Open Access Article
Open Access ArticleResveratrol regulates skeletal muscle fibers switching through the AdipoR1-AMPK-PGC-1α pathway
Qinyang
Jiang†
a,
Xiaofang
Cheng†
a,
Yueyue
Cui
a,
Qin
Xia
a,
Xueyu
Yan
 a,
Mingyuan
Zhang
a,
Ganqiu
Lan
a,
Jiaqi
Liu
b,
Tizhong
Shan
*b and
Yanna
Huang
a,
Mingyuan
Zhang
a,
Ganqiu
Lan
a,
Jiaqi
Liu
b,
Tizhong
Shan
*b and
Yanna
Huang
 *a
*a
aCollege of Animal Science and Technology, Guangxi University, Nanning, Guangxi 530004, China. E-mail: huangyn@gxu.edu.cn; Tel: +86-771-3235635
bCollege of Animal Science, Zhejiang University, Hangzhou, Zhejiang 310058, China. E-mail: tzshan@zju.edu.cn
First published on 16th May 2019
Abstract
This study was conducted to investigate the effect and underlying mechanism of Resveratrol (RES) in regulating skeletal muscle fiber-type switching. We found that RES had no effect on the body weight and food intake of Kunming mice (KM mice) that were orally administered with 400 mg kg−1 d−1 RES for 12 weeks. Notably, the RES administration significantly increased the expression of myosin heavy chain (MyHC) 1, MyHC2a, and MyHC2x in the extensor digitorum longus (EDL) and soleus (SOL) muscles. Furthermore, the muscle immunostaining of the results showed that the RES treatment led to the myofiber type transition from glycolytic to oxidative in muscles. The mRNA and protein levels of the adiponectin receptor (AdipoR), AMP-activated protein kinase (AMPK), peroxisome proliferator-activated receptor-γ coactivator-1α (PGC-1α) in EDL and SOL were drastically increased after RES treatment. Moreover, the plasma Adiponectin (AdipoQ) protein levels were higher in the RES-treated mice compared to the control mice. Moreover, the in vitro results further demonstrated that the 20 μM RES treatment increased the expression of AdipoR1, AdipoR2, AMPK, PGC-1α and MyHC1, but decreased the expression of MyHC2b in C2C12 myoblasts. Furthermore, mechanistic studies revealed that silencing the AdiopR1, not the AdiopR2, abolished the effect of RES on the expression of AMPK and PGC-1α in the C2C12 cells. These results indicated that RES could regulate skeletal fiber switching through the AdiopR1-AMPK-PGC-1α pathway. This work may provide a new strategy for enhancing endurance and relieving muscle diseases caused by oxidative muscle fiber deficiency.
1 Introduction
Muscle fibers are the basic muscle units. Based on the myosin heavy chain (MyHC) polymorphism, skeletal muscle fiber types in mammals are divided into four types: MyHC1 (Type I), MyHC2a (Type IIA), MyHC2x (Type IIX), MyHC2b (Type IIB).1 Types I and IIA are considered to be oxidative fibers, containing relatively large mitochondria and myoglobin contents.2 Type I muscle fiber is also known as slow fiber. Type IIB is considered to consist of glycolytic fibers, containing greater amounts of glycogen and glucose. Skeletal muscle fiber plays an important role in determining the performance of athletes.3 Elite endurance athletes have a high percentage of oxidative muscle fibers in their skeletal muscles.4 In addition, having more oxidative muscle fibers provides more resistance to the dystrophic pathology in Duchenne muscular dystrophy patients.5 Various factors such as exercise, nutrition, hormone levels, and external environmental stimuli can affect the mutual transformation of muscle fiber types.2,6,7 Therefore, increasing the proportion of oxidative type I muscle fiber is the main way to enhance endurance and relieve muscle diseases caused by oxidative muscle fiber deficiency.Resveratrol (RES), a natural polyphenol, is extracted from polygonum cuspidatum, peanut, grape seed and other plants. Numerous studies have revealed its extensive biological activity including anti-inflammatory8 and anti-oxidative effect,9 extending the lifespan of diverse species,10,11 preventing early atherosclerotic events12 and treating obesity and diabetes.13,14 It has been reported that RES plays an important role in the regulation of muscle fiber type transformation toward more oxidative muscle fibers.13,15,16 However, the underlying mechanism is not well understood.
