Open Access Article
Open Access ArticleSustainable bioproduction of the blue pigment indigoidine: Expanding the range of heterologous products in R. toruloides to include non-ribosomal peptides†
Maren
Wehrs
abc,
John M.
Gladden
 cd,
Yuzhong
Liu
ac,
Lukas
Platz
ac,
Jan-Philip
Prahl
ae,
Jadie
Moon
ac,
Gabriella
Papa
ae,
Eric
Sundstrom
ae,
Gina M.
Geiselman
ad,
Deepti
Tanjore
ae,
Jay D.
Keasling
acfghij,
Todd R.
Pray
ae,
Blake A.
Simmons
cd,
Yuzhong
Liu
ac,
Lukas
Platz
ac,
Jan-Philip
Prahl
ae,
Jadie
Moon
ac,
Gabriella
Papa
ae,
Eric
Sundstrom
ae,
Gina M.
Geiselman
ad,
Deepti
Tanjore
ae,
Jay D.
Keasling
acfghij,
Todd R.
Pray
ae,
Blake A.
Simmons
 ac and
Aindrila
Mukhopadhyay
ac and
Aindrila
Mukhopadhyay
 *ack
*ack
aBiological Systems and Engineering Division, Lawrence Berkeley National Laboratory, Berkeley, CA 94720, USA. E-mail: amukhopadhyay@lbl.gov
bInstitut für Genetik, Technische Universität Braunschweig, Braunschweig, Germany
cJoint BioEnergy Institute, Lawrence Berkeley National Laboratory, Emeryville, CA 94608, USA
dBiological and Engineering Sciences Center, Sandia National Laboratories, 7011 East Avenue, Livermore, California 94551, USA
eAdvanced Biofuels and Bioproducts Process Development Unit, Lawrence Berkeley National Laboratory, Emeryville, CA 94608, USA
fDepartment of Plant and Microbial Biology, University of California, Berkeley, CA 94720, USA
gDepartment of Bioengineering, University of California, Berkeley, CA 94720, USA
hDepartment of Chemical and Biomolecular Engineering, University of California, Berkeley, CA 94720, USA
iThe Novo Nordisk Foundation Center for Biosustainability, Technical University of Denmark, Denmark
jSynthetic Biochemistry Center, Institute for Synthetic Biology, Shenzhen Institutes for Advanced Technologies, Shenzhen, China
kEnvironmental Genomics and Systems Biology Division, Lawrence Berkeley National Laboratory, Berkeley, CA 94720, USA
First published on 28th May 2019
Abstract
Non-ribosomal peptides (NRPs) constitute a diverse class of valuable secondary metabolites, with potential industrial applications including use as pharmaceuticals, polymers and dyes. Current industrial production of dyes is predominantly achieved via chemical synthesis, which can involve toxic precursors and generate hazardous byproducts. Thus, alternative routes of dye production are highly desirable to enhance both workplace safety and environmental sustainability. Correspondingly, biological synthesis of dyes from renewable carbon would serve an ideal green chemistry paradigm. Therefore, we engineered the fungal host Rhodosporidium toruloides to produce the blue pigment indigoidine, an NRP with potential applications in the dye industry, using various low-cost carbon and nitrogen sources. To demonstrate production from renewable carbon sources and assess process scalability we produced indigoidine in 2 L bioreactors, reaching titers of 2.9 ± 0.8 g L−1 using a sorghum lignocellulosic hydrolysate in a batch process and 86.3 ± 7.4 g L−1 using glucose in a high-gravity fed-batch process. This study represents the first heterologous production of an NRP in R. toruloides, thus extending the range of microbial hosts that can be used for sustainable, heterologous production of NRPs. In addition, this is the first demonstration of producing an NRP using lignocellulose. These results highlight the potential of R. toruloides for the sustainable, and scalable production of NRPs, with the highest reported titer of indigoidine or any heterologously produced NRP to date.
Introduction
Public awareness of eco-safety and health concerns associated with the production and use of certain toxic compounds within the commercial sector has given rise to a growing demand for environmentally sustainable natural products. Secondary metabolites such as polyketides and non-ribosomal peptides (NRPs) have generated significant interest because of their potential use in a wide range of industries including pharmaceuticals, polymers, flavors and fragrances, and natural dyes.1–3 Natural dyes have been used for centuries to color textiles,4 cosmetics, and food ingredients.5 Current industrial production of dyes is predominantly achieved via chemical synthesis, which can involve toxic precursors and generate hazardous chemicals as byproducts of the process. Microbial production of dyes has the potential to circumvent the issues with chemical synthesis and can also provide higher production levels and purity than can be achieved by isolating these dyes from natural sources.6,7Indigoidine is a natural blue pigment produced by several bacteria via known biosynthetic gene clusters.8–12 This 3′,3′-bipyridyl pigment is formed through condensation of two molecules of L-glutamine catalyzed by a non-ribosomal peptide synthetase (NRPS).8,10 The secondary metabolite class of NRPs includes molecules with a range of pharmaceutical applications, such as immunosuppressants, antibiotics, anticancer drugs and antiviral compounds.1 However, the low NRP production levels from native hosts and their complex chemical structures impede mass production by purification from biological material or chemical synthesis, respectively. Furthermore, despite the availability of biosynthetic tools for metabolic engineering and pathway discovery, optimization of NRP production in their natural hosts remains challenging.
One of the greatest challenges in the deployment of any bioproduct into the market, native or non-native to the production host, is engineering the host to produce at high titer, rate, and yield (TRY) in industrial settings.13,14 Strain development of a production host for industrial applications typically involves many years of costly research and resources.15 Therefore, selection of a host organism that naturally has the ability to produce high TRY of a class of bioproducts in industrially relevant settings would be of great economic and environmental value.
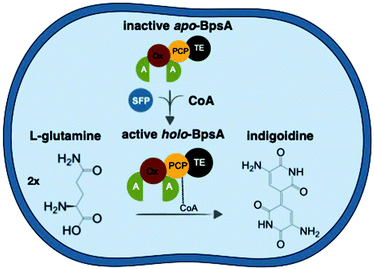
The basidiomycete Rhodosporidium toruloides, also known as Rhodotorula toruloides, is emerging as a robust and metabolically flexible host for bioproduction. This oleaginous red yeast natively produces high amounts of lipids, carotenoids and industrially relevant enzymes.16R. toruloides features many host characteristics vital for commercial-scale production, such as the capacity to grow to high cell densities,17 the ability to utilize a wide range of nitrogen and carbon sources18 and tolerance to inhibitory compounds found in unrefined substrates.19 In addition to its native suitable industrial features, recent developments in the establishment of metabolic engineering tools and -omics techniques for R. toruloides20–24 have not only enabled optimization of native bioproducts but also the ability to produce non-native bioproducts. While R. toruloides has successfully been engineered to produce heterologous products from pathways that natively have high carbon flux, like fatty acid-derived products25 and non-native terpenes,26R. toruloides has not been explored for the production heterologous NRPs. Here, we develop R. toruloides as a platform host for the production of this class of bioproducts. To explore NRP production, we genetically engineered R. toruloides to express the NRPS BpsA from Streptomyces lavendulae that converts native metabolite pools of glutamine into the blue pigment indigoidine. Using colorimetric production assays, we analyzed indigoidine production under different cultivation conditions and at different scales. We show that R. toruloides is capable of converting various low-cost carbon and nitrogen sources to indigoidine and demonstrate the importance of carbon–nitrogen ratio for efficient production. To our knowledge, this is not only the first demonstration of heterologous NRP production in R. toruloides, but also the highest titer of an NRP ever reported in any microbial host. Our findings emphasize the potential of R. toruloides to serve as a robust platform host for sustainable, heterologous production of many bioproducts, now including NRPs (Fig. 1).
Experimental
Strain construction
The haploid R. toruloides strain IFO0880 served as parental strain for this study. BpsA and sfp sequences were codon-optimized for expression in R. toruloides. Gene synthesis and plasmid construction of pgen335.sfp.bpsA were performed by Genscript (Piscataway NJ). Pgen335.sfp.bpsA contains a Nourseothricin selection marker. All genes were expressed from native promoters: BpsA was expressed using Tef1p, while Sfp expression was driven by Acp1p. All strains and plasmids used in this study have been deposited in the public instance of the JBEI Registry27 along with their corresponding information and are available upon request through the Joint BioEnergy Institute Strain Registry (https://public-registry.jbei.org/folders/419). Pgen335.sfp.bpsA was introduced into R. toruloides recipient strain by Agrobacterium tumefaciens-mediated transformation (ATMT) as previously described.21Media and cultivation conditions
Overnight cultures of R. toruloides were grown in 5 mL standard rich glucose medium (YPD, 1% (w/v) Bacto yeast extract, 2% (w/v) Bacto peptone, 2% (w/v) Dextrose) from a single colony at 30 °C, shaking at 200 rpm, unless otherwise stated. These overnight cultures were used to inoculate production cultures to OD800 of 0.15, which were cultivated at 25 °C, shaking at 200 rpm, unless indicated otherwise. All productions were carried out in three or four technical replicates. For correlation of OD800 to commonly used OD600 the reader is referred to Fig. S1.†All experiments were performed in synthetic defined medium (SD, 1.7 g L−1 yeast nitrogen base without amino acids and ammonium sulfate (Becton Dickinson, Franklin Lakes NJ), 0.79 g L−1 complete supplement mixture without yeast nitrogen base (Sunrise Science Products, San Diego CA) with 100 g L−1 sugar and 10.6 g L−1 urea as nitrogen base) unless stated otherwise. The synthetic defined media was buffered with 10 mM citrate buffer at pH 5, except for media used in experiments to characterize nitrogen sources and ratio, which was buffered with 10 mM phosphate buffer at pH 6. Culture tubes used in this study were 55 ml rimless culture tubes (25 × 150 mm, product number 9820-25 Corning®).
The pretreatment configuration of the lignocellulosic feedstocks, eucalyptus and switchgrass, and the characterization of resulting hydrolysates are described in detail elsewhere.28 The hydrolysates were mixed in a 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio (v/v) with 10× synthetic defined media (17 g L−1 yeast nitrogen base without amino acids and ammonium sulfate (Becton Dickinson, Franklin Lakes NJ), 7.9 g L−1 complete supplement mixture without yeast nitrogen base (Sunrise Science Products, San Diego CA) 79.5 g L−1 urea as nitrogen base) and buffered with 10 mM citrate buffer at pH 5.
1 ratio (v/v) with 10× synthetic defined media (17 g L−1 yeast nitrogen base without amino acids and ammonium sulfate (Becton Dickinson, Franklin Lakes NJ), 7.9 g L−1 complete supplement mixture without yeast nitrogen base (Sunrise Science Products, San Diego CA) 79.5 g L−1 urea as nitrogen base) and buffered with 10 mM citrate buffer at pH 5.
Indigoidine extraction
Indigoidine was extracted using a previously developed protocol by Yu et al. with slight modifications.9 Briefly, 0.5 mL of culture was centrifuged at 21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 3 min and the supernatant removed. For cell lysis and simultaneous extraction of indigoidine, 100 μL of acid washed beads (0.5 mm, BioSpec, Bartlesville OK) and 2 mL DMSO + 2% Tween® 20 were added to the cell pellet and vortexed twice for 1 min using Mini-Beadbeater-96 (Biospec, Bartlesville OK) at 3600 rpm. After centrifugation at 21
000g for 3 min and the supernatant removed. For cell lysis and simultaneous extraction of indigoidine, 100 μL of acid washed beads (0.5 mm, BioSpec, Bartlesville OK) and 2 mL DMSO + 2% Tween® 20 were added to the cell pellet and vortexed twice for 1 min using Mini-Beadbeater-96 (Biospec, Bartlesville OK) at 3600 rpm. After centrifugation at 21![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 3 min, the indigoidine concentration was determined by measuring the OD612 of the supernatant using a BioTek Synergy 4 plate reader (Biotek, Winooski VT), preheated to 25 °C and applying a standard curve, we have developed previously.29
000g for 3 min, the indigoidine concentration was determined by measuring the OD612 of the supernatant using a BioTek Synergy 4 plate reader (Biotek, Winooski VT), preheated to 25 °C and applying a standard curve, we have developed previously.29
Measurement of absorption spectra of indigoidine in supernatant at different pH and oxidation states
To test the effect of varying pH on the pigment, the pH of 500 μL of cell culture supernatant was adapted to indicated values using 1 M HCl or 1 M NaOH. Subsequently, 100 μL of the supernatant were transferred to a well of a 96-black-well-clear-bottom plate. Absorption spectra were measured using a BioTek Synergy 4 plate reader, preheated to 25 °C.Sugar quantification
Sugar concentrations were quantified on a 1200 series HPLC (Agilent Technologies, location) equipped with an Aminex H column (Bio-Rad, Hercules CA). To remove cells, samples were filtered through 0.45 μm filters (VWR, Radnor, PA) and 5 μL of each sample was injected onto the column, preheated to 50 °C. The column was eluted with 4 mM sulfuric acid (H2SO4) at a flow rate of 600 μL min−1 for 20 minutes. The eluents were monitored by a refractive index detector, and concentrations were calculated by peak area comparison to known standards.Separation-free process coupling pretreatment, saccharification and cultivation
The one-pot process using sorghum biomass was performed according to Sundstrom et al. using the same biomass with minor modifications.19 Briefly, the ionic liquid pretreatment and enzymatic hydrolysis was performed in a 10 L Parr reactor (Parr Instrument Company, Moline, IL, USA). Twenty-five percent biomass loading was achieved by using 600 g of biomass in a 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 90 ratio of cholinium lysinate [Ch][Lys] and water. The pretreatment was carried out at 140 °C for 1 h, stirring at 50 rpm using three arm, self-centering anchor with PTFE wiper blades.
90 ratio of cholinium lysinate [Ch][Lys] and water. The pretreatment was carried out at 140 °C for 1 h, stirring at 50 rpm using three arm, self-centering anchor with PTFE wiper blades.
Following pretreatment, the reactor was cooled down and the pH was adjusted to pH 5 with 50% v/v H2SO4. The IL-treated biomass was diluted with DI water to achieve a solid loading of 20% w/w. The cellulase complex Cellic® CTec2 and HTec2 (Novozymes, Franklinton, NC, USA) (ratio of 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 v/v) was used at loading of 10 mg g−1 biomass dosing 31.9 mL of the (hemi) cellulolytic enzymatic cocktails (i.e. 53 mL kg−1 biomass). The reaction vessel was mixed at 30 rpm and heated to 50 °C for 72 h. The amount of sugar generated by hydrolysis was monitored by taking an aliquot from the slurry at specific times and analyzed by HPLC for sugar quantification as described (Fig. S2†). Enzymatic digestibility was defined as the glucose yield based on the maximum potential glucose from glucan in biomass. In the calculation of cellulose conversion to glucose, it was considered a cellulose
1 v/v) was used at loading of 10 mg g−1 biomass dosing 31.9 mL of the (hemi) cellulolytic enzymatic cocktails (i.e. 53 mL kg−1 biomass). The reaction vessel was mixed at 30 rpm and heated to 50 °C for 72 h. The amount of sugar generated by hydrolysis was monitored by taking an aliquot from the slurry at specific times and analyzed by HPLC for sugar quantification as described (Fig. S2†). Enzymatic digestibility was defined as the glucose yield based on the maximum potential glucose from glucan in biomass. In the calculation of cellulose conversion to glucose, it was considered a cellulose![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) glucose ratio of 1
glucose ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.11.30 Prior to use, the hydrolysate was pasteurized at 80 °C for 3 h in a BINDER FED 720 heating chamber (BINDER GmbH, Tuttlingen, Germany).
1.11.30 Prior to use, the hydrolysate was pasteurized at 80 °C for 3 h in a BINDER FED 720 heating chamber (BINDER GmbH, Tuttlingen, Germany).
The subsequent indigoidine production was performed using 2 L Sartorius BIOSTAT B® fermentation system (Sartorius AG., Goettingen, Germany), each agitated with two Rushton impellers, with an initial working volume of 1 L unfiltered sorghum hydrolysate containing 7.95 g L−1 urea and 50 mL seed culture pre-cultured overnight in a 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 mixture of YP10%D (1% (w/v) Bacto yeast extract, 2% (w/v) Bacto peptone, 10% (w/v) Dextrose) and filtered hydrolysate (0.2 μm filter), adjusted to pH 7. The bioreactor cultivations were inoculated at pH 7 and adjusted to maintain below pH 7.5 using HCl throughout the process (initially 1 N HCl, switched to 6 N HCl after 48 h). 1 mL 30% cefotaxime was added to the batch medium to inhibit contamination. Process values were monitored and recorded using the integrated Sartorius software (BioPAT MFCS/win). Dissolved oxygen was controlled to remain above a set point of 30% by varying agitation from 400–900 rpm. Cultivation temperature was held constant at 25 °C.
50 mixture of YP10%D (1% (w/v) Bacto yeast extract, 2% (w/v) Bacto peptone, 10% (w/v) Dextrose) and filtered hydrolysate (0.2 μm filter), adjusted to pH 7. The bioreactor cultivations were inoculated at pH 7 and adjusted to maintain below pH 7.5 using HCl throughout the process (initially 1 N HCl, switched to 6 N HCl after 48 h). 1 mL 30% cefotaxime was added to the batch medium to inhibit contamination. Process values were monitored and recorded using the integrated Sartorius software (BioPAT MFCS/win). Dissolved oxygen was controlled to remain above a set point of 30% by varying agitation from 400–900 rpm. Cultivation temperature was held constant at 25 °C.
Fed-batch experiments at 2 L bioreactor scale
Fed-batch experiments were performed using 2 L Sartorius BIOSTAT B® fermentation system (Sartorius AG., Goettingen, Germany), each agitated with two Rushton impellers, with an initial working volume of 1 L synthetic defined medium (SD, 1.7 g L−1 yeast nitrogen base without amino acids and ammonium sulfate (VWR International), 0.79 g L−1 complete supplement mixture without yeast nitrogen base (Sunrise Science Products) with 100 g L−1 glucose and 10.6 g L−1 urea as nitrogen base) and 50 mL seed culture pre-cultured overnight in the same media.The bioreactor cultivations were inoculated at pH 5. Initially, the pH was not controlled, and pH increased to pH 8 after 40 h. To avoid oxidation of newly produced indigoidine, the pH was then adjusted to pH 7 using HCl (initially 1 N HCl, switched to 6 N HCl after 48 h of cultivation). A 600 g L−1 glucose solution containing 63.6 g L−1 urea was used as carbon and nitrogen feed. 1 mL 30% cefotaxime was added to the batch medium to inhibit contamination. Process values were monitored and recorded using the integrated Sartorius software (BioPAT MFCS/win). Feeding parameters were implemented using customized LabVIEW Virtual Instruments (National Instruments, Austin, TX).
Dissolved oxygen was controlled to 30% by varying agitation between 400–900 rpm. Cultivation temperature was held constant at 25 °C. Batch glucose was allowed to drop to approx. 20 g L−1 before starting continuous feed addition at 0.063 mL min−1 (86 h EFT). The feed rate was later modified to 0.189 mL min−1 (96 h EFT), 0.130 mL min−1 (116 h EFT), and 0.145 mL min−1 (125 h EFT) in an attempt to maintain glucose concentrations between 20 g L−1 and 100 g L−1. Feed addition was stopped after measuring glucose concentrations higher than 100 g L−1 (144 h EFT).
Indigoidine purification and chemical analysis
Purification of indigoidine was performed using a modified protocol from Yu et al.9 To obtain pure indigoidine for further chemical analysis, the culture broth was centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 5 min to separate the cells from the media. After removal of the supernatant, the cell pellet was resuspended in an equal volume of dimethylformamide (DMF) and cell lysis was performed by subjecting the cells to an using ultrasonic bath (Model 5510, Branson Ultrasonics, Fremont CA) for 30 min to facilitate extraction of indigoidine into the solvent. THF can be used for extraction of indigoidine as an alternative to DMF. The extraction process was repeated with fresh solvent until the supernatant was colorless and the organic solution was combined, evaporated in vacuo, and the resultant dark solid was washed twice each with water, methanol, ethyl acetate and hexane (15 mL × 2) using centrifugation. Finally, the product was dried under high vacuum to yield 400 mg dry indigoidine from 400 mL culture broth with a concentration of 2 g L−1 indigoidine (Fig. S8†). Solution 1H nuclear magnetic resonance (NMR) spectra were recorded on a Bruker AVQ-400 (400 MHz) spectrometer (Billerica, MA) operating with an Avance electronics console (Las Vegas, NV).
000g for 5 min to separate the cells from the media. After removal of the supernatant, the cell pellet was resuspended in an equal volume of dimethylformamide (DMF) and cell lysis was performed by subjecting the cells to an using ultrasonic bath (Model 5510, Branson Ultrasonics, Fremont CA) for 30 min to facilitate extraction of indigoidine into the solvent. THF can be used for extraction of indigoidine as an alternative to DMF. The extraction process was repeated with fresh solvent until the supernatant was colorless and the organic solution was combined, evaporated in vacuo, and the resultant dark solid was washed twice each with water, methanol, ethyl acetate and hexane (15 mL × 2) using centrifugation. Finally, the product was dried under high vacuum to yield 400 mg dry indigoidine from 400 mL culture broth with a concentration of 2 g L−1 indigoidine (Fig. S8†). Solution 1H nuclear magnetic resonance (NMR) spectra were recorded on a Bruker AVQ-400 (400 MHz) spectrometer (Billerica, MA) operating with an Avance electronics console (Las Vegas, NV).
Results and discussion
R. toruloides as a platform host for production of the non-ribosomal peptide indigoidine
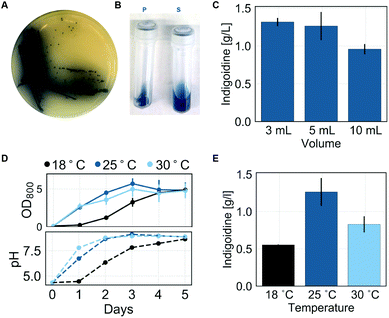
To establish the indigoidine pathway in R. toruloides, we genomically integrated codon-optimized sequences of the B. subtilis PPTase sfp, previously shown to successfully activate the S. lavendulae apo-BpsA in S. cerevisiae,29 and the single module type NRPS gene bpsA into R. toruloides IFO0880 via A. tumefaciens-mediated transformation (ATMT).22Transformants that successfully expressed these two genes were dark blue due to high-level production of indigoidine (Fig. 2A). In addition to strong pigmentation of the resultant colonies, the surrounding agar showed dark blue tinting that increased in intensity over time, indicating secretion of indigoidine from the cells. This behavior was maintained in liquid cultures, where the pigmentation appeared in both the cell pellet and the supernatant (Fig. 2B).
After initial demonstration of indigoidine formation in R. toruloides, we optimized indigoidine production by assessing the impact of physical cultivation parameters on production efficiency. A single transformant (hereafter referred to as BlueBelle) was selected for the subsequent optimization. Physical cultivation parameters, including oxygen availability and temperature of the culture, are known to affect the productivity of microbial systems. The filling volume, and correspondingly the headspace of a culture, is recognized to impact oxygen transfer from the gaseous into the liquid phase.31–33 Thus, to characterize the effect of oxygen transfer and cultivation temperature on the efficiency of indigoidine production, we compared indigoidine production of BlueBelle cultivated in 55 ml culture tubes using three different filling volumes (3 mL, 5 mL and 10 mL corresponding to 5%, 9% and 18% filling volume) and three different temperatures (18 °C, 25 °C and 30 °C) after 3 days in standard synthetic defined media with a starting concentration of 100 g L−1 glucose and 5 g L−1 of ammonium sulfate. Oxygenation is a step required for the formation of indigoidine from glutamine.10 Hence, we expected the superior oxygen transfer associated with lower filling volume of culture tubes to positively affect pigment production. We detected no significant difference in pigment production for 3 mL and 5 mL cultures. However, as expected, indigoidine production was reduced by 24% in 10 mL cultures compared to 3 mL and 5 mL cultures (Fig. 2C).
The optimal production temperature of a microbial bioprocess can cause a trade-off between optimal growth temperature and optimal temperature for pathway efficiency, the latter often correlating with production of correctly folded proteins. To determine the optimal temperature for indigoidine production and to establish whether the production of indigoidine is linked with microbial growth, we measured microbial growth by means of OD800 measurements of 5 mL production cultures of BlueBelle grown at three different temperatures (18 °C, 25 °C and 30 °C) and quantified indigoidine production after three days of production (Fig. 2D and E). We observed no significant difference in the growth profile between cultures grown at 25 °C and 30 °C, whereas the growth rate was decreased for cultures at 18 °C (Fig. 2D). While all cultures reached a similar final OD800 independent of cultivation temperature, cultures grown at 25 °C and 30 °C reached the final OD800 after 3 days of cultivation and cultures grown at 18 °C did not reach the final OD800 until 5 days after start of the cultivation. Interestingly, the culture pH tracked with microbial growth: the pH of all cultures increased throughout the course of production and reached a similar final value of 8.9. Similar growth and pH profiles were observed for wild-type cultures cultivated under the same conditions (Fig. S3†).
Overall, we observed that cultivation temperature impacts indigoidine production more strongly than culture volumes for the conditions tested (Fig. 2E). Amongst the tested temperatures, cultivation at 25 °C resulted in the highest indigoidine titer after 3 days. In comparison to cultures grown at 25 °C, the indigoidine titer achieved in cultures grown at 30 °C and 18 °C was reduced by 34% and 57%, respectively. Thus, while BlueBelle shows no significant growth impairment when cultivated at 30 °C compared to 25 °C, the production of indigoidine is significantly decreased at higher temperatures.
Assuming that the production of indigoidine is directly coupled with biomass at lower temperatures, the differences in indigoidine titer at 18 °C compared to 25 °C could be caused by the differences of biomass accumulation at these temperatures. To test this hypothesis, we quantified indigoidine after 5 days of cultivation when all cultures had reached similar OD800. We found that the indigoidine titer produced by cultures grown at 18 °C increased during this period in comparison to cultures grown at higher temperatures and reached similar final titers of indigoidine as that produced by cultures grown at 25 °C (Fig. S4†). These findings indicate that, at lower temperatures, indigoidine production is linked to microbial growth.
We observed a change in the hue of indigoidine culture over the course of the dye production. While the supernatant of all production cultures is blue at initial phases of growth, the color changed to green with increasing pH for all cultures, independent of growth temperature, and to yellow at later stages of the production for cultures grown at 25 °C and 30 °C (Fig. S5†). To better characterize the pH dependent colorimetric properties of this compound, we performed pH adjustments of supernatant from a production culture and recorded the absorption spectra of the resultant solutions (Fig. S6†). To avoid prior changes in pigment hue due to changes in the culture pH, we utilized the supernatant of a production culture cultivated at 25 °C for 24 hours at neutral pH (Fig. 2D).
We observed distinct changes in the color of the solution with changes in the pH: the hue of the supernatant ranged from red in acidic conditions to blue at neutral pH, and green under alkaline conditions (Fig. S6A and S6D†). To determine the origin of these colors and confirm the aforementioned observations, we recorded UV-Vis spectra of these solutions with pH ranging from 9 to 2 in intervals of 1. We detected three absorption maxima in these solutions: 470 nm, 500 nm and 610 nm corresponding to perception of orange, red and blue (Fig. S6A†). To investigate the reversibility of the process, we re-adjusted the pH from pH 2 to pH 7 using 1 M NaOH. Indeed, we observed a color change of the solution from red to blue (Fig. S6B†). Both the red and blue colors fade over time in the presence of air, resulting in an orange solution with absorption maxima at 470 nm. Similarly, an absorption band at 470 nm was detected in the UV-Vis spectrum of the supernatant at pH 9 that appeared yellow (Fig. S6A†). Thus, we hypothesized the orange pigment to be a product of oxidation. To confirm this hypothesis, we oxidized the various products at different pH (e.g. red pigment at pH 2, blue pigment at pH 7 and orange pigment at pH 9) using hydrogen peroxide (Fig. S6C†). We observed a change in color towards yellow/orange for all three solutions upon addition of the oxidizing reagent, which coincided with the disappearance of the absorption maxima at 610 nm (blue pigment). These observations are in agreement with studies performed by Kuhn et al. to characterize the chemical properties of indigoidine and are summarized in Fig. S6D.†34 ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Addition of the reducing agent dithionite to the various products at different pH (e.g. red pigment at pH 2, blue pigment at pH 7 and orange pigment at pH 9) resulted in loss of color (Fig. S6C†). This observation is in agreement with previous work, describing the formation of a reduced, colorless but fluorescent form, leuco-indigoidine.35,36
Addition of the reducing agent dithionite to the various products at different pH (e.g. red pigment at pH 2, blue pigment at pH 7 and orange pigment at pH 9) resulted in loss of color (Fig. S6C†). This observation is in agreement with previous work, describing the formation of a reduced, colorless but fluorescent form, leuco-indigoidine.35,36
Importance of nitrogen source for indigoidine production
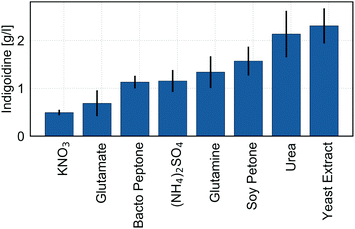
In addition to enhancing physical cultivation parameters like oxygen availability and cultivation temperature, optimization of the production medium composition presents a powerful means to improve a microbial process. The capacity to efficiently utilize multiple, low-cost feedstocks is critical to maintain sustainability and increase economic feasibility of a bioprocess due to potential changes in substrate cost and availability. NRPs in general, and the blue 3′,3′-bipyridyl target molecule indigoidine specifically, are molecules with high nitrogen content, increasing the importance of nitrogen supplied to the culture medium.To determine the most suitable nitrogen source for the production of indigoidine in R. toruloides, we cultivated BlueBelle for three days in minimal media containing 100 g L−1 glucose and individual widely-available nitrogen sources, and then quantified indigoidine. To avoid secondary effects originating from carbon–nitrogen ratio, each nitrogen source was normalized to the elemental nitrogen content to contain 1.06 g L−1 nitrogen and added accordingly (Table S1†). In general, indigoidine production varied significantly based on nitrogen source, covering a 4-fold range from the least suitable nitrogen source (potassium nitrate, KNO3) to the most suitable, which was urea and yeast extract (Fig. 3), emphasizing the importance of the choice of nitrogen source for the production of indigoidine.
The commonly used inorganic nitrogen source ammonium sulfate resulted in comparatively low indigoidine production (Fig. 3). In agreement with studies examining various nitrogen sources for lipid production in R. toruloides,37 this observation indicates the presence of nitrogen catabolite repression (NCR) elicited by extracellular ammonium. The presence of extracellular ammonium has been suggested to repress various catabolic enzymes required for nitrogen assimilation, including glutamate dehydrogenase which catalyzes the reversible reaction of glutamate to alpha-ketoglutarate,37 and this could lead to reduced flux towards the final product, indigoidine.
Other studies have shown that R. toruloides can readily assimilate amino acids as nitrogen source for lipid production under nitrogen-limited conditions.38 To allow utilization of nitrogen for cellular metabolism, any nitrogen source first has to be converted to glutamate or glutamine, which serve as nitrogen donors for all other nitrogen containing compounds in the cell.39 Interestingly, exogenous addition of the amino acids glutamine or glutamate, precursors tin the pathway to indigoidine, did not yield high production of indigoidine (Fig. 3). This observation could originate from slow assimilation or a regulatory response that reduces native flux toward these compounds under non-nitrogen limited conditions. These conclusions are in general agreement with differential transcriptomic and proteomic analyses of R. toruloides under different nitrogen regimes, illustrating the effect of nitrogen availability on central carbon metabolism and lipid metabolism.40
In contrast to the amino acids glutamine and glutamate, use of urea as a nitrogen source resulted in a high titer of indigoidine, indicating that the use of urea as sole nitrogen source elicits faster uptake and assimilation. This conclusion is in agreement with previous reports describing faster uptake of urea and a faster metabolic response as compared to ammonium or glutamate, partly due to upregulation of urease activity when urea is used as nitrogen source.38 Additionally, the enhanced production of indigoidine could result from an increased accumulation of the precursor glutamine. To use urea as a nitrogen donor, the compound is first hydrolyzed by a urease or decarboxylated by an urea amidolyase to yield ammonia, which in turn is used as substrate to form glutamate and glutamine from alpha-ketoglutarate.40,41 Indeed, early biochemical studies revealed that enzymes required for L-glutamine synthesis from ammonia show higher activities when urea was used as nitrogen source compared to glutamine.38
The use of yeast extract as a nitrogen source resulted in similarly high titers of indigoidine (Fig. 3). Yeast extract is a complex nitrogen source, predominantly prepared by autolysis of yeast cells, and thus contains all soluble cell compounds, such as peptide and amino acids, vitamins, trace elements and nucleic acids.42 However, the poorly defined composition, batch-to-batch variability and high cost of commercially available yeast extracts present constraints for industrial processes.42,43 Based on these factors, we optimized the process using urea as the defined nitrogen source.
Nitrogen limitation is known to enhance accumulation of storage lipids in oleaginous yeasts.37 The effects of varying nitrogen concentrations and nitrogen limitation on production of lipids and fatty acid-derived products have been thoroughly studied in R. toruloides.44 To better characterize the effect of nitrogen availability on the production of the TCA cycle derived, heterologous product indigoidine, we cultivated BlueBelle in minimal medium containing 100 g L−1 glucose at different carbon/nitrogen (C/N) ratios, ranging from C/N 2 to C/N 160, and determined indigoidine titer as well as culture pH after 3 days (Fig. 4).
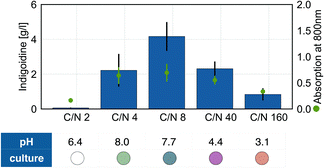 | ||
| Fig. 4 Impact of C/N ratio on the indigoidine production, microbial growth and culture pH after 3 days of cultivation using 100 g L−1 glucose and varying amounts of urea as carbon and nitrogen source respectively. Differently colored circles in the table represent depictions of the culture hue. Images of the culture broth can be found in Fig. S7.† Error bars represent SD of 3 replicates. | ||
As the C/N ratio presents an important factor known to affect carbon flux distribution into either storage lipids or by-products in oleaginous yeast,45–47 specifically citrate in R. toruloides,40 we expected to find an interdependency between C/N ratio and indigoidine production. Indeed, amongst the conditions tested, we found a C/N ratio of 8 to result in the highest indigoidine production after 3 days. A deviation from this ratio to either lower or higher C/N ratio, e.g. C/N 4 or C/N 40 respectively, resulted in lower titers. Growth was mostly stalled at the lowest C/N ratio tested (C/N 2) as well as the highest C/N ratio tested (C/N 160), possibly due to inhibitory effects originating from high amounts of urea added to the medium (42.5 g L−1)48 and nitrogen limitation, respectively.
