Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Muconic acid production from methane using rationally-engineered methanotrophic biocatalysts†
Calvin A.
Henard‡
 *a,
Ilya R.
Akberdin§
*a,
Ilya R.
Akberdin§
 bc,
Marina G.
Kalyuzhnaya
bc,
Marina G.
Kalyuzhnaya
 b and
Michael T.
Guarnieri
b and
Michael T.
Guarnieri
 *a
*a
aNational Bioenergy Center, National Renewable Energy Laboratory, Golden, CO, USA. E-mail: Michael.Guarnieri@nrel.gov; Calvin.Henard@unt.edu
bBiology Department, San Diego State University, San Diego, CA, USA
cFederal Research Center Institute of Cytology and Genetics SB RAS, Novosibirsk, Russia
First published on 25th November 2019
Abstract
Here, we demonstrate bioconversion of methane to muconic acid, a dicarboxylic acid that can be upgraded to an array of platform chemicals, by three gammaproteobacterial methanotrophs. All engineered methanotrophs expressing a heterologous dihydroxyshikimate dehydratase, protocatechuate decarboxylase, and catechol dioxygenase produced muconic acid from methane, with the highest titer (12.4 mg MA per L), yield (2.8 mg MA per g CH4), and specific productivity (1.2 mg MA per g dcw, 48 hr) synthesized by Methylotuvimicrobium buryatense, Methylococcus capsulatus, and Methylotuvimicrobium alcaliphilium, respectively. Methylotuvimicrobium alcaliphilum genome-scale model-guided strain engineering predicted that disruption of the pyruvate dehydrogenase or shikimate dehydrogenase would significantly enhance flux to the heterologous muconic acid pathway in this organism. However, knock-out of these targets caused a growth defect, and coupled with similar muconic acid titers (∼1 mg L−1), resulted in minimal flux enhancement to muconic acid in these genetically-modified strains. The shikimate dehydrogenase mutant's ability to grow without aromatic amino acid supplementation revealed that M. alcaliphilum likely encodes an unidentified enzyme or pathway with shikimate biosynthetic capacity, which prevents maximal flux through the synthetic muconic acid pathway. This study expands the suite of products that can be generated from methane using methanotrophic biocatalysts, lays the foundation for green production of muconic acid-derived polymers from methane, and highlights the need for further analysis of methanotroph biosynthetic potential to guide refinement of metabolic models and strain engineering.
Introduction
Muconic acid (MA) is a dicarboxylic acid that can be upgraded to produce high-value commodity chemicals, including terepthalic acid, which in turn serves as a precursor for the production of polyester PET.1 Additionally, MA is readily converted to adipic acid, a “top 50” bulk chemical, via hydrogenation, with high yield and specificity.2,3 Adipic acid represents a multi-billion-dollar global market, with nylon-6,6 accounting for >85% of global adipic acid utilization.3 The dicarboxylic functionality of adipic acid also affords a wide variety of upgrading strategies including lactonization, diolization, and ketonization.1 As such, adipic acid can be readily converted to other large-market, high-value molecules, such as plasticizers, lubricants, engineering resins, and polyurethanes. Conventional adipic acid production has been estimated to account for nearly 10% of global N2O emissions.4 Though recent technologies have been deployed to help abate these emissions,5–8 conventional routes to adipic acid production almost exclusively employ non-renewable, petroleum feedstocks, specifically ketone/alcohol (KA) oil generated via oxidation of cyclohexanone and cyclohexanol with nitric acid.4 Thus, there is an increasing need to develop sustainable, green routes to produce MA and derivatives thereof.Recently, a number of alternative bio-based MA production routes have been proposed, utilizing lignocellulosic aromatics and sugars as substrates for wild-type and genetically modified microbial biocatalysts.1–3,8 MA biosynthesis naturally occurs during the catabolism of aromatic compounds in a subset of microorganisms, primarily saprophilic bacteria and fungi, which have ready access to plant-derived aromatics. Production of MA via heterologous pathway incorporation has been demonstrated in heterotrophic bacteria and yeast by exploiting naturally occurring intermediary metabolic pathways involved in the detoxification or catabolism of aromatic compounds such as toluene and benzoate,1–3,9–11 through catechol and protocatechuate intermediates. Alternatively, MA can be synthesized in a three-step process from the 3-dehydroshikimate (DHS) intermediate produced in the shikimate pathway,3 which is essential for the synthesis of aromatic amino acids. Lastly, MA can also be generated by converting the anthranilate intermediate of tryptophan biosynthesis into catechol by the anthranilate 1,2-dioxygenase.12 The above strategies have been successfully deployed in an array of model heterotrophic microbes, however, to date, there has been no demonstration of MA bioproduction from C1 carbon substrates.