Adiponectin (AdipoQ) is known as one of the important adipocytokines. Adiponectin is closely related to the development of insulin resistance and type 2 diabetes,17 and negatively correlated with the development of atherosclerosis.18 Krause et al. (2008) found that Adiponectin was expressed in the skeletal muscle fibers of mouse and was able to regulate the muscle fiber types.19 Iwabu et al. (2010) found that Adiponectin and adiponectin receptor 1 (AdipoR1) regulate PGC-1α and mitochondria by Ca2+ and AMPK/SIRT1, and then increase the oxidative type I myofibers in the skeletal muscle.20 However, whether RES could regulate the skeletal muscle fiber types switching through the adiponectin pathway remains unclear. Therefore, in this study, we investigated the effects of RES on the expression of AdipoR and muscle fiber type-related genes. In addition, whether RES regulates muscle fiber type switching through AdipoR1 or AdipoR2 was also determined.
2 Method
2.1 Animals and sample collection
Male KM mice (n = 20, License No. SCXK Gui 2014-0002) of 21 days of age were used as experimental animals. The mice were housed in the animal facility with 25 °C, 50% relative humidity, 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 12 light–dark cycle. The mice had free access to a commercial diet and water during the experimental period. The basal diet used in this study was purchased from the Jiangsu Xietong Organism Company in China. The nutrients for the formula feeds of mice were formulated according to the National Standards of China, GB14924.3-2010. All of the mice were randomly divided into two groups: the dimethyl sulfoxide (DMSO) solvent group (control group, n = 10) and the 400 mg kg−1 day−1 RES (Solarbio, Beijing, China) group (n = 10) dissolved in DMSO by oral gavage, as previously described.21 The experimental period lasted for 12 weeks. This study was performed in strict accordance with the NIH guidelines for the care and use of laboratory animals (NIH Publication No. 85-23 Rev. 1985) and was approved by the Institutional Animal Care and Use Committee of Guangxi University (Nanning, China).
12 light–dark cycle. The mice had free access to a commercial diet and water during the experimental period. The basal diet used in this study was purchased from the Jiangsu Xietong Organism Company in China. The nutrients for the formula feeds of mice were formulated according to the National Standards of China, GB14924.3-2010. All of the mice were randomly divided into two groups: the dimethyl sulfoxide (DMSO) solvent group (control group, n = 10) and the 400 mg kg−1 day−1 RES (Solarbio, Beijing, China) group (n = 10) dissolved in DMSO by oral gavage, as previously described.21 The experimental period lasted for 12 weeks. This study was performed in strict accordance with the NIH guidelines for the care and use of laboratory animals (NIH Publication No. 85-23 Rev. 1985) and was approved by the Institutional Animal Care and Use Committee of Guangxi University (Nanning, China).
At the end of the experiment, blood samples were collected from the mice after 12 h of fasting. All of the mice were sacrificed by neck dislocation. The heart, liver, spleen, lung, kidney, tibialis anterior muscle (TA), gastrocnemius muscle (GAS), extensor digitorum longus (EDL), soleus muscle (SOL), brown adipose tissue (BAT) and white adipose tissue (WAT) were collected for analysis.
2.2 Plasma adiponectin level measurement
The plasma adiponectin levels were measured by a commercial ELISA kit (Westang Biotechnology Systems, Shanghai, China).2.3 Cryosectioning, immunostaining and image capture
Fresh muscle samples, including the EDL and SOL muscles from mice, were embedded in an optimal cutting temperature (O.C.T) compound (Sakura Finetek), followed by snap freezing in a dry ice cooled bath containing isopentane. A Leica CM 1850 cryostat was used to cut the muscles into 10 μm thick sections. The sections were placed on Superfrost Plus glass slides (Electron Microscopy Sciences). A blocking buffer (2% bovine serum albumin, 5% horse serum, 0.2% Triton X-100 and 0.1% sodium azide in PBS) was used to block the tissue sections for 30 min. The incubated tissues with primary antibodies NOQ (Abcam) and MF20 (Developmental Studies Hybridoma Bank, Iowa, USA) were diluted using the blocking buffer for 1 h at room temperature, then incubated with secondary antibodies and Hoechst stain (diluted in PBS) for 30 min at room temperature, and finally mounted with Dako fluorescence mounting media (Glostrup, Denmark). The fluorescence images were captured by a Leica-TCS-SP8MP fluorescence microscope (Mannheim, Germany). The immunostaining and image capture were done as previously described.222.4 Cell culture and treatment
The C2C12 myoblasts were cultured in Dulbecco's modified Eagle's medium (DMEM) containing 10% fetal bovine serum (FBS) at 37 °C and 5% CO2. The C2C12 cells were induced to undergo myogenic differentiation with a differentiation medium containing 2% horse serum for 6 days. For the RES treatment, C2C12 cells were treated with 20 μM resveratrol or a solvent (DMSO).2.5 Knockdown of AdipoRs with AdipoRs-shRNA plasmids
Both AdipoR1 and AdipoR2 shRNA plasmids were bought from GeneCopoeia (GeneCopoeia, China). The AdipoR1-shRNA and AdipoR2-shRNA plasmids were each transfected into C2C12 cells at a confluence of 70% with Lipofectamine™ LTX & Plus Reagent (Thermofisher Scientific, Shanghai, China), according to the manufacturer's instruction. C2C12 cells transfected with a corresponding empty vector of the AdipoRs-shRNA plasmid served as the control groups. The C2C12 cells were transfected with different shRNA plasmids and were incubated in a culture medium with or without 20 μM resveratrol for 48 h. The experimental group of RNAi is listed in Table 1. At the end of the experiment, cells of different groups were collected for further analysis.| Condition | Group | ||||
|---|---|---|---|---|---|
| Control | AdipoR1-shRNA group | AdipoR2-shRNA group | |||
| “+” = treated with corresponding condition; “−” = treated without corresponding condition. | |||||
| RES | − | − | + | − | + |
| AdipoR1-shRNA plasmid | − | + | + | − | − |
| AdipoR2-shRNA plasmid | − | − | − | + | + |
2.6 Total RNA isolation and real-time PCR
The total RNA from tissues or cells was extracted and purified using RNAiso (TAKARA, Dalian, China), according to the manufacturer's instructions. The total RNA was dissolved in RNase-free H2O and it's concentration and purity were measured with a spectrophotometer (Q5000, USA). The ratios of the absorbance of all samples at 260 and 280 nm were in the range of 1.8–2.0. The reverse transcription of 1 μg of the total RNA from different tissues or from cells in the culture to cDNA was prepared with the PrimeScript™ II 1st Strand cDNA Synthesis Kit (TAKARA, Dalian, China). The real-time quantitative polymerase chain reaction amplification was performed using the SYBR® Premix Ex Taq II (Tli RNaseH Plus) with a CFX-96 Real Time PCR detection system (Bio-Rad, USA) according to the primers shown in Table 2, as previously described.23 The relative amount of gene expression was analyzed using the 2−ΔΔCT method and 18S ribosomal RNA as a control for normalization.| Gene | Primer name | Sequences (5′–3′) | Fragments size (bp) |
|---|---|---|---|
| AdipoQ | Forward | TGTTCCTCTTAATCCTGCCCA | 144 |
| Reverse | CCAACCTGCACAAGTTCCCTT | ||
| AdipoR1 | Forward | ACGTTGGAGAGTCATCCCGTAT | 133 |
| Reverse | CTCTGTGTGGATGCGGAAGAT | ||
| AdipoR2 | Forward | GGAGTGTTCGTGGGCTTAGG | 140 |
| Reverse | GCAGCTCCGGTGATATAGAGG | ||
| AMPK | Forward | CACCCTGAAAGAGTACCGT | