for the next imaging cycle. The analysis is continued etching layer by layer through the sample. All the consecutive 2D images are then stacked together to produce a 3D chemical distribution of the sample. Alternatively, in more recently developed instruments, for example the J105 and nanoSIMS, a single beam can be employed to dynamically erode the sample while 2D images are obtained along the depth of the sample.One of the first demonstrations was the 3D imaging of single HeLa cells using the J105 SIMS instrument with a 40 keV C60+ beam.40 Frozen hydrated cells were used in order to preserve the intact molecular structure of the cells. Signature ions such as the PC headgroup m/z 184 and DNA base adenine m/z 136 were detected and used to generate a 3D image. Subcellular features were observed, particularly the nucleus and endoplasmic reticulum. A similar instrumental approach was used to visualize the 3D distribution of TiO2 NPs inside single Tetrahymena cells.41 The study showed that TiO2 NPs were incorporated into the cells via the "mouth" of the cells and mainly accumulated in the food vacuoles. In another study, to investigate the organization and interaction between cholesterol and sphingolipids on membrane and subcellular compartments, Yeager et al.42 showed the 3D localization of these lipids in single Madin-Darby canine kidney cells using nanoSIMS and an 8 keV Cs+ gun. The cells were labelled with 18O-cholesterol and 15N-sphingolipids, fixed with glutaraldehyde and osmium tetroxide, and coated with an iridium layer before imaging with transmission electron microscopy (TEM) and nanoSIMS. The TEM imaging helps to visualize the morphology of the cells. The 3D images clearly showed distinguished enriched intracellular regions of cholesterol and sphingolipids, particularly 18O-cholesterol was concentrated in the tubular projections along the analysis depth, whereas 15N-sphingolipids were localized in small pockets which were not overlapping with the 18O-cholesterol area.
Although emerging as an attractive imaging modality, several existing challenges in 3D imaging restrict its widespread applications in the biological field. The main challenge is the accurate reconstruction of 2D images into a 3D image. This occurs in the data treatment step which originally does not account for the topography and matrix effects existing in the samples, and the uneven ionization of biological and organic materials and inorganic or metallic substrates. As a result, a distortion is observed in the generated 3D image, especially near the samples/substrate interface. Data reshaping is necessary in order to include all these factors. Different reshaping methods have been applied based on the recognition of the sample/substrate interface43 or the correlation of the sample with topographical information, which can be measured before and in parallel with SIMS imaging.44 However, the experimental setup for the correlative imaging also presents difficulty and complication. In addition, challenges in handling very huge data for the treatment of 3D MS data and long experimental time for high resolution imaging are common and typical in this modality. Nonetheless, new technologies and developments have been continued to improve and implement 3D imaging with the hope of further expanding the technique to the biological and medical fields. Detailed discussion on the challenges and perspectives of 3D imaging can be found in selected literature.45,46
Pharmaceuticals
In biomedicine related to pharmaceuticals, imaging of drugs and targeted biomolecules has become an important target in the field of SIMS imaging with a goal to obtain a thorough knowledge about the properties of the drugs, their cellular targets, their bio-transformation inside a biological system, and to understand their side effects. The questions can, at least in part, be answered with SIMS imaging. It should be noted that in pharmaceutical research, 3D imaging has been a favorable approach as it is very useful for visualizing the uptake and accumulation of drugs in subcellular compartments. 3D ToF-SIMS was applied to study the uptake and distribution of the antiarrhythmic drug, amiodarone and its main metabolite, desethylamiodarone, in single lung macrophages.47 A dual beam configuration was utilized in the analysis including an LMIG Bi3+ for imaging, and a GCIB Ar2000+ for etching. The cells were incubated with the drug and afterwards freeze-dried before SIMS analysis. Amiodarone was found to be distributed homogeneously inside the cells but was absent in the nucleus, whereas nucleus markers such as a fragment of adenine at m/z 136.1 and a fragment of ribose at m/z 81. PCs were found to be co-localized with the drug as expected because amiodarone is highly lipophilic. The 3D image showed that the drug as well as its metabolites were distributed throughout the entire cells but were not present in the nucleus. In a subsequent study, amiodarone was also imaged with high spatial resolution nanoSIMS and electron microscopy (EM) in order to observe its distribution in specific organelles in lung macrophages.48 For the nanoSIMS measurements, the cells were fixed with a mixture of tannic acid and glutaraldehyde which helped seal the cellular membrane and prevent the diffusion of the drug. Amiodarone was found to mainly cause accumulation of phospholipids in multilamellar lysosomes. This supports the speculation that amiodarone induces phospholipidosis, a lysosomal disorder caused by an excess accumulation of phospholipids, leading to cellular functional disorders.Another example is the study of the localization of the antibiotics ampicillin (AMP) and tetracycline (TET) in single Gram-negative bacteria Escherichia coli (E. coli). This was carried out with 3D ToF-SIMS and a C60+ primary ion beam.49 A spatial resolution of ∼300 nm was achieved. The cells were analyzed using the freeze-drying approach. There was a nonlinear relationship between the detected signal of the antibiotics and their amount exposed to the E. coli. The SIMS image showed the localization of the antibiotics within individual bacteria. AMP was mainly found within the first 400 nm subsurface layer of the cells, which was the periplasmic location where the main targets of AMP, the penicillin binding proteins, were found. On the other hand, TET was observed beyond the 400 nm subsurface layer. In addition, the study showed a low susceptibility of TET in the E. coli expressing the TET-specific efflux pump, which was consistent with a result measured by an antimicrobial activity assay showing that the TET-efflux pump contributed to the reduction of TET susceptibility in E. coli.
In drug delivery technology, a nanocarrier is designed to contain a platinum drug for cancer treatment.50 It contains a polymer backbone coupled with a fluorescence dye Cy5.5 and labeled with 15N, and a substrate peptide for matrix metalloproteinase (MMP) (Fig. 3B). The nanocarrier is designed to specifically target tumors by moving to the environment where the tumor associated proteases, MMP, are located. The proteases then cleave the substrate peptide of the carrier inducing the nanocarrier to undergo a morphological change. This leads to a microscale arrangement of the nanoparticles locking them in the tumor environment. The Pt drug subsequently dissociated from the carrier and bound to its target, the DNA, in the nucleus. To evaluate the distribution of the nanocarrier and the drug in tumor tissue, a combination of nanoSIMS and structured illumination microscopy (SIM) was utilized. Live fluorescence microscopy was performed to track the fluorophore of the nanocarrier injected into mice for up to 5 days. SIM microscopy also enabled identification of subcellular organelles with respect to the localization of the nanocarriers. Additionally, nanoSIMS imaging allowed the simultaneous detection of the distribution of the nanocarrier and Pt drug in correlation to endogenous ions such as 31P, a typical ion found in DNA and nucleus (Fig. 3B). The data showed that a much stronger signal of the drug was obtained in the tissue containing the nanocarrier compared to the control. Moreover, the Pt drug was found to accumulate in the nucleus and to not co-localize with the nanocarrier, meaning that the Pt drug effectively dissociated from the nanocarriers and reached the DNA target in the nucleus. More applications of SIMS in pharmaceutical analysis can be found in selected review literature.51,52
Brain diseases and pathology
Brain diseases, including neurodegenerative disorders, are the consequence of the death or progressive degeneration and alteration of the functions of neurons. Despite being one of the most severe disease classes, knowledge on their underlying molecular mechanisms has been limited. SIMS has been increasingly recognized as a valuable diagnostic tool as it provides specific chemical information related to these diseases with subcellular resolution. In addition, SIMS can serve either as a non-target and label-free technique for screening purposes, when pre-knowledge of the diseases and samples is limited, or as a targeted imaging method when specific biomarkers are used.A method using ToF-SIMS and 25 keV Bi3+ has been developed to detect amyloid-β (Aβ), a characteristic protein aggregate in Alzheimer's's disease (AD), in transgenic mouse brain by the use of antibody conjugated liposomes, namely immunoliposomes.53 The liposome, about 200 nm in size, consists of a lipid bilayer with 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 lipid molecules that allowed significant amplification of the protein signal. The liposome was coupled to an antibody, 6E10, which binds to only a specific protein, Aβ. To distinguish the signal of the immunoliposome from that of the endogenous lipids, the liposome was made with deuterated lipids. Specific binding of the immunoliposomes to the Aβ aggregate was first confirmed using a quartz crystal microbalance with dissipation, QCM-D, monitoring and fluorescence microscopy on a model surface of Aβ and AD mouse brain tissue. The mouse brain was incubated with the immunoliposomes and imaged with ToF-SIMS after being fixed and freeze dried. The SIMS images showed a clear signal of the deuterated lipids representing Aβ aggregates which were found to localize in the cortex and hippocampus. The localization was in good agreement with the results obtained by fluorescence microscopy imaging. The method is promising for the study of the structural relationship between Aβ aggregates and endogenous lipids; however, further improvements in sample preparation are needed in order to preserve the distribution of lipids.
000 lipid molecules that allowed significant amplification of the protein signal. The liposome was coupled to an antibody, 6E10, which binds to only a specific protein, Aβ. To distinguish the signal of the immunoliposome from that of the endogenous lipids, the liposome was made with deuterated lipids. Specific binding of the immunoliposomes to the Aβ aggregate was first confirmed using a quartz crystal microbalance with dissipation, QCM-D, monitoring and fluorescence microscopy on a model surface of Aβ and AD mouse brain tissue. The mouse brain was incubated with the immunoliposomes and imaged with ToF-SIMS after being fixed and freeze dried. The SIMS images showed a clear signal of the deuterated lipids representing Aβ aggregates which were found to localize in the cortex and hippocampus. The localization was in good agreement with the results obtained by fluorescence microscopy imaging. The method is promising for the study of the structural relationship between Aβ aggregates and endogenous lipids; however, further improvements in sample preparation are needed in order to preserve the distribution of lipids.
To investigate the effect of traumatic brain injury on the molecular structure of the brain, Tian et al.54 developed a dynamic reactive ionization method for ToF-SIMS with a 20 keV (CO2)3500+ GCIB via treatment of the brain tissue with 1-ethyl-3-[(dimethylamino) propyl] carbodiimide hydrochloride and phospholipase C (EDC/PLC). The EDC/PLC sample treatment was shown to suppress the signal of highly abundant phospholipids, especially PCs, and enhance the signal of high mass cardiolipin (CL) (m/z 1300–1530) and gangliosides (GM1, GD1, and GT1) (m/z > 1500) in the brain. The entire brain sections were imaged to observe the localization of cardiolipin and gangliosides in the EDC/PLC treated brain. It was found that the CLs with longer carbon chains and more double bonds were dominant in the cortex, whereas those with shorter carbon chains and less double bonds localized in the hippocampus. In contrast, the gangliosides with a 18:0 fatty acid distributed equally between the cortex and hippocampus while the ones with a 20:0 fatty acid were localized in the hippocampus at higher abundance. The localization of these lipids was observed within the substructures inside the hippocampus with a spatial resolution of 8 μm. The injured brain section was imaged showing that there was a loss of CLs and GT1 in the injured brain compared to the control. The most significant loss was polyunsaturated CLs in the cornu ammonis of the hippocampus and thalamus. These regions are known to be involved in memory formation, seizure, and neurodegeneration.54 This indicated that there might be a relationship between CLs and cognition.
Cisplatin has been one of the common chemotherapy drugs for cancer treatment; however, the drug leads to various side effects in patients.55 To understand the chemical effects of cisplatin at the cellular level, ToF-SIMS with a 40 keV (CO2)6000+ GCIB was utilized to image freeze-dried PC12 cells treated with cisplatin.56 It was found that cisplatin caused significant changes in the lipid composition of the cell surface – a dramatic decrease in the amount of PCs was noticed, along with their salt adducts with sodium and potassium ions, and cholesterol, but significant elevation of diacylglycerols (DAGs) was observed. The results were explained based on the signal transduction pathways of sphingomyelinases and the induction of apoptosis by cisplatin which are involved in the synthesis and metabolism of membrane lipids.
Cocaine and methylphenidate are both known psychostimulants; however, they exhibit opposite cognitive effects.57,58 While cocaine causes deficits in attention, memory and behavior, methylphenidate shows improvement in attention, focus and learning. The reason behind this was explored by a study of the effects of these drugs on the molecular structure of Drosophila brains using ToF-SIMS imaging.59 Two sets of flies were treated with these drugs in parallel for three days and then embedded, frozen, and cut into thin slices. The frozen hydrated brain sections were then analyzed with SIMS using a 40 keV Ar4000+ GCIB. It was shown that the lipid composition of the fly brains, both the localization and concentration, was altered dramatically after the drug treatments (Fig. 3C). The most noticeable aspect is that these drugs induced opposite trends. For example a significant increase in the abundance of lamellar shaped PCs and a decrease in conical shaped PEs and PIs were observed in the cocaine-treated brain. In contrast, PCs decreased and PEs and PIs elevated considerably in the methylphenidate-treated brain (Fig. 3C). The data suggest that the lipid structure of the brain is involved in the molecular mechanism of the drugs’ effects on the brain and might be involved in cognition.
SIMS analysis has great potential for cell imaging, including localization of pharmaceuticals in cells, and neuro-biological research because it provides comprehensive molecular information about the properties and effects of endogenous and exogenous elements with high spatial resolution.
Correlative SIMS and other imaging techniques
In order to complement the information that cannot be obtained by SIMS, particularly the morphological structures and the localizations of proteins and organelles in biological samples, different groups have developed techniques that correlate the data recorded by SIMS with those by other techniques, such as fluorescence microscopy, EM, atomic force microscopy (AFM), X-ray spectroscopy, and MALDI. We describe some of these correlative approaches, elaborate on the additional information provided by each and give examples of the biological questions answered by them.NanoSIMS and super-resolution microscopy
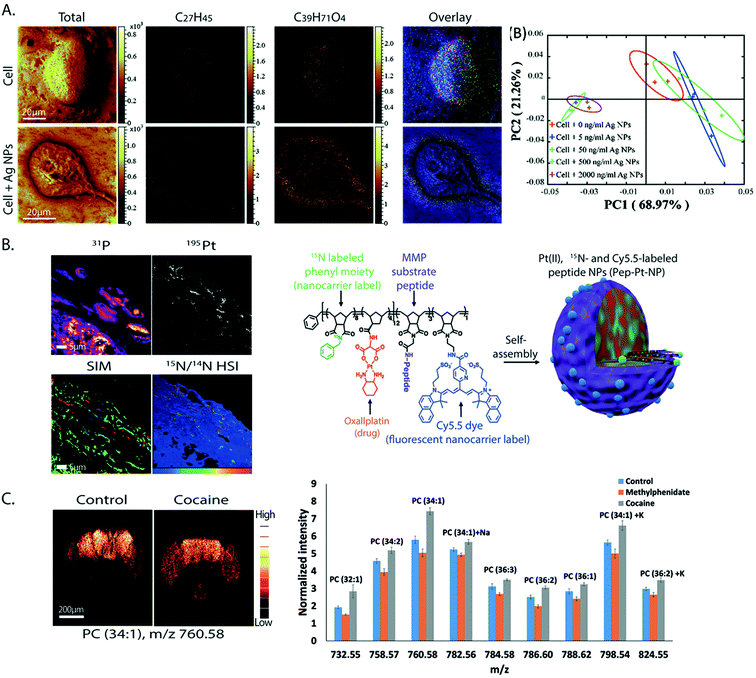
From all the SIMS techniques, nanoSIMS provides unsurpassed lateral resolution. NanoSIMS has been combined with super-resolution stimulated emission depletion (STED) microscopy to correlate the protein turnover to specific molecular targets and subcellular structures of hippocampal neurons, which were obtained by nanoSIMS measurements and STED microscopy, respectively.6015N-Leucine was incorporated into the newly synthesized proteins for tracking the protein turnover. Different synaptic proteins such as synaptophysin 1, bassoon, and the mitochondrial marker TOMM20 were labeled and visualized by STED microscopy. The correlated data showed that the protein turnover in the synapse was higher than that in the axonal area (Fig. 4A). A combination of nanoSIMS and fluorescence microscopy has been employed in various studies such as an investigation of the effect of erythropoietin on neurogenesis in the brain and aging of synaptic vesicles.61,62 | ||
| Fig. 4 Correlative imaging of SIMS with other imaging modalities. (A) Correlative SIMS and STED to study the protein turnover in hippocampal neurons. Top row, from left to right: fluorescence microscopy image of the Golgi marker TGN38 (green), synaptic vesicle marker synaptophysin 1 (red), and endoplasmic reticulum marker calnexin (blue) and an overlay of these proteins. Bottom row, from left to right: STED image of calnexin, nanoSIMS ratio image of 15N/14N showing the localization of the turnover, and a chart showing the turnover rate in different organelles. Reproduced from ref. 60 with permission. (B) Correlation of TEM and nanoSIMS imaging to study the distribution of dopamine loading inside single vesicles of PC12 by treatment with 13C-L-DOPA and reserpine. Top row, from left to right: the TEM image, nanoSIMS image of 12C14N, and ratio image of 13C14N/12C14N showing the dopamine enrichment in the vesicles (indicated by red arrows). Bottom row, from left to right: schematic of a dense core vesicle; 3D surface plots of TEM signals and nanoSIMS signal of 13C14N. Reproduced from ref. 63 with permission. (C) Correlation of SIMS and MALDI imaging to visualize the molecular distribution of human cerebellum. From left to right: the SIMS image (red: cholesterol at m/z 386 and green: PC headgroup at m/z 184); MALDI image (green: m/z 3926, blue: m/z 1756 and red: m/z 2817); and tissue stained with H&E (light pink: grey matter, pink: white matter, and purple: granular layer). Reproduced from ref. 68 with permission. | ||
NanoSIMS and electron microscopy