Methanotrophic bacteria are promising hosts for carbon sequestration and conversion of C1 carbon streams to diverse target molecules, including fuel and chemical intermediates.13,14 These bacteria are key global biochemical regulators capable of utilizing CH4 as a carbon and energy source and possess core metabolic machinery enabling metabolic engineering strategies paralleling those found in conventional heterotrophic hosts.15,16 These characteristics have led to intensified industrial interest for deployment of methanotrophs as production biocatalysts for bioconversion of gaseous waste streams, including anaerobic digestion-derived biogas,17,18 which is primarily comprised of CH4 and CO2. Indeed, recent efforts have demonstrated the potential utility of methanotrophic biocatalysts for production of platform chemicals from diverse waste streams.19–25 Here, we present the first heterologous production of MA from CH4 using methanotrophic bacteria as a production chassis. Further, we leveraged the M. alcaliphilum 20Z genome-scale model26 to guide rational metabolic engineering to increase carbon flux to the MA pathway, which revealed a yet unidentified shikimate biosynthetic enzyme or pathway in this organism. These results underscore that further elucidation of methanotroph central metabolism is required to accurately model and predict methanotrophic biosynthetic capacity.
Results and discussion
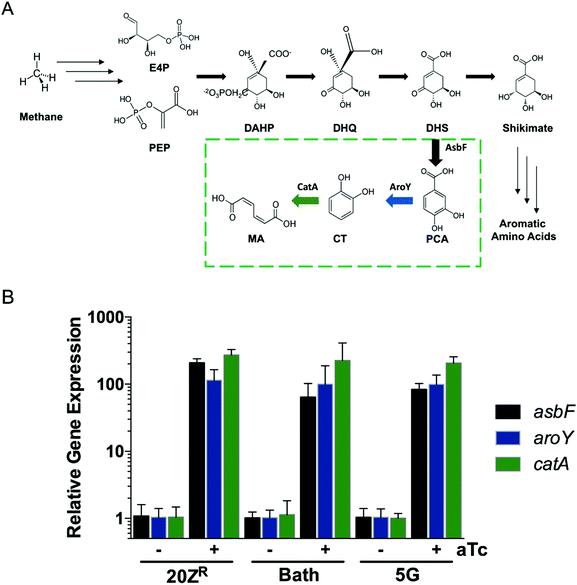
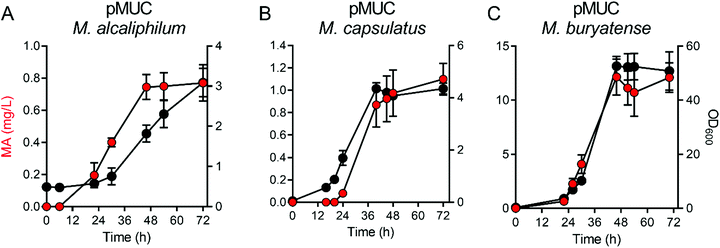
Methanotrophs are not known to naturally produce MA. However, these bacteria encode genes requisite for de novo aromatic amino acid biosynthesis and can be cultivated on minimal media lacking amino acid supplementation, indicating that the upstream metabolic machinery for shikimate pathway-derived MA production is present. Leveraging this metabolic machinery, we incorporated a MA biosynthetic pathway encoding the Bacillus thuringiensis AsbF dihydroxyshikimic acid dehydratase, Enterobacter cloacae AroY protocatechuate decarboxylase, and an Acinetobacter sp. CatAP76A catechol dioxygenase variant27 in order to shunt the aromatic amino acid precursor dihydroxyshikimic acid to MA in methanotrophic bacteria (Fig. 1A). The resultant vector, pMUC, was introduced into Methylotuvimicrobium alcaliphilum 20ZR, Methylotuvimicrobium buryatense 5G, and Methylococcus capsulatus Bath, industrially-promising gammaproteobacterial methanotrophs with established metabolic engineering tools, including genome-scale metabolic models.26,28–30 Quantitative reverse-transcription PCR (qRT-PCR) analysis of the engineered strains harboring the biosynthetic pathway under the control of the inducible tetracycline promoter/operator22 indicated that this promoter is functional in all three methanotrophic strains, with significant expression of AsbF, AroY, and CatA-encoding genes detected after induction (Fig. 1B).We evaluated growth and MA production by the pMUC strains in 0.5 L continuous-stirred tank reactors (CSTRs) with constant gas supply (20% CH4 v/v in air, 1 vvm; 2% CO2 was also supplied to M. capsulatus Bath). All strains excreted MA during active growth, coincident with carbon flux through the shikimate pathway to provide proteinogenic aromatic amino acids (Fig. 2). We did not detect the MA precursors protocatechuic acid and catechol in the culture medium, contrary to prior studies using yeast or bacterial biocatalysts expressing a similar heterologous MA biosynthetic pathway.2,3,31M. alcaliphilum and M. capsulatus reached cell densities of OD600 = 6–8 after 48 h, excreting MA at titers of 0.75 mg L−1 and 0.97 mg L−1 MA, respectively (Fig. 2A and B, Table 1). We previously increased the growth potential of M. buryatense in CSTR cultivation via media optimization, achieving maximum OD600 ∼ 50 culture densities.21,22 Under these optimized cultivation conditions, M. buryatense excreted 12.4 mg L−1 MA into the culture medium following 48 h of cultivation (Fig. 2C and Table 1), which is comparable with other proof-of-concept product titers (10–100 mg L−1) produced by engineered methanotrophs19,22–24,32 Although we were able to increase the MA titer by high-density cell cultivation, yields (0.65–2.7 mg MA per g CH4) and specific productivities (0.68–1.2 mg MA per g DCW) were comparable between the engineered methanotrophs, with M. capsulatus exhibiting the highest yield. M. alcaliphilum exhibited the highest specific productivity (Table 1), which served as the basis for down-selection and further evaluation.
| Strain | Titer (mg MA per L/48 h) | CH4 consumed (g L−1/48 h) | Maximum yield (mg MA per g CH4, 24–48 h) | Specific productivity (mg MA per g DCW, 48 h) |
|---|---|---|---|---|
| The data represent the mean ± standard deviation of two biological replicate bioreactors. | ||||
| M. alcaliphilum pMUC | 0.75 ± 0.08 | 1.35 ± 0.33 | 0.65 ± 0.12 | 1.2 ± 0.01 |
| M. capsulatus pMUC | 0.97 ± 0.21 | 0.74 ± 0.07 | 2.8 ± 0.04 | 0.68 ± 0.16 |
| M. buryatense pMUC | 12.4 ± 2.26 | 12.5 ± 0.14 | 1.2 ± 0.22 | 1.1 ± 0.20 |
We leveraged our previously developed M. alcaliphilum genome-scale model (GSM)26 to identify genetic targets for increased carbon flux to the MA pathway. Based on the GSM, the maximum theoretical specific productivity of MA production is 0.2 g MA per g DCW. Disruption of the shikimate dehydrogenase (encoded by MALCv4_3465) and the pyruvate dehydrogenase (encoded by MALCv4_1357-9) were predicted by the model to significantly increase flux through the heterologous MA pathway (Table 2). This agrees with other studies where disruption of the shikimate dehydrogenase is required in heterotrophic hosts to achieve heterologous MA biosynthesis from aromatic amino acid precursors by inhibiting flux through the shikimate pathway and increasing DHS substrate availability,1,3,31 while disruption of pyruvate dehydrogenase would be predicted to increase availability of phosphoenolpyruvate and erythrose-4-phosphate shikimate pathway precursors. We introduced the heterologous MA biosynthetic pathway into our previously generated M. alcaliphilum pyruvate dehydrogenase knock-out strain20 and evaluated MA production in CSTRs (Table 2). The pMUC Δpdh strain produced similar MA titers but displayed a growth defect compared to the pMUC strain; thus a 1.5-fold increase in specific productivity was observed, albeit much lower than the 23.2-fold flux increase predicted by the GSM (Table 2). The discrepancy between predicted and actual flux values could be in part due to the high lactic acid flux observed in this strain (Table 2), presumably because of increased pyruvate availability and conversion by the native lactate dehydrogenase.