Interestingly, the color of the culture also changed with varying C/N ratio (Fig. 4), indicating changes in pH (Fig. S6†) during the cultivation triggered by carbon or nitrogen limitations. This observation was further confirmed by pH measurements. After 3 days of cultivation, the cultures in low C/N ratio, thus not nitrogen limited, showed a green or blue hue, corresponding to pH values of 8.0 for C/N 4 and 7.7 for C/N 8, respectively. In contrast, cultures grown in media containing a high C/N ratio, thus under nitrogen limitation, showed a purple to red hue, indicating a low pH, which was confirmed by pH measurements. This observation could originate directly from the varying concentrations of urea in these solutions49 or from modified metabolic activities. To test whether the increase in pH in cultures containing a low C/N ratio is solely a result of high urea concentrations, we cultivated BlueBelle in rich media with the same concentrations of the complex nitrogen sources yeast extract and peptone containing either 20 g L−1 (YPD) or 100 g L−1 glucose (YP10D) (Fig. S7†). Here, as well, we found that the pH increased over time for media having a lower C/N ratio (YPD). This observation renders the hypothesis that the varying concentrations of urea are the sole cause for the differences in culture pH unlikely. However, the observed decrease in culture pH with increasing C/N ratio together with the decrease in production, is consistent with the current understanding of the regulation of lipid accumulation in R. toruloides.40,44,50 Generally, nitrogen limitation was found to enhance lipid accumulation in oleaginous as well as non-oleaginous yeasts.51 Under these conditions, oleaginous yeasts produce large amounts of TCA cycle intermediates, foremost citrate.37,52 During nitrogen starvation the activity of the isocitrate dehydrogenase is decreased, which is caused by low levels of its allosteric regulator AMP, resulting from an increased activity of AMP deaminase to ensure a stable pool of internal ammonium required for cell viability.40,53 Consequently, mitochondrial citrate is not further metabolized via the TCA cycle resulting in accumulation and subsequent secretion into the cytosol where it serves as substrate to ATP citrate lyase to form acetyl-CoA for fatty acid biosynthesis.53,54 Thus, the decrease in production together with the decrease in culture pH with increasing C/N ratio likely originates from the metabolic flux shift to lipid biosynthesis. This shift results in the stalling of flux through the TCA cycle towards the precursor of indigoidine, alpha-ketoglutarate, and is accompanied by enhanced secretion of citrate.38
Further, we observed a delayed increase in indigoidine production after 7 days of cultivation at C/N 40 (Fig. S7†), possibly due to remobilization of storage lipids accumulated during earlier stages of the cultivation as a result of the onset carbon depletion at late stages of the cultivation.55,56 This observation further supports the hypothesis, that higher C/N ratios favor storage lipid accumulation over completion of the TCA cycle and underlines the importance of a customized carbon-to-nitrogen ratio for efficient production of different products. Further experiments are needed to fully understand the regulatory mechanisms underlying the balance between lipid biogenesis and TCA cycle progression.
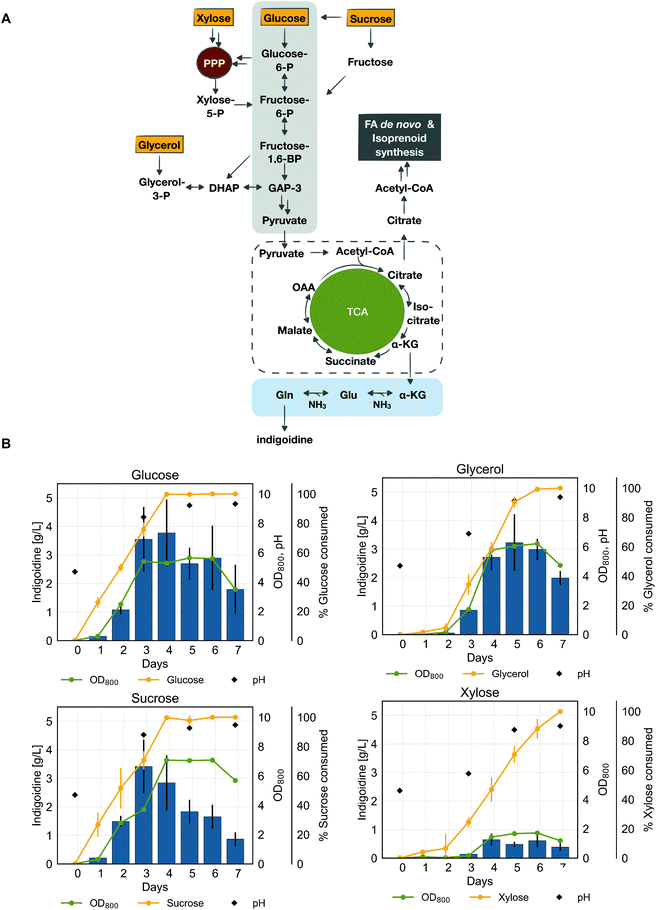
Utilization of various carbon sources for indigoidine production
Various carbon sources, such as glucose, glycerol and biomass derived sugars can feed into the TCA cycle of R. toruloides via different yet integrated pathways, and can be used to produce native products such as lipids and carotenoids as well as heterologous bioproducts with relatively high initial titers such as fatty alcohols and terpenes, all of which funnel mainly through the metabolite acetyl-CoA.26,57–59 It remains unknown whether TCA cycle intermediates can be used to produce heterologous bioproducts with similar efficiency to the natural products without engineering the carbon flux through central metabolism. Thus, we sought to characterize the ability of R. toruloides to efficiently convert four commonly-used carbon sources into indigoidine: glucose, xylose, sucrose and glycerol.To gain a more detailed understanding of the efficiency of the conversion of these carbon sources, we examined sugar utilization, cell growth, culture pH, and indigoidine production over the course of seven days. Generally, we found that indigoidine production correlated linearly with biomass formation for all the carbon sources tested (Fig. 5). Similar to observations made in the experiment investigating indigoidine production at different cultivation temperatures (Fig. 2D), we found the culture pH to increase with biomass density over the course of the cultivation, reaching similar final values (pH ∼ 9) in all conditions tested.
Using glucose and sucrose as carbon sources resulted in similar production profiles, especially during the first 3 days of cultivation (Fig. 5, left panel). During this initial phase of production, biomass formation coincided with sugar consumption and indigoidine production. While sugar consumption rate remained comparable for these carbon sources and both sucrose and glucose were fully consumed after 4 days, the growth profiles show differences in the later stages of production (after 4 days). The use of sucrose as carbon source resulted in a further increase in biomass formation from day 3 to day 4, reaching a maximum cell density (OD800) of 7.1 on day 4, whereas growth stalled after 3 days when grown on glucose. Interestingly, in both cases, indigoidine titers decreased in the later stages of production.
Unlike sucrose and glucose, we observed a delay in consumption of glycerol and xylose. This observation indicates the need of adaptation to allow utilization of these carbon sources. For glycerol, this observation is consistent with reports describing carbon catabolite repression after cultivation in glucose containing medium (Fig. 5, right panel).59 With the exception of the initial adaptation phase needed for glycerol, the production profile of indigoidine on glycerol as a carbon source is highly similar to the production on glucose and sucrose. Similarly, biomass formation coincided with sugar consumption and indigoidine production, reaching a lower maximum indigoidine titer of 3.2 g L−1 in glycerol compared to 3.8 g L−1 in glucose. However, biomass accumulation in glycerol was higher compared to growth on glucose (OD800 of 6.21 compared to 5.64). These results are consistent with previous reports60 that compared growth and lipid production of R. toruloides from glycerol and glucose.
Of all the carbon sources tested, xylose resulted in the lowest growth and indigoidine titer. Similarly to growth on glycerol, xylose was steadily consumed after an initial adaptation phase, however at a slower rate than any of the other carbon sources as previously reported.26 Xylose is metabolized via the pentose-phosphate-pathway, which results in loss of carbon through CO2 and thus less biomass and product formation.59
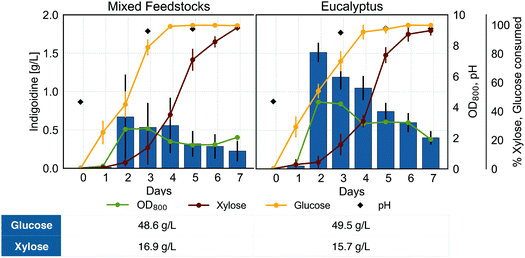
Production of the NRP indigoidine from lignocellulosic biomass
The ability to convert various carbon sources from lignocellulosic biomass to a bioproduct is an important requirement for a sustainable bioprocess from renewable feedstocks. To demonstrate the potential of R. toruloides to produce indigoidine from a mix of carbon liberated from lignocellulosic biomass, we cultivated BlueBelle in hydrolysates derived from several lignocellulosic feedstocks and used as a sole carbon source. We examined growth, sugar utilization, indigoidine production, and culture pH using hydrolysates from eucalyptus as well as from mixed feedstocks (switchgrass and eucalyptus) predominantly containing glucose and xylose (Fig. 6), which were thoroughly characterized in an earlier study.28In the hydrolysate cultivations, efficient xylose consumption was delayed by 2 days of cultivation, indicative of a required adaptation phase for efficient utilization of xylose as observed when using xylose as sole carbon source for indigoidine production (Fig. 5, bottom right). Similar to growth on single carbon sources, we found that the indigoidine titer corresponded to biomass accumulation, reaching maximum titers of 1.51 g L−1 and 0.67 g L−1 for eucalyptus and mixed feedstocks, respectively. In contrast to growth on single carbon sources, the maximum indigoidine titer on hydrolysates was reached earlier, after 2 days of growth for both liquors tested, and decreased afterwards, when the culture was not yet depleted of all carbon sources. The early decrease in detectable indigoidine when grown on biomass-derived carbon sources compared to a refined, single carbon source could be due to inhibitors originating from the lignocellulosic biomass.61 This hypothesis is supported by the observation of marked differences in growth and sugar consumption between the two hydrolysates despite having very similar amounts of sugars and total carbon, as different biomass was used for the preparation while maintaining the same deconstruction and saccharification protocol.28
Considering the lower amount of total sugar and specifically lower concentrations of glucose in the hydrolysate obtained from mixed feedstocks and eucalyptus (Fig. 6) as compared to the 100 g L−1 of sugar used in the single carbon cultivations, the lower titers obtained from the lignocellulosic hydrolysates are not surprising. Overall, successful production of indigoidine highlights the potential of R. toruloides to convert lignocellulosic biomass and presents the first report of non-ribosomal peptide production from renewable feedstocks.