NanoSIMS has also been combined with EM. EM provides information on the morphological structure of cells and tissues with a very high spatial resolution (low nm scale). A combination of nanoSIMS and TEM has been used to investigate the distribution of the neurotransmitter dopamine inside the dense core vesicles (Fig. 4B).63 It was shown that the distribution of dopamine inside the dense core can be altered after a treatment with reserpine, a drug for depleting dopamine from vesicles, and that the equilibration between intravesicular compartments is slow in closed vesicles.EM can also be used to obtain elemental information in the energy-dispersive X-ray (EDX) microanalysis mode or electron energy loss spectroscopy (EELS) mode. These EM methods have been combined with nanoSIMS to study neuromelanin granules in the substantia nigra.64 EDX/EELS provided the ultrastructure and general elemental composition, whereas nanoSIMS added information about specific trace elements and isotopes. However, the correlation of SIMS with EM can be challenging, as the sample preparation must be compatible to both techniques. Furthermore, care must be taken to correlate the same regions for analysis. To overcome these problems, an instrument combining both TEM and SIMS has been developed.65 In addition, image fusion techniques have recently been tested and improved for their application in correlative imaging. For example, a Laplacian pyramid fusion method, a method for combining the image information from different sources in order to provide comprehensive visual information with improved clarity and accuracy compared to the individual sources, significantly improved the fusion between nanoSIMS and TEM images.66
ToF-SIMS and other imaging techniques
SIMS has also been combined with other MSI techniques, for example with nanoparticle assisted laser desorption ionization (NP-LDI) to study the intact lipids in mouse brain tissue.67 The brain samples were first sprayed with Au NPs to facilitate molecular ionization before being imaged with ToF-SIMS and a Ar4000+ GCIB, followed by NP-LDI analysis. Both approaches found common but also newly detected species, thus complementing each other. Another study also employed ToF-SIMS and GCIB combined with MALDI to study the molecular distribution in human cerebellum tissues (Fig. 4C).68 ToF-SIMS is ideal for detecting lipids, drugs, and metabolites with a m/z of up to 2000 Da, whereas MALDI allows the analysis of intact lipids, peptides, and proteins. By combining these two techniques, one can achieve the best of both worlds in an extended mass range of detection. ToF-SIMS has also been combined with nanoSIMS where ToF-SIMS was used as a screening tool and nanoSIMS was then performed to study the structures of interest with higher spatial details. This was applied to study the preservation conditions and to look at the subcellular distribution of OsO4 in chemically fixed adrenal cells. The lysosome was identified as a main accumulation region of OsO4.69Veith and colleagues combined SIMS with fast micro X-ray fluorescence spectroscopy (μ-TRF) to detect CeO2 particles in biological tissue sections.70 μ-TRF allows a fast elemental analysis of big tissues, such as lung tissue. Once a region of interest was defined by μ-TRF, ToF-SIMS was used to analyze the organic and inorganic molecules at higher lateral resolution. Finally, for 3D SIMS imaging, since the sputtering of the sample is not uniform and depends on different parameters, it is not easy to estimate the 3D-volume of analysis by SIMS alone. Thus, SIMS could be combined with AFM, which helped to measure the 3D topography of the sample surface. Combining both techniques, Terlier et al.71 demonstrated an improvement in the accuracy of 3D-reconstruction as well as in the correlation between the chemical information and the physical properties of the sample surface.
Conclusions and perspectives
SIMS imaging has been demonstrated as a powerful and valuable analytical technique for research in neurobiology and cell biology. It offers the unique advantages of providing information-rich data, with both target and non-target analytical approaches, chemical specificity, high spatial resolution, and high sensitivity of detection. SIMS has been and is still undergoing fast and continuous technological developments to facilitate its increasing applications in the biological field in general, and in neurobiology and cell biology in particular.Regarding the detection capability, primary ion sources with a higher energy, smaller beam size and softer ionization capable of detecting intact biomolecules in an extended mass range are still a main focus of development. Spatial resolution is expected to not only be sufficient for a single cell but also reach beyond to the single- and sub-organelle level. In addition, methods for improving the ionization efficiency, including improving secondary ion yield and sensitivity, such as the modifications of sample surfaces with reactive reagents are highly desired.
For chemical identification, there have been considerable developments with tandem MS SIMS for elucidating molecular structures in complex biological samples; however, this area is still in the stage of proof of principle. Further effort is needed to improve the spatial resolution and detection mass range, which are the current limitations of the technique, before it will find wide use in bioimaging.
Development of targeted imaging methodologies especially for large biomolecules such as proteins has been a new goal for SIMS imaging. The main challenge for this is the sensitivity and selectivity of detection to obtain both a high signal-to-noise ratio for the molecules of interest and high spatial resolution. Although there have been a few significant studies in the development of effective labeling probes for SIMS, further investigation is desirable to allow simultaneous imaging of different proteins and lipids at subcellular spatial resolution.
Finally, SIMS imaging produces very large datasets which contain a full spectrum of hundreds of peaks for an individual pixel of an entire imaged area. Proper data handling and reliable statistical analysis are in high demand. Development of a standardization method for data treatment, including criteria of sample quantity, data normalization and pre-treatment, statistical analysis, as well as more databases for mass peak assignment have been ongoing and this will contribute to be a major part of the efforts to enable SIMS imaging to become a routine analytical method in neurobiology and cell biology.
Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank Professor S. O. Rizzoli for very helpful comments on the review. We acknowledge the grants from DFG (SFB1286/B1) and VR (Swedish Research Council) to N. T. N. Phan. We thank many colleagues and collaborators that have contributed to the work cited in this review.References
- J. Fletcher, Latest Applications of 3D ToF-SIMS Bio-imaging, Biointerphases, 2015, 10(1), 018902 CrossRef PubMed.
- J. Hanrieder, N. T. N. Phan, M. E. Kurczy and A. G. Ewing, Imaging Mass Spectrometry in Neuroscience, ACS Chem. Neurosci., 2013, 4(5), 666–679 CrossRef CAS PubMed.
- H. Jungnickel, P. Laux and A. Luch, Time-of-Flight Secondary Ion Mass Spectrometry (ToF-SIMS): A New Tool for the Analysis of Toxicological Effects on Single Cell Level, Toxics, 2016, 4(1), 5 CrossRef PubMed.
- J. C. Vickerman and D. Briggs, ToF-SIMS: Surface Analysis by Mass Spectrometry, IM Pulication, 2013 Search PubMed.
- N. J. Popczun, L. Breuer, A. Wucher and N. Winograd, On the SIMS Ionization Probability of Organic Molecules, J. Am. Soc. Mass Spectrom., 2017, 28(6), 1182–1191 CrossRef CAS PubMed.
- D. Weibel, S. Wong, N. Lockyer, P. Blenkinsopp, R. Hill and J. C. Vickerman, A C60 Primary Ion Beam System for Time of Flight Secondary Ion Mass Spectrometry: Its Development and Secondary Ion Yield Characteristics, Anal. Chem., 2003, 75(7), 1754–1764 CrossRef CAS.
- F. Kollmer, Cluster primary ion bombardment of organic materials, Appl. Surf. Sci., 2004, 231–232, 153–158 CrossRef CAS.
- N. Toyoda, J. Matsuo, T. Aoki, I. Yamada and D. B. Fenner, Secondary Ion Mass Spectrometry with Gas Cluster Ion Beams, Appl. Surf. Sci., 2003, 203, 214–218 CrossRef.
- H. Tian, D. Maciążek, Z. Postawa, B. J. Garrison and N. Winograd, CO2 Cluster Ion Beam, an Alternative Projectile for Secondary Ion Mass Spectrometry, J. Am. Soc. Mass Spectrom., 2016, 27(9), 1476–1482 CrossRef CAS PubMed.
- S. S. Rabbani, A. Barber, J. S. Fletcher, N. P. Lockyer and J. C. Vickerman, Enhancing Secondary Ion Yields in Time of Flight-Secondary Ion Mass Spectrometry Using Water Cluster Primary Beams, Anal. Chem., 2013, 85(12), 5654–5658 CrossRef PubMed.
- A. Benninghoven, Chemical Analysis of Inorganic and Organic Surfaces and Thin Films by Static Time-of-Flight Secondary Ion Mass Spectrometry (TOF-SIMS), Angew. Chem., Int. Ed. Engl., 1994, 33(10), 1023–1043 CrossRef.
- M. Steinhauser and C. Lechene, Quantitative Imaging of Subcellular Metabolism with Stable Isotopes and Multi-Isotope Imaging Mass Spectrometry, Semin. Cell Dev. Biol., 2013, 24(8–9), 661–667 CrossRef CAS PubMed.
- M. E. Kurczy, P. D. Piehowski, S. A. Parry, M. Jiang, G. Chen and A. G. Ewing, et al., Which is More Important in Bioimaging SIMS Experiments-The Sample Preparation or The Nature of The Projectile?, Appl. Surf. Sci., 2008, 255(4), 1298–1304 CrossRef CAS.
- M. Passarelli and N. Winograd, Lipid Imaging with Time-of-Flight Secondary Ion Mass Spectrometry (ToF-SIMS), Biochim. Biophys. Acta, Mol. Cell Biol. Lipids, 2011, 1811(11), 976–990 CrossRef CAS.
- M. Kraft and H. Klitzing, Imaging Lipids with Secondary Ion Mass Spectrometry, Biochim. Biophys. Acta, Mol. Cell Biol. Lipids, 2014, 1841(8), 1108–1119 CrossRef CAS PubMed.
- J. S. Fletcher, S. Rabbani, A. Henderson, P. Blenkinsopp, S. P. Thompson and N. P. Lockyer, et al., A New Dynamic in Mass Spectral Imaging of Single Biological Cells, Anal. Chem., 2008, 80(23), 9058–9064 CrossRef CAS PubMed.
- Q. P. Vanbellingen, N. Elie, M. J. Eller, S. Della-Negra, D. Touboul and A. Brunelle, Time-of-flight Secondary Ion Mass Spectrometry Imaging of Biological Samples with Delayed Extraction for High Mass and High Spatial Resolutions, Rapid Commun. Mass Spectrom., 2015, 29(13), 1187–1195 CrossRef CAS PubMed.
- R. N. S. Sodhi, Time-of-flight Secondary Ion Mass Spectrometry (TOF-SIMS):—Versatility in Chemical and Imaging Surface Analysis, Analyst, 2004, 129, 483–487 RSC.