| Strain | Predicted MA flux ratio | Observed max MA flux ratio | Observed max LA flux ratio | Biomass (g DCW, 96 h) |
|---|---|---|---|---|
| The data represent the mean ± standard deviation of two biological replicate bioreactors. | ||||
| 20ZR pMUC | 1 | 0.91 ± 0.13 | 0.84 ± 0.22 | 0.69 ± 0.008 |
| 20ZR pMUC Δpdh | 23.2 | 1.5 ± 0.07 | 4.0 ± 0.06 | 0.35 ± 0.02 |
| 20ZR pMUC ΔaroE | 29.4 | 1.0 ± 0.11 | 1.4 ± 0.53 | 0.31 ± 0.04 |
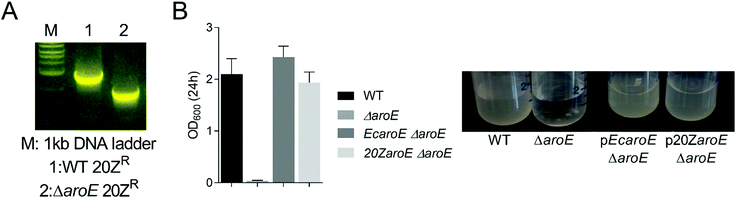
We hypothesized that knock-out of MALCv4_3465 would result in an aromatic amino acid auxotrophic strain similar to that observed in Escherichia coli. We successfully removed the locus via marker exchange mutagenesis (Fig. 3A). However, a MALCv4_3465 mutant (ΔaroE) was successfully isolated in the absence of shikimate or aromatic amino acid supplementation, suggesting an alternate shikimate biosynthetic pathway is present in M. alcaliphilum. To assess the potential for increased DHS substrate availability in the M. alcaliphilum ΔaroE strain, we introduced pMUC and analyzed MA production is CSTRs. Similar to the pMUC Δpdh strain, the pMUC ΔaroE strain displayed a slight growth defect but a similar MA specific productivity relative to the isogenic pMUC strain (Table 2). The GSM predicted that removal of the shikimate dehydrogenase would increase flux through the MA pathway by 29.4-fold and result in aromatic amino acid auxotrophy with no carbon flux to biomass. Although deletion of MALCv4_3465 decreased M. alcaliphilum growth, the ΔaroE strain's ability to grow in the absence of shikimate or aromatic amino acid supplementation provides additional support for the presence of an alternative shikimate biosynthetic enzyme encoded by M. alcaliphilum. To confirm accurate gene annotation of MALCv4_3465 as a shikimate dehydrogenase, we tested the ability of MALCv4_3465 to complement an E. coli shikimate dehydrogenase KO strain that does not grow on M9 minimal medium without aromatic amino acid supplementation.33 Successful complementation of the E. coli ΔaroE non growth phenotype in M9 medium was achieved via plasmid-based expression of the E. coli aroE gene or MALCv4_3465, supporting that MALCv4_3465 encodes a functional shikimate dehydrogenase (Fig. 3B), and provides an additional line of evidence that M. alcaliphilum encodes an alternative shikimate biosynthetic enzyme. However, alternative AroE or YdiB homologs34 were not identified in the M. alcaliphilum 20ZR genome via homology query. Based on previous metabolic engineering efforts to produce MA from non-aromatic substrates in heterotrophic hosts, increasing flux to the DHS substrate via gene disruption, pathway overexpression, and/or generation of feedback-insensitive shikimate pathway enzymes are requisite for efficient MA production from non-aromatic precursors.35 Thus, a complete understanding of shikimate biosynthetic pathways is needed to more accurately predict competing metabolic pathways and guide future rational metabolic engineering pursuits to increase MA production metrics in M. alcaliphilum.
Notably, the highest yield observed in this study (2.7 mg MA per g CH4) is comparable to initial MA yields produced by heterotrophic biocatalysts expressing a similar heterologous MA biosynthetic pathway.3,31 MA yields from glucose in these heterotrophic hosts have been significantly improved (12.9 mg MA per g glucose) via additional genetic modifications for increased flux to the MA pathway, including removal of aromatic amino acid negative feedback regulation and disruption/modification of the shikimate dehydrogenase that was attempted in M. alcaliphilum here.3,35–38 These results indicate a substantial yield enhancement can be achieved if an alternative shikimate biosynthetic route can be identified and disrupted. Future strain engineering efforts for efficient MA production from CH4 will thus entail identification and disruption of the unidentified shikimate biosynthetic enzyme, as well as homologous overexpression of transketolase and/or phosphoketolase for increased erythrose-4-phosphate substrate availability, and expression of either a native or heterologous feedback insensitive 2-dehydro-3-deoxyphosphoheptonate (DAHP) aldolase variant to increase flux through the shikimate pathway.
Conclusions
CH4 is the second most abundant greenhouse gas (GHG), with increased warming potential relative to CO2. Nearly 60% of CH4 emissions are derived from anthropogenic sources, including shale gas fracking, landfill gases, and industrial agriculture operations. Further, CH4 is the primary component of natural gas and anaerobic digestion-derived biogas, and recent reports have indicated CH4 represents a high-volume, low-cost substrate that is well-suited for biomanufacturing.13,39 Importantly, the conversion of natural gas and biogas can simultaneously offset petroleum usage while mitigated GHGs with massive economic and environmental impacts.40 In contrast to heterotrophic microbial conversion strategies employing biomass-derived substrates, the utilization of CH4 as a bioconversion substrate does not compete with food production. Thus, strategies that convert CH4 to high-value intermediates present an opportunity to mitigate and sequester GHG emissions, while concurrently valorizing CH4. Additionally, CH4 biocatalysis offers a means to concurrently liquefy and upgrade CH4, enabling its utilization in conventional transportation and industrial manufacturing infrastructure. To this end, recent efforts have successfully generated methanotrophic biocatalysts with diverse production capacity for fuel and chemical intermediates.17,19–25The work presented here provides proof-of-concept for production of MA from CH4. Methanotrophic production of MA from CH4 presents a novel, green pathway to biosynthesize a key platform intermediate capable of displacing petroleum in the production of high-volume commodity chemicals and biopolymers. However, a series of biological and process limitations will need to be addressed to achieve economically-viable production. A primary biological driver is the identification and disruption of alternative shikimate biosynthetic genes, which will enable increased flux to MA precursors. Genome-wide mutagenic approaches offer a promising route to identify auxotrophic phenotypes, implicating genes with redundant functionality. Lastly, conventional gas fermentation challenges remain a hurdle; gas mass transfer limitations, in particular, will need to be addressed in order to achieve economically-viable production rates.
Materials and methods
Bacterial strains
Methylotuvimicrobium (previously Methylomicrobium) alcaliphilum 20ZR,26Methylotuvimicrobium buryatense 5GB1,28 and Methylococcus capsulatus Bath (Table S1†) were cultivated in either nitrate mineral salts (NMS) medium (Bath) or NMS medium supplemented with 1.5% NaCl and carbonate buffer as previously described.41–43 Strains were routinely maintained on solid medium in stainless-steel gas chambers (Schuett-biotec GmbH) containing 20% (v/v) methane in air.Muconic acid biosynthetic pathway construction
Plasmids and primers used in this study are presented in Table S2.† The muconic acid biosynthetic pathway was constructed by ligating codon-optimized asbF-encoding dehydroshikimate dehydratase from Bacillus thuringensis, aroY-encoding protocatechuate decarboxylase from Enterobacter cloacae, and the catA-encoding catechol dioxygenase P76A variant from Acinetobacter baylyi (codon-optimized gene sequences are in Table S3†) into the pCAH01 inducible expression vector22 to generate pMUC using 2× Gibson Assembly Mix from New England Biolabs (Ipswich, MA), following the manufacturers protocol. The pMUC vector sequence was confirmed by sequence analysis (Genewiz, South Plainfield, NJ). This vector was transformed into the methanotrophic strains via biparental mating using E. coli S17 as previously described.28,42 Positive transformants selected on solid medium containing 100 μg mL−1 of kanamycin were confirmed by PCR and sequence analysis.Quantitative reverse transcription PCR
Total RNA was isolated from cultures grown for 48 h on solid medium with and without pMUC induction with 0.5 μg mL−1 anhydrotetracycline inducer using the Monarch Total RNA miniprep kit (New England Biolabs) followed by removal of contaminating DNA by treatment with Turbo DNase (Life Technologies, Carlsbad, CA. cDNA was synthesized from 1 μg total RNA using the SuperScript VILO cDNA synthesis kit (ThermoFisher) following the manufacturer's protocol. Real-time PCR was performed by using SYBR IQ master mix and a QuantStudio Real-Time PCR System (ThermoFisher). The primers used for the qRT-PCR are listed in Table S2.† The cycle threshold (Ct) value for each gene was determined and normalized to the rpoD gene encoding the RNA polymerase housekeeping sigma factor. Relative expression between induced and uninduced control samples was calculated by using the ΔΔCt method.Methane fermentations