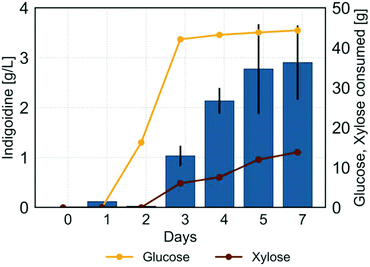
The use of lignocellulosic hydrolysates as carbon source in many cases includes a solid–liquid separation and several washing steps after the pretreatment process,62 which results in loss of lignocellulosic biomass and increased process cost. To demonstrate the feasibility of indigoidine production in R. toruloides from lignocellulosic hydrolysates in a more sustainable manner, we performed a separation-free biomass deconstruction and conversion process using sorghum that couples pretreatment, saccharification and indigoidine production in a single unit operation. R. toruloides was cultivated under this regime in a 2 L bioreactor as previously described by Sundstrom et al. for the production of the terpene bioproduct bisabolene.19 HPLC analysis showed that the prepared hydrolysate contained 44.3 g L−1 glucose and 13.8 g L−1 xylose (Fig. S2†). To avoid additional cellular stress due to high pH as well as oxidation of the pigment in the culture broth as observed in shake flasks, the pH of the culture was maintained at 7. In order to allow R. toruloides to adapt to the ionic liquid cholinium lysinate used for pretreatment and any other inhibitors present in the hydrolysate, the seed culture used to inoculate the bioreactor contained a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mix of hydrolysate and rich medium.
1 mix of hydrolysate and rich medium.
Unlike previous experiments performed in shake flasks, glucose utilization was delayed by a day when grown in the bioreactor using the one-pot lignocellulosic hydrolysate (Fig. 7). This observation suggests a need to adapt to the one-pot environment, including lignin-derived aromatic compounds, some of which can be inhibitory.61 Indeed, Yaegashi et al. found that R. toruloides engineered for bisabolene production preferably utilized the aromatic compound p-coumaric acid in a mock medium also containing glucose and xylose.26 Further, they showed that other lignin-related aromatic compounds can be readily consumed by R. toruloides including ferulic acid, even though some were inhibitory.26 Additionally, the ionic liquid used for pretreatment, cholinium lysinate, is known to inhibit microbial growth at higher concentrations. As the seed culture was pre-grown in diluted hydrolysate, the delay in growth observed after transfer to the bioreactor could have been caused by the need for an adaptation phase to the higher concentration of ionic liquid present in the hydrolysate.
After an initial lag, glucose was mostly consumed within 3 days of cultivation. Similar to observations made in previous experiments, efficient xylose consumption was delayed until the majority of glucose was consumed. Indigoidine production commenced with almost complete consumption of glucose at day 3 of the cultivation and increased to a final titer of 2.91 g L−1. Overall, the indigoidine yield from sugar achieved in the separation-free bioreactor process was 0.045 g/g, which was 18% higher than the one achieved at shake flask level using the best single carbon source glucose (0.038 g/g) and 95% higher compared to using the best hydrolysate derived from eucalyptus (0.023 g/g).
Fed-batch process with pH control results in increase of indigoidine titer
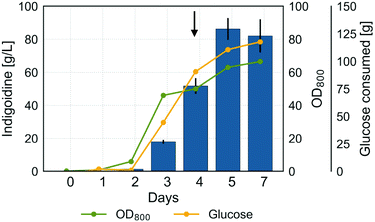
Following cultivation optimization in shake flasks and demonstration of sustainable indigoidine production using lignocellulosic feedstocks, we sought to further enhance indigoidine production in R. toruloides and to demonstrate the feasibility of process scale-up. In previous experiments, we demonstrated a positive correlation between biomass accumulation and indigoidine production. Further, we established the importance of the C/N ratio for efficient indigoidine production and pH for optimal growth of R. toruloides. Thus, we hypothesized that maintaining a neutral pH and ensuring an excess of carbon and nitrogen at an adequate ratio would allow for higher accumulation of biomass while maintaining efficient indigoidine production. To test this hypothesis, we performed a fed-batch cultivation in a 2 L bioreactor at pH 7.Similar to the experiments performed in shake flasks, indigoidine production tracked with biomass accumulation under these conditions (Fig. 8). However, unlike experiments performed in shake flasks, efficient glucose consumption was delayed by 48 h, indicating the need to adapt to the bioreactor environment. After an initial lag phase with slow glucose consumption and growth, the carbon source was utilized rapidly in the next cultivation phase accompanied by an increase in indigoidine production and biomass formation (days 2 to 4). In the last phase of the cultivation, after 4 days, indigoidine production increased further, while microbial growth slowed down significantly. A maximum indigoidine titer of 86.3 ± 7.4 g L−1 was achieved after 5 days (116 h) of cultivation with an overall productivity and yield of 0.73 g L−1 h−1 and 0.91 gindigoidine/gglucose, respectively (99.4 g indigoidine net produced and 109.6 g glucose net consumed).
In addition to the production of a target compound, the practicability of separating the compound from the culture broth and the achieved quality of the product after purification have an important bearing for the economic success of a biotechnological process. To demonstrate the feasibility of separation and purification of indigoidine, we performed indigoidine extraction and purification to obtain a dry powder and performed NMR analysis to confirm the structure and purity (Fig. S8†). To our knowledge, these results represent the highest reported production of NRPs in the scientific literature. The culture reached a final OD800 of 66, corresponding to a dry cell weight (DCW) of 27.6 ± 4.9 g l−1. The obtained DCW values are similar to those previously reported for high-gravity fed-batch cultivation for bisabolene production using R. toruloides (25 g L−1 after 135 h of production).26 However, they remain below the DCW values reported for lipid overproduction (106.5 g L−1 after 134 h of production), likely due the high lipid content accounting for 67.5% glipid/gbiomass or 71.9 g L−1 of DCW.17
The high titers observed in our study compare favorably with bioproduction of other NRPs. Industrial production of the antibiotic penicillin has been achieved in P. chrysogenum but is an example of producing a native fungal NRP in its natural fungal host.63 There are no directly comparable systems that both utilize a bacterial NRP in a fungal host and rely on conversion of a crude hydrolysate in bioreactors. The results in the present study far outperforms earlier reports of Indigoidine production in both bacterial (8.81 ± 0.21 g l−1 in E. coli in shake flasks)64 and fungal systems (0.98 g L−1 in S. cerevisiae in 2 L bioreactor).29 Bacterial NRPs have been mainly examined in bacterial systems and are typically small-scale batch-mode proof-of-concept studies, conducted in the model bacterium Escherichia coli,65,66 or hosts that natively express NRPSs such as Streptomyces sp.67 Most recently, a study in S. cerevisiae used the bpsA based indigoidine production to examine the impact of carbon catabolite repression on the production of heterologous TCA cycle derived products.29 However, as a Crabtree-positive yeast that cannot efficiently utilize the major classes of carbon present in lignocellulosic biomass, S. cerevisiae may not be an ideal system for the sustainable production of this NRP.68–70
Each production system, comprised of carbon source, a host organism and a production pathway, is unique and requires specific process parameters to ensure optimal performance. Thus, we lack a suitable Indigoidine production system against which we can benchmark our study. However individual aspects of the present production system compare well with existing processes.
Conclusions
This study represents the necessary innovation required to establish an industrially relevant platform for the production of non-ribosomal peptides – a large category of natural products with a broad range of applications. Specifically, we demonstrate the sustainable production of indigoidine using the basidiomycete yeast R. toruloides. The goal of this study was to develop a platform that could be used for the industrially relevant, sustainable, high-titer production of this promising dye candidate.To this end, several important biotechnological aspects, including the demonstration of fed-batch bioreactor processes, designed specifically to improve production, were successful, reaching indigoidine titers of 86.3 ± 7.4 g L−1 and yield 0.91 gindigoidine/gglucose from glucose and urea. Additionally, we showed that R. toruloides can convert lignocellulosic hydrolysates and various other carbon and nitrogen sources to indigoidine, and that the efficiency of indigoidine production is partly dependent on the carbon-to-nitrogen ratio.