- J. Nuñez, R. Renslow, J. B. Cliff and C. R. Anderton, NanoSIMS for Biological Applications: Current Practices and Analyses, Biointerphases, 2018, 13(3), 03B301 CrossRef PubMed.
- J. L. Guerquin-Kern, T. D. Wu, C. Quintana and A. Croisy, Progress in Analytical Imaging of The Cell by Dynamic Secondary Ion Mass Spectrometry (SIMS Microscopy), Biochim. Biophys. Acta, Gen. Subj., 2005, 1724(3), 228–238 CrossRef CAS PubMed.
- J. Malherbe, F. Penen, M.-P. Isaure, J. Frank, G. Hause and D. Dobritzsch, et al., A New Radio Frequency Plasma Oxygen Primary Ion Source on Nano Secondary Ion Mass Spectrometry for Improved Lateral Resolution and Detection of Electropositive Elements at Single Cell Level, Anal. Chem., 2016, 88(14), 7130–7136 CrossRef CAS PubMed.
- D. F. Smith, A. Kiss, F. E. Leach, E. W. Robinson, L. Paša-Tolić and R. M. A. Heeren, High Mass Accuracy and High Mass Resolving Power FT-ICR Secondary Ion Mass Spectrometry for Biological Tissue Imaging, Anal. Bioanal. Chem., 2013, 405(18), 6069–6076 CrossRef CAS PubMed.
- A. Carado, J. Kozole, M. Passarelli, N. Winograd and A. Loboda, J. Wingate. Cluster SIMS With a Hybrid Quadrupole Time-of-Flight Mass Spectrometer, Appl. Surf. Sci., 2008, 255(4), 1610–1613 CrossRef CAS.
- N. T. N. Phan, M. Munem, A. G. Ewing and J. S. Fletcher, MS/MS Analysis and Imaging of Lipids Across Drosophila Brain Using Secondary Ion Mass Spectrometry, Anal. Bioanal. Chem., 2017, 409(16), 3923–3932 CrossRef CAS PubMed.
- G. L. Fisher, A. L. Bruinen, N. O. Potočnik, J. S. Hammond, S. R. Bryan and P. E. Larson, et al., A New Method and Mass Spectrometer Design for TOF-SIMS Parallel Imaging MS/MS, Anal. Chem., 2016, 88(12), 6433–6440 CrossRef CAS PubMed.
- N. O. Potočnik, G. L. Fisher, A. Prop and R. M. A. Heeren, Sequencing and Identification of Endogenous Neuropeptides with Matrix-Enhanced Secondary Ion Mass Spectrometry Tandem Mass Spectrometry, Anal. Chem., 2017, 89(16), 8223–8227 CrossRef PubMed.
- M. K. Passarelli, A. Pirkl, R. Moellers, D. Grinfeld, F. Kollmer and R. Havelund, et al., The 3D OrbiSIMS—Label-free Metabolic Imaging with Subcellular Lateral Resolution and High Mass-resolving Power, Nat. Methods, 2017, 14(12), 1175–1183 CrossRef CAS PubMed.
- A. Haase, H. F. Arlinghaus, J. Tentschert, H. Jungnickel, P. Graf and A. Mantion, et al., Application of Laser Postionization Secondary Neutral Mass Spectrometry/Time-of-Flight Secondary Ion Mass Spectrometry in Nanotoxicology: Visualization of Nanosilver in Human Macrophages and Cellular Responses, ACS Nano, 2011, 5(4), 3059–3068 CrossRef CAS PubMed.
- L. Breuer L, H. Tian, A. Wucher and N. Winograd, Molecular SIMS Ionization Probability Studied with Laser Postionization: Influence of the Projectile Cluster, J. Phys. Chem. C, 2018, 123(1), 565–574 CrossRef.
- M. Heeger, B. J. Tyler, M. Körsgen and H. F. Arlinghaus, Laser Postionization of Neutral Molecules Sputtered Using Bismuth and Argon Cluster Primary Ions, Biointerphases, 2018, 13(3), 03B412 CrossRef PubMed.
- M. Angelo, S. C. Bendall, R. Finck, M. B. Hale, C. Hitzman and A. D. Borowsky, et al., Multiplexed ion beam imaging of human breast tumors, Nat. Med., 2014, 20, 436–442 CrossRef CAS.
- G. Thiery-Lavenant, C. Guillermier, M. Wang and C. Lechene, Detection of immunolabels with multi-isotope imaging mass spectrometry (MIMS), Surf. Interface Analysis., 2014, 46(S1), 147–149 CrossRef CAS PubMed.
- I. C. Vreja, S. Kabatas, S. K. Saka, K. Kröhnert, C. Höschen and F. Opazo, et al., Secondary-Ion Mass Spectrometry of Genetically Encoded Targets, Angew. Chem., Int. Ed., 2015, 54(19), 5784–5788 CrossRef CAS PubMed.
- S. Kabatas, P. Agüi-Gonzalez, K. A. Saal, S. Jähne, F. Opazo and S. O. Rizzoli, et al., Boron-Containing Probes for Non-optical High-Resolution Imaging of Biological Samples, Angew. Chem., Int. Ed., 2019, 58, 3438–3443 CrossRef CAS PubMed.
- S. G. Ostrowski, C. T. Van Bell, N. Winograd and A. G. Ewing, Mass Spectrometric Imaging of Highly Curved Membranes during Tetrahymena Mating, Science, 2004, 305(5680), 71–73 CrossRef CAS PubMed.
- M. K. Passarelli, A. G. Ewing and N. Winograd, Single-Cell Lipidomics: Characterizing and Imaging Lipids on the Surface of Individual Aplysia californica Neurons with Cluster Secondary Ion Mass Spectrometry, Anal. Chem., 2013, 85(4), 2231–2238 CrossRef CAS PubMed.