100 mL seed cultures were inoculated from plate biomass and grown in 150 mL bubble columns with a continuous gas flow (20% CH4, in air, 1 volume of gas per volume of liquid per minute (vvm)). M. capsulatus Bath cultures were also supplied with 2% CO2 in the gas mixture. Gas fermentations were performed in a 0.5 L Biostat-Q plus bioreactor (Sartorius, Gottingen, Germany) with a culture volume of 300 mL. The M. buryatense bioreactor medium was supplemented with 8× KNO3 (80 mM), 2× phosphate buffer (4.6 mM), and 4× trace element solution to support high cell growth, as previously described.22,44 Seed cultures were diluted to a final OD600 = 0.1–0.5. Mixing was achieved by using a bottom marine impeller at 500 rpm. A continuous flow rate of 1 vvm 20% CH4 in air (plus 2% CO2 for M. capsulatus Bath) was maintained for the duration of cultivation. At indicated intervals, growth was measured spectrophotometrically at 600 nm and a 1 mL sample was taken for HPLC analysis. The culture supernatant was filtered using a 0.2 μm syringe filter and then a 0.1 mL injection was separated using a model 1260 HPLC (Agilent, Santa Clara, CA) and a cation H HPx-87H column (Bio-Rad). A 0.6 mL min−1 flow rate at 55 °C with 0.01 N sulfuric acid as the mobile phase was used. Refractive index and diode array detectors were used for compound detection. Muconic acid concentrations were calculated by regression analysis compared to known standards with the minimum detection limit of 0.2 mg L−1 muconic acid. The percent CH4 in the off-gas was measured every 10 min for the duration of bioreactor fermentations via mass spectrometry. Percent CH4 consumption was converted to weight based on CH4 density (0.000656 g mL−1) and the flow rate (60 mL CH4 per min). Dry Cell Weight (DCW) was either measured directly after freeze-drying or estimated from the final OD of the cell culture using the following equation: DCW = OD × (0.35 ± 0.04) g L−1.26 Muconic acid yields were calculated using consumed substrate data and represented as mg muconic acid produced per g CH4 consumed.M. alcaliphilum shikimate dehydrogenase mutant construction
Plasmids and primers used for amplification of the shikimate dehydrogenase flanking regions and the pK18mobsacB vector for construction of the shikimate dehydrogenase (aroE) knockout are shown in Tables S1 and S2.† 1000 bp genomic fragments flanking the shikimate dehydrogenase (MALCv4_3465) gene were amplified by PCR and cloned into the pK18mobsacB plasmid via Gibson assembly. The resulting plasmid was introduced to the 20ZR strain by biparental conjugation as described previously.28 Positive transformants selected on medium supplemented with kanamycin (100 μg mL−1) were subsequently plated on medium containing 2.5% sucrose for counterselection to remove the kanamycin resistance marker. Sucrose-resistant, kanamycin-sensitive colonies were PCR-genotyped for removal of the MALCv4_3465 locus.E. coli shikimate dehydrogenase mutant complementation
The E. coli shikimate dehydrogenase aroE and M. alcaliphilum 20ZR MALCv4_3465 genes with native ribosomal binding sites were amplified from the corresponding genomic DNA using primers appending XbaI and HindIII restriction sites to the 5′ and 3′ ends (Table S2†), respectively. Genes were directionally cloned into the pBluescript II KS+ vector. Expression constructs were sequence verified and transformed into the JW3242 ΔaroE721::kan parent strain via electroporation. Positive transformants selected on LB medium containing kanamycin and ampicillin were grown in M9 minimal medium supplemented with 0.5 mM IPTG for shikimate dehydrogenase induction. Bacterial growth of the complementing strains in M9 medium was compared to wild-type E. coli BW25113 and JW3242 ΔaroE721::kan parent strains, the latter of which does not grow on M9 medium.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We would like to thank Holly Rohrer of the National Renewable Energy Laboratory for technical assistance. Funding provided by the U.S. Department of Energy Office of Energy Efficiency and Renewable Energy Bioenergy Technologies Office DE-FOA-0001085. This work was authored in part by Alliance for Sustainable Energy, LLC, the manager and operator of the National Renewable Energy Laboratory for the U.S. Department of Energy under Contract No. DE-AC36-08GO28308. The views expressed in the article do not necessarily represent the views of the DOE or the U.S. Government. The U.S. Government retains and the publisher, by accepting the article for publication, acknowledges that the U.S. Government retains a non-exclusive, paid-up, irrevocable, worldwide license to publish or reproduce the published form of this work, or allow others to do so, for U.S. Government purposes.References
- N.-Z. Xie, H. Liang, R.-B. Huang and P. Xu, Biotechnol. Adv., 2014, 32, 615–622 CrossRef CAS PubMed.
- C. Weber, C. Brückner, S. Weinreb, C. Lehr, C. Essl and E. Boles, Appl. Environ. Microbiol., 2012, 78, 8421–8430 CrossRef CAS PubMed.