In addition to containing the same chromophore as the industrially dominant blue dye indigo, indigoidine can act as powerful radical scavenger and strong antioxidant.10 Further, indigoidine may have potential use as a colorimetric redox state and pH sensor, rendering it a natural blue dye with various industrially important properties. Due to the structural similarity to indigo in terms of amine functional groups that result in strong intermolecular hydrogen bonds and π–π stacking interactions, indigoidine may be suitable for similar industrial applications demonstrated for indigo and its derivatives, including biodegradable organic semiconductors and transistors.71–73 However, the nitrogen content of indigoidine is higher compared to indigo, potentially resulting in different chemical and physical properties, expanding the application space of this class of compounds.
While indigoidine itself shows immense potential for industrial applications, the high titers achieved in this study further underline the potential to explore the basidiomycete R. toruloides as a production host for other classes of heterologous products besides NRPs. The successful demonstration of activating a heterologous NRPS via phosphopantetheinylation suggests that this yeast has potential to produce other classes of compounds that require this step, such as polyketides (PK) or NRP-PK hybrids.74
Overall, our study presents the first demonstration of the microbial production of a heterologous NRPS-derived product, the blue pigment indigoidine, at titers approaching 100 g L−1. These results suggest that R. toruloides may be key to enabling industrial production of a wide range of valuable NRP and PK bioproducts. Further, our results demonstrate the ability of R. toruloides to facilitate the critical phosphopantetheinylation step required to access to broad category of compounds derived from NRPs and other secondary metabolites. Our results along with the emergence of effective synthetic biology tools and its native industrial features emphasize the potential of R. toruloides as highly versatile microbial production host.
Author contributions
MW, JG and AM conceptualized the project and designed the study. MW, LP, JPP, YL, JM, GP and GMG conducted the experiments and collected data. MW, LP, JPP, YL, ES and GP performed data analysis. MW, JG and AM composed the manuscript. JG, DT, TP, BS, JDK and AM provided critical feedback on the manuscript and provided resources. All authors read and approved the manuscript.Conflicts of interest
J.D.K. has financial interests in Amyris, Lygos, Constructive Biology, Demetrix, Napigen, and Maple Bio.Acknowledgements
We thank James Kirby, Joonhoon Kim, Jeff Skerker, Jon Magnussen, Harsha Magurudeniya and Alberto Rodriguez for valuable discussions during the preparation of this manuscript. This work was part of the DOE Joint BioEnergy Institute (http://www.jbei.org) supported by the U.S. Department of Energy, Office of Science, Office of Biological and Environmental Research, through contract DE-AC02-05CH11231 between Lawrence Berkeley National Laboratory and the U. S. Department of Energy. The work at the Advanced Biofuels and Bioproducts Process Development Unit was supported by the US Department of Energy's BioEnergy Technology Office (BETO) in the Office of Energy Efficiency and Renewable Energy, through the contract with LBNL. The United States Government retains and the publisher, by accepting the article for publication, acknowledges that the United States Government retains a non-exclusive, paid-up, irrevocable, worldwide license to publish or reproduce the published form of this manuscript, or allow others to do so, for United States Government purposes. The Department of Energy will provide public access to these results of federally sponsored research in accordance with the DOE Public Access Plan (http://energy.gov/downloads/doe-public-access-plan).References
- M. A. Martínez-Núñez and V. E. L. y López, Sustainable Chem. Processes, 2016, 4, 13 CrossRef.
- S. Yuzawa, J. D. Keasling and L. Katz, J. Antibiot., 2017, 70, 378–385 CrossRef CAS PubMed.
- P. S. Vankar, Reson., 2000, 5, 73–80 CrossRef CAS.
- S. Kadolph, Delta Kappa Gamma Bulletin, 2008.
- A. C. Dweck, Int. J. Cosmet. Sci., 2002, 24, 287–302 CrossRef CAS PubMed.
- T. M. Hsu, D. H. Welner, Z. N. Russ, B. Cervantes, R. L. Prathuri, P. D. Adams and J. E. Dueber, Nat. Chem. Biol., 2018, 14, 256–261 CrossRef CAS PubMed.
- R. J. N. Frandsen, P. Khorsand-Jamal, K. T. Kongstad, M. Nafisi, R. M. Kannangara, D. Staerk, F. T. Okkels, K. Binderup, B. Madsen, B. L. Møller, U. Thrane and U. H. Mortensen, Sci. Rep., 2018, 8, 12853 CrossRef PubMed.
- H. Takahashi, T. Kumagai, K. Kitani, M. Mori, Y. Matoba and M. Sugiyama, J. Biol. Chem., 2007, 282, 9073–9081 CrossRef CAS PubMed.
- D. Yu, F. Xu, J. Valiente, S. Wang and J. Zhan, J. Ind. Microbiol. Biotechnol., 2013, 40, 159–168 CrossRef CAS PubMed.
- S. Reverchon, C. Rouanet, D. Expert and W. Nasser, J. Bacteriol., 2002, 184, 654–665 CrossRef CAS PubMed.
- R. Kuhn, M. P. Starr, D. A. Kuhn, H. Bauer and H. J. Knackmuss, Arch. Mikrobiol., 1965, 51, 71–84 CAS.
- M. P. Starr, G. Cosens and H. J. Knackmuss, Appl. Microbiol., 1966, 14, 870–872 CAS.
- M. Wehrs, D. Tanjore, T. Eng, J. Lievense, T. R. Pray and A. Mukhopadhyay, Trends Microbiol., 2019 DOI:10.1016/j.tim.2019.01.006.
- V. Chubukov, A. Mukhopadhyay, C. J. Petzold, J. D. Keasling and H. G. Martín, NPJ Syst. Biol. Appl., 2016, 2, 16009 CrossRef PubMed.
- J. Nielsen and J. D. Keasling, Cell, 2016, 164, 1185–1197 CrossRef CAS PubMed.
- Y.-K. Park, J.-M. Nicaud and R. Ledesma-Amaro, Trends Biotechnol., 2018, 36, 304–317 CrossRef CAS PubMed.
- Y. Li, Z. Zhao and F. Bai, Enzyme Microb. Technol., 2007, 41, 312–317 CrossRef CAS.
- J. Xu, J. Econ. Dyn. Control, 2017, 80, 54–74 CrossRef.
- E. Sundstrom, J. Yaegashi, J. Yan, F. Masson, G. Papa, A. Rodriguez, M. Mirsiaghi, L. Liang, Q. He, D. Tanjore, T. R. Pray, S. Singh, B. Simmons, N. Sun, J. Magnuson and J. Gladden, Green Chem., 2018, 20, 2870–2879 RSC.
- C. M. J. Koh, Y. Liu, Moehninsi, M. Du and L. Ji, BMC Microbiol., 2014, 14, 50 CrossRef PubMed.
- S. Zhang, J. M. Skerker, C. D. Rutter, M. J. Maurer, A. P. Arkin and C. V. Rao, Biotechnol. Bioeng., 2016, 113, 1056–1066 CrossRef CAS PubMed.
- X. Lin, Y. Wang, S. Zhang, Z. Zhu, Y. J. Zhou, F. Yang, W. Sun, X. Wang and Z. K. Zhao, FEMS Yeast Res., 2014, 14, 547–555 CrossRef CAS PubMed.
- A. M. B. Johns, J. Love and S. J. Aves, Front. Microbiol., 2016, 7, 1666 Search PubMed.
- L. C. Nora, M. Wehrs, J. Kim, J.-F. Cheng, A. Tarver, B. A. Simmons, J. Magnuson, M. Harmon-Smith, R. Silva-Rocha, J. M. Gladden, A. Mukhopadhyay, J. M. Skerker and J. Kirby, BioRxiv, 2019, 592774 Search PubMed.
- S. Fillet, C. Ronchel, C. Callejo, M.-J. Fajardo, H. Moralejo and J. L. Adrio, Appl. Microbiol. Biotechnol., 2017, 101, 7271–7280 CrossRef CAS PubMed.
- J. Yaegashi, J. Kirby, M. Ito, J. Sun, T. Dutta, M. Mirsiaghi, E. R. Sundstrom, A. Rodriguez, E. Baidoo, D. Tanjore, T. Pray, K. Sale, S. Singh, J. D. Keasling, B. A. Simmons, S. W. Singer, J. K. Magnuson, A. P. Arkin, J. M. Skerker and J. M. Gladden, Biotechnol. Biofuels, 2017, 10, 241 CrossRef PubMed.
- T. S. Ham, Z. Dmytriv, H. Plahar, J. Chen, N. J. Hillson and J. D. Keasling, Nucleic Acids Res., 2012, 40, e141 CrossRef PubMed.