- E. B. Monroe, J. C. Jurchen, J. Lee, S. S. Rubakhin and J. V. Sweedler, Vitamin E Imaging and Localization in the Neuronal Membrane, J. Am. Chem. Soc., 2005, 127(35), 12152–12153 CrossRef CAS PubMed.
- A. C. Burduşel, O. Gherasim, A. M. Grumezescu, L. Mogoantă L, A. Ficai and E. Andronescu, Biomedical Applications of Silver Nanoparticles: An Up-to-Date Overview, Nanomaterials, 2018, 8(9), 681 CrossRef PubMed.
- X. Hua, H. W. Li and Y. T. Long, Investigation of Silver Nanoparticle Induced Lipids Changes on a Single Cell Surface by Time-of-Flight Secondary Ion Mass Spectrometry, Anal. Chem., 2018, 90(2), 1072–1076 CrossRef CAS PubMed.
- J. S. Fletcher, S. Rabbani, A. Henderson, N. P. Lockyer and J. C. Vickerman, Three-dimensional Mass Spectral Imaging of HeLa-M Cells–sample Preparation, Data Interpretation and Visualisation, Rapid Commun. Mass Spectrom., 2011, 25(7), 925–932 CrossRef CAS PubMed.
- T. B. Angerer and J. S. Fletcher, 3D Imaging of TiO2 nanoparticle accumulation in Tetrahymena pyriformis, Surf. Interface Anal., 2014, 46(S1), 198–203 CrossRef CAS.
- A. N. Yeager, P. K. Weber and M. L. Kraft, Three-dimensional imaging of cholesterol and sphingolipids within a Madin-Darby canine kidney cell, Biointerphases, 2016, 11(2), 02A309 CrossRef PubMed.
- J. S. Fletcher, Latest applications of 3D ToF-SIMS bio-imaging, Biointerphases, 2015, 10(1), 018902 CrossRef PubMed.
- M. A. Robinson, D. J. Graham and D. G. Castner, ToF-SIMS Depth Profiling of Cells: Z-correction, 3D Imaging, and Sputter Rate of Individual NIH/3T3 Fibroblasts, Anal. Chem., 2012, 84(11), 4880–4885 CrossRef CAS PubMed.
- J. S. Fletcher, J. C. Vickerman and N. Winograd, Label free biochemical 2D and 3D imaging using secondary ion mass spectrometry, Curr. Opin. Chem. Biol., 2011, 15(5), 733–740 CrossRef CAS PubMed.
- J. S. Fletcher, Latest applications of 3D ToF-SIMS bio-imaging, Biointerphases, 2015, 10(1), 018902 CrossRef PubMed.
- M. K. Passarelli, C. F. Newman, P. S. Marshall, A. West, I. S. Gilmore and J. Bunch, et al., Single-Cell Analysis: Visualizing Pharmaceutical and Metabolite Uptake in Cells with Label-Free 3D Mass Spectrometry Imaging, Anal. Chem., 2015, 87(13), 6696–6702 CrossRef CAS PubMed.
- H. Jiang, M. K. Passarelli, P. M. G. Munro, M. R. Kilburn, A. West and C. T. Dollery, et al., High-resolution Sub-cellular Imaging by Correlative NanoSIMS and Electron Microscopy of Amiodarone Internalisation by Lung Macrophages as Evidence for Drug-induced Phospholipidosis, Chem. Commun., 2017, 53(9), 1506–1509 RSC.
- H. Tian, D. A. Six, T. Krucker, J. A. Leeds and N. Winograd, Subcellular Chemical Imaging of Antibiotics in Single Bacteria Using C60-Secondary Ion Mass Spectrometry, Anal. Chem., 2017, 89(9), 5050–5057 CrossRef CAS PubMed.
- M. T. Proetto, C. E. Callmann, J. Cliff, C. J. Szymanski, D. Hu and S. B. Howell, et al., Tumor Retention of Enzyme-Responsive Pt(II) Drug-Loaded Nanoparticles Imaged by Nanoscale Secondary Ion Mass Spectrometry and Fluorescence Microscopy, ACS Cent. Sci., 2018, 4(11), 1477–1484 CrossRef CAS PubMed.
- Y. P. Kim, H. K. Shon, S. K. Shin and T. G. Lee, Probing nanoparticles and nanoparticle-conjugated biomolecules using time-of-flight secondary ion mass spectrometry, Mass Spectrom. Rev., 2015, 34(2), 237–247 CrossRef CAS.
- Q. P. Vanbellingen, A. Castellanos, M. Rodríguez-Silva, I. Paudel, J. W. Chambers and F. A. Fernandez-Lima, Analysis of Chemotherapeutic Drug Delivery at The Single Cell Level Using 3D-MSI-TOF-SIMS, J. Am. Soc. Mass Spectrom., 2016, 27(12), 2033–2040 CrossRef CAS.
- L. Carlred, V. Vukojević, B. Johansson, M. Schalling, F. Höök and P. Sjövall, Imaging of Amyloid-β in Alzheimer's Disease Transgenic Mouse Brains with ToF-SIMS Using Immunoliposomes, Biointerphases, 2016, 11(2), 02A312 CrossRef.
- H. Tian, L. J. Sparvero, A. A. Amoscato, A. Bloom, H. Bayır and V. E. Kagan, et al., Gas Cluster Ion Beam Time-of-Flight Secondary Ion Mass Spectrometry High-Resolution Imaging of Cardiolipin Speciation in the Brain: Identification of Molecular Losses after Traumatic Injury, Anal. Chem., 2017, 89(8), 4611–4619 CrossRef CAS PubMed.
- T. A. Ahles, J. C. Root and E. L. Ryan, Cancer- and Cancer Treatment-associated Cognitive Change: an Update on The State of The Science, J. Clin. Oncol., 2012, 30(30), 3675–3686 CrossRef CAS PubMed.
- A. S. Mohammadi, X. Li and A. G. Ewing, Mass Spectrometry Imaging Suggests That Cisplatin Affects Exocytotic Release by Alteration of Cell Membrane Lipids, Anal. Chem., 2018, 90(14), 8509–8516 CrossRef CAS.
- D. Jovanovski, S. Erb and K. K. Zakzanis, Neurocognitive Deficits in Cocaine Users: a Quantitative Review of the Evidence, J. Clin. Exp. Neuropsychol., 2005, 27(2), 189–204 CrossRef.
- A. F. Arnsten and A. G. Dudley, Methylphenidate Improves Prefrontal Cortical Cognitive Function Through Alpha2 Adrenoceptor and Dopamine D1 Receptor Actions: Relevance to Therapeutic Effects in Attention Deficit Hyperactivity Disorder, Behav. Brain Funct., 2005, 1(1), 2 CrossRef PubMed.
- M. H. Philipsen, N. T. N. Phan, J. S. Fletcher, P. Malmberg and A. G. Ewing, Mass Spectrometry Imaging Shows Cocaine and Methylphenidate Have Opposite Effects on Major Lipids in Drosophila Brain, ACS Chem. Neurosci., 2018, 9(6), 1462–1468 CrossRef CAS PubMed.
- S. K. Saka, A. Vogts, K. Kröhnert, F. Hillion, S. O. Rizzoli and J. T. Wessels, Correlated Optical and Isotopic Nanoscopy, Nat. Commun., 2014, 5, 3664 CrossRef.
- I. Hassouna, C. Ott, L. Wüstefeld, N. Offen, R. A. Neher and M. Mitkovski, et al., Revisiting Adult Neurogenesis and The Role of Erythropoietin for Neuronal and Oligodendroglial Differentiation in The Hippocampus, Mol. Psychiatry, 2016, 21(12), 1752–1767 CrossRef CAS.
- X. Hua, C. Szymanski, Z. Wang, Y. Zhou, X. Ma and J. Yu, et al., Chemical Imaging of Molecular Changes in a Hydrated Single Cell by Dynamic Secondary Ion Mass Spectrometry and Super-resolution Microscopy, Integr. Biol., 2016, 8(5), 635–644 CrossRef CAS.
- J. Lovrić, J. Dunevall, A. Larsson, L. Ren, S. Andersson and A. Meibom, et al., Nano Secondary Ion Mass Spectrometry Imaging of Dopamine Distribution Across Nanometer Vesicles, ACS Nano, 2017, 11(4), 3446–3455 CrossRef.
- A. Biesemeier, O. Eibl, S. Eswara, J. N. Audinot, T. Wirtz and G. Pezzoli, et al., Elemental Mapping of Neuromelanin Organelles of Human Substantia Nigra: Correlative Ultrastructural and Chemical Analysis by Analytical Transmission Electron Microscopy and Nano-secondary Ion Mass Spectrometry, J. Neurochem., 2016, 138(2), 339–353 CrossRef CAS.
- L. Yedra, S. Eswara, D. Dowsett and T. Wirtz, In-situ Isotopic Analysis at Nanoscale using Parallel Ion Electron Spectrometry: A Powerful New Paradigm for Correlative Microscopy, Sci. Rep., 2016, 6, 28705 CrossRef CAS PubMed.
- F. Vollnhals, J. N. Audinot, T. Wirtz, M. Mercier-Bonin, I. Fourquaux and B. Schroeppel, et al., Correlative Microscopy Combining Secondary Ion Mass Spectrometry and Electron Microscopy: Comparison of Intensity-Hue-Saturation and Laplacian Pyramid Methods for Image Fusion, Anal. Chem., 2017, 89(20), 10702–10710 CrossRef CAS PubMed.
- A. S. Mohammadi, N. T. N. Phan, J. S. Fletcher and A. G. Ewing, Intact Lipid Imaging of Mouse Brain Samples: MALDI, Nanoparticle-laser Desorption Ionization, and 40 keV Argon Cluster Secondary Ion Mass Spectrometry, Anal. Bioanal. Chem., 2016, 408(24), 6857–6868 CrossRef CAS.
- G. B. Eijkel, B. K. Kaletaş, I. M. Van Der Wiel, J. M. Kros, T. M. Luider and R. M. A. Heeren, Correlating MALDI and SIMS Imaging Mass Spectrometric Datasets of Biological Tissue Surfaces, Surf. Interface Anal., 2009, 41(8), 675–685 CrossRef CAS.
- J. Lovrić, P. Malmberg, B. R. Johansson, J. S. Fletcher and A. G. Ewing, Multimodal Imaging of Chemically Fixed Cells in Preparation for NanoSIMS, Anal. Chem., 2016, 88(17), 8841–8848 CrossRef PubMed.
- L. Veith, D. Dietrich, A. Vennemann, D. Breitenstein, C. Engelhard and U. Karst, et al., Combination of Micro X-ray Fluorescence Spectroscopy and Time-of-flight Secondary Ion Mass Spectrometry Imaging for The Marker-free Detection of CeO2 Nanoparticles in Tissue Sections, J. Anal. At. Spectrom., 2018, 33(3), 491–501 RSC.
- T. Terlier, J. Lee, K. Lee and Y. Lee, Improvement of the Correlative AFM and ToF-SIMS Approach Using an Empirical Sputter Model for 3D Chemical Characterization, Anal. Chem., 2018, 90(3), 1701–1709 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2019 |