- K. A. Curran, J. M. Leavitt, A. S. Karim and H. S. Alper, Metab. Eng., 2013, 15, 55–66 CrossRef CAS PubMed.
- K. Sato, M. Aoki and R. Noyori, Science, 1998, 281, 1646–1647 CrossRef CAS PubMed.
- M. Zhao, D. Huang, X. Zhang, M. A. G. Koffas, J. Zhou and Y. Deng, Metab. Eng., 2018, 47, 254–262 CrossRef CAS PubMed.
- K. Raj, S. Partow, K. Correia, A. N. Khusnutdinova, A. F. Yakunin and R. Mahadevan, Metab. Eng. Commun., 2018, 6, 28–32 CrossRef PubMed.
- T. Babu, E. J. Yun, S. Kim, D. H. Kim, K. H. Liu, S. R. Kim and K. H. Kim, Process Biochem., 2015, 50, 2066–2071 CrossRef CAS.
- D. R. Vardon, M. A. Franden, C. W. Johnson, E. M. Karp, M. T. Guarnieri, J. G. Linger, M. J. Salm, T. J. Strathmann and G. T. Beckham, Energy Environ. Sci., 2015, 8, 617–628 RSC.
- J. Becker, M. Kuhl, M. Kohlstedt, S. Starck and C. Wittmann, Microb. Cell Fact., 2018, 17, 115 CrossRef PubMed.
- T. Sonoki, K. Takahashi, H. Sugita, M. Hatamura, Y. Azuma, T. Sato, S. Suzuki, N. Kamimura and E. Masai, ACS Sustainable Chem. Eng., 2018, 6, 1256–1264 CrossRef CAS.
- D. Salvachua, C. W. Johnson, C. A. Singer, H. Rohrer, D. J. Peterson, B. A. Black, A. Knapp and G. T. Beckham, Green Chem., 2018, 20, 5007–5019 RSC.
- X. Sun, Y. Lin, Q. Huang, Q. Yuan and Y. Yan, Appl. Environ. Microbiol., 2013, 79, 4024–4030 CrossRef CAS PubMed.
- J. M. Clomburg, A. M. Crumbley and R. Gonzalez, Science, 2017, 355 Search PubMed.
- R. J. Conrado and R. Gonzalez, Science, 2014, 343, 621–623 CrossRef CAS PubMed.
- L. Chistoserdova, M. G. Kalyuzhnaya and M. E. Lidstrom, Annu. Rev. Microbiol., 2009, 63, 477–499 CrossRef CAS PubMed.
- M. G. Kalyuzhnaya, S. Yang, O. N. Rozova, N. E. Smalley, J. Clubb, A. Lamb, G. A. N. Gowda, D. Raftery, Y. Fu, F. Bringel, S. Vuilleumier, D. A. C. Beck, Y. A. Trotsenko, V. N. Khmelenina and M. E. Lidstrom, Nat. Commun., 2013, 4, 2785 CrossRef CAS PubMed.
- C. A. Henard and M. T. Guarnieri, in Methane biocatalysis: paving the way to sustainability, ed. M. G. Kalyuzhnaya and X.-H. Xing, Springer International Publishing, Cham, 2018, pp. 117–132 Search PubMed.
- M. G. Kalyuzhnaya, A. W. Puri and M. E. Lidstrom, Metab. Eng., 2015, 29, 142–152 CrossRef CAS PubMed.
- D. T. N. Nguyen, O. K. Lee, S. Hadiyati, A. N. Affifah, M. S. Kim and E. Y. Lee, Metab. Eng., 2019, 54, 170–179 CrossRef CAS PubMed.
- C. A. Henard, T. G. Franklin, B. Youhenna, S. But, D. Alexander, M. G. Kalyuzhnaya and M. T. Guarnieri, Front. Microbiol., 2018, 9, 2610 CrossRef PubMed.
- C. A. Henard, H. K. Smith and M. T. Guarnieri, Metab. Eng., 2017, 41, 152–158 CrossRef CAS PubMed.
- C. A. Henard, H. Smith, N. Dowe, M. G. Kalyuzhnaya, P. T. Pienkos and M. T. Guarnieri, Sci. Rep., 2016, 6, 21585 CrossRef CAS PubMed.
- S. Garg, H. Wu, J. M. Clomburg and G. N. Bennett, Metab. Eng., 2018, 48, 175–183 CrossRef CAS PubMed.
- A. D. Nguyen, I. Y. Hwang, O. K. Lee, D. Kim, M. G. Kalyuzhnaya, R. Mariyana, S. Hadiyati, M. S. Kim and E. Y. Lee, Metab. Eng., 2018, 47, 323–333 CrossRef CAS PubMed.