- C. Li, D. Tanjore, W. He, J. Wong, J. L. Gardner, V. S. Thompson, N. A. Yancey, K. L. Sale, B. A. Simmons and S. Singh, BioEnergy Res., 2015, 8, 982–991 CrossRef CAS.
- M. Wehrs, J.-P. Prahl, J. Moon, Y. Li, D. Tanjore, J. D. Keasling, T. Pray and A. Mukhopadhyay, Microb. Cell Fact., 2018, 17, 193 CrossRef PubMed.
- C. Li, B. Knierim, C. Manisseri, R. Arora, H. V. Scheller, M. Auer, K. P. Vogel, B. A. Simmons and S. Singh, Bioresour. Technol., 2010, 101, 4900–4906 CrossRef CAS PubMed.
- C. E. Gómez Sánchez, A. Martínez-Trujillo and G. Aguilar Osorio, Lett. Appl. Microbiol., 2012, 55, 444–452 CrossRef PubMed.
- H. J. Henzler and M. Schedel, Bioprocess Eng., 1991, 7, 123–131 CrossRef CAS.
- W. Klöckner and J. Büchs, Trends Biotechnol., 2012, 30, 307–314 CrossRef PubMed.
- R. Kuhn, H. Bauer and H.-J. Knackmuss, Chem. Ber., 1965, 98, 2139–2153 CrossRef CAS PubMed.
- M. Müller, S. Ausländer, D. Ausländer, C. Kemmer and M. Fussenegger, Metab. Eng., 2012, 14, 325–335 CrossRef PubMed.
- W. Heumann, D. Young and C. Gottlich, Biochim. Biophys. Acta, Gen. Subj., 1968, 156, 429–431 CrossRef CAS.
- C. T. Evans and C. Ratledge, Microbiology, 1984, 130, 1693–1704 CAS.
- C. T. Evans and C. Ratledge, Microbiology, 1984, 130, 1705–1710 CAS.
- E. G. ter Schure, N. A. W. van Riel and C. T. Verrips, FEMS Microbiol. Rev., 2000, 24, 67–83 CrossRef CAS PubMed.
- Z. Zhu, S. Zhang, H. Liu, H. Shen, X. Lin, F. Yang, Y. J. Zhou, G. Jin, M. Ye, H. Zou and Z. K. Zhao, Nat. Commun., 2012, 3, 1112 CrossRef PubMed.
- M. T. Castañeda, S. Nuñez, F. Garelli, C. Voget and H. De Battista, J. Biotechnol., 2018, 280, 11–18 CrossRef PubMed.
- S. Klotz, A. Kuenz and U. Prüße, Green Chem., 2017, 19, 4633–4641 RSC.
- J. Zhang, J. Reddy, B. Buckland and R. Greasham, Biotechnol. Bioeng., 2003, 82, 640–652 CrossRef CAS PubMed.
- H. Liu, X. Zhao, F. Wang, Y. Li, X. Jiang, M. Ye, Z. K. Zhao and H. Zou, Yeast, 2009, 26, 553–566 CrossRef CAS PubMed.
- M. Tai and G. Stephanopoulos, Metab. Eng., 2013, 15, 1–9 CrossRef CAS PubMed.
- A. Beopoulos, J. Cescut, R. Haddouche, J.-L. Uribelarrea, C. Molina-Jouve and J.-M. Nicaud, Prog. Lipid Res., 2009, 48, 375–387 CrossRef CAS PubMed.
- S. Papanikolaou and G. Aggelis, Lipid Technol., 2009, 21, 83–87 CrossRef CAS.
- M. Brabender, M. S. Hussain, G. Rodriguez and M. A. Blenner, Appl. Microbiol. Biotechnol., 2018, 102, 2313–2322 CrossRef CAS PubMed.
- H. B. Bull, K. Breese, G. L. Ferguson and C. A. Swenson, Arch. Biochem. Biophys., 1964, 104, 297–304 CrossRef CAS PubMed.
- H. Shen, X. Zhang, Z. Gong, Y. Wang, X. Yu, X. Yang and Z. K. Zhao, Appl. Microbiol. Biotechnol., 2017, 101, 3801–3809 CrossRef CAS PubMed.
- I. Kolouchová, O. Maťátková, K. Sigler, J. Masák and T. Řezanka, Folia Microbiol., 2016, 61, 431–438 CrossRef PubMed.
- M. Kavšček, G. Bhutada, T. Madl and K. Natter, BMC Syst. Biol., 2015, 9, 72 CrossRef PubMed.
- M. Athenaki, C. Gardeli, P. Diamantopoulou, S. S. Tchakouteu, D. Sarris, A. Philippoussis and S. Papanikolaou, J. Appl. Microbiol., 2018, 124, 336–367 CrossRef CAS PubMed.
- C. Ratledge and J. P. Wynn, Adv. Appl. Microbiol., 2002, 51, 1–51 CAS.
- R. Dulermo, H. Gamboa-Meléndez, R. Ledesma-Amaro, F. Thévenieau and J.-M. Nicaud, Biochim. Biophys. Acta, 2015, 1851, 1202–1217 CrossRef CAS PubMed.
- G. Singh, S. Sinha, K. K. Bandyopadhyay, M. Lawrence and D. Paul, Microb. Cell Fact., 2018, 17, 182 CrossRef PubMed.
- S. T. Coradetti, D. Pinel, G. M. Geiselman, M. Ito, S. J. Mondo, M. C. Reilly, Y.-F. Cheng, S. Bauer, I. V. Grigoriev, J. M. Gladden, B. A. Simmons, R. B. Brem, A. P. Arkin and J. M. Skerker, eLife, 2018, 7, e32110 CrossRef PubMed.
- S. Fillet, J. Gibert, B. Suárez, A. Lara, C. Ronchel and J. L. Adrio, J. Ind. Microbiol. Biotechnol., 2015, 42, 1463–1472 CrossRef CAS PubMed.
- R. R. Bommareddy, W. Sabra, G. Maheshwari and A.-P. Zeng, Microb. Cell Fact., 2015, 14, 36 CrossRef PubMed.
- J. Xu, X. Zhao, W. Wang, W. Du and D. Liu, Biochem. Eng. J., 2012, 65, 30–36 CrossRef CAS.
- L. J. Jönsson and C. Martín, Bioresour. Technol., 2016, 199, 103–112 CrossRef PubMed.
- J. Shi, J. M. Gladden, N. Sathitsuksanoh, P. Kambam, L. Sandoval, D. Mitra, S. Zhang, A. George, S. W. Singer, B. A. Simmons and S. Singh, Green Chem., 2013, 15, 2579 RSC.
- R. P. Elander, Appl. Microbiol. Biotechnol., 2003, 61, 385–392 CrossRef CAS PubMed.
- F. Xu, D. Gage and J. Zhan, J. Ind. Microbiol. Biotechnol., 2015, 42, 1149–1155 CrossRef CAS PubMed.
- H. Zhang, L. Fang, M. S. Osburne and B. A. Pfeifer, Methods Mol. Biol., 2016, 1401, 121–134 CrossRef CAS PubMed.
- J. Li and P. Neubauer, New Biotechnol., 2014, 31, 579–585 CrossRef CAS PubMed.
- L. Huo, J. J. Hug, C. Fu, X. Bian, Y. Zhang and R. Müller, Nat. Prod. Rep., 2019 10.1039/c8np00091c.
- O. Frick and C. Wittmann, Microb. Cell Fact., 2005, 4, 30 CrossRef PubMed.
- H. G. Crabtree, Biochem. J., 1929, 23, 536–545 CrossRef CAS PubMed.
- M. Klimacek, S. Krahulec, U. Sauer and B. Nidetzky, Appl. Environ. Microbiol., 2010, 76, 7566–7574 CrossRef CAS PubMed.
- E. D. Głowacki, G. Voss and N. S. Sariciftci, Adv. Mater., 2013, 25, 6783–6800 CrossRef PubMed.
- M. Irimia-Vladu, E. D. Głowacki, P. A. Troshin, G. Schwabegger, L. Leonat, D. K. Susarova, O. Krystal, M. Ullah, Y. Kanbur, M. A. Bodea, V. F. Razumov, H. Sitter, S. Bauer and N. S. Sariciftci, Adv. Mater., 2012, 24, 375–380 CrossRef CAS PubMed.
- M. Sytnyk, E. D. Głowacki, S. Yakunin, G. Voss, W. Schöfberger, D. Kriegner, J. Stangl, R. Trotta, C. Gollner, S. Tollabimazraehno, G. Romanazzi, Z. Bozkurt, M. Havlicek, N. S. Sariciftci and W. Heiss, J. Am. Chem. Soc., 2014, 136, 16522–16532 CrossRef CAS PubMed.
- T. A. Keating and C. T. Walsh, Curr. Opin. Chem. Biol., 1999, 3, 598–606 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9gc00920e |
| This journal is © The Royal Society of Chemistry 2019 |