- S. Garg, J. M. Clomburg and R. Gonzalez, J. Ind. Microbiol. Biotechnol., 2018, 45, 379–391 CrossRef CAS PubMed.
- I. R. Akberdin, M. Thompson, R. Hamilton, N. Desai, D. Alexander, C. A. Henard, M. T. Guarnieri and M. G. Kalyuzhnaya, Sci. Rep., 2018, 8, 2512 CrossRef PubMed.
- L. Han, P. Liu, J. Sun, Y. Wu, Y. Zhang, W. Chen, J. Lin, Q. Wang and Y. Ma, Sci. Rep., 2015, 5, 13435 CrossRef CAS PubMed.
- A. W. Puri, S. Owen, F. Chu, T. Chavkin, D. A. C. Beck, M. G. Kalyuzhnaya and M. E. Lidstrom, Appl. Environ. Microbiol., 2015, 81, 1775–1781 CrossRef PubMed.
- A. de la Torre, A. Metivier, F. Chu, L. M. L. Laurens, D. A. C. Beck, P. T. Pienkos, M. E. Lidstrom and M. G. Kalyuzhnaya, Microb. Cell Fact., 2015, 14, 188 CrossRef PubMed.
- C. Lieven, L. A. H. Petersen, S. B. Jørgensen, K. V. Gernaey, M. J. Herrgard and N. Sonnenschein, Front. Microbiol., 2018, 9, 2947 CrossRef PubMed.
- C. W. Johnson, D. Salvachúa, P. Khanna, H. Smith, D. J. Peterson and G. T. Beckham, Metab. Eng. Commun., 2016, 3, 111–119 CrossRef PubMed.
- L. T. Nguyen and E. Y. Lee, Biotechnol. Biofuels, 2019, 12, 147 CrossRef PubMed.
- T. Baba, T. Ara, M. Hasegawa, Y. Takai, Y. Okumura, M. Baba, K. A. Datsenko, M. Tomita, B. L. Wanner and H. Mori, Mol. Syst. Biol., 2006, 2, 2006.0008 CrossRef PubMed.
- G. Michel, A. W. Roszak, V. Sauvé, J. Maclean, A. Matte, J. R. Coggins, M. Cygler and A. J. Lapthorn, J. Biol. Chem., 2003, 278, 19463–19472 CrossRef CAS PubMed.
- N. J. H. Averesch and J. O. Krömer, Front. Bioeng. Biotechnol., 2018, 6, 32 CrossRef PubMed.
- J. M. Leavitt, J. M. Wagner, C. C. Tu, A. Tong, Y. Liu and H. S. Alper, Biotechnol. J., 2017, 12(10) DOI:10.1002/biot.201600687.
- M. Suastegui, J. E. Matthiesen, J. M. Carraher, N. Hernandez, N. Rodriguez Quiroz, A. Okerlund, E. W. Cochran, Z. Shao and J.-P. Tessonnier, Angew. Chem., Int. Ed., 2016, 55, 2368–2373 CrossRef CAS PubMed.
- W. Niu, K. M. Draths and J. W. Frost, Biotechnol. Prog., 2002, 18, 201–211 CrossRef CAS PubMed.
- A. D. Comer, M. R. Long, J. L. Reed and B. F. Pfleger, Metab. Eng. Commun., 2017, 5, 26–33 CrossRef PubMed.
- Biogas Opportunities Roadmap, Department of Energy, https://www.energy.gov/downloads/biogas-opportunities-roadmap.
- R. Whittenbury, K. C. Phillips and J. F. Wilkinson, J. Gen. Microbiol., 1970, 61, 205–218 CrossRef CAS PubMed.
- D. S. Ojala, D. A. C. Beck and M. G. Kalyuzhnaya, Methods Enzymol., 2011, 495, 99–118 Search PubMed.
- T. Tapscott, M. T. Guarnieri and C. A. Henard, Appl. Environ. Microbiol., 2019, 85(11) DOI:10.1128/AEM.00340-19.
- Q. Fei, A. W. Puri, H. Smith, N. Dowe and P. T. Pienkos, Biotechnol. Biofuels, 2018, 11, 129 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9gc03722e |
| ‡ Current affiliation: Department of Biological Sciences and BioDiscovery Institute, University of North Texas, Denton, TX, USA. |
| § Current affiliation: BIOSOFT.RU, LLC, Novosibirsk, Russian Federation. |
| This journal is © The Royal Society of Chemistry 2019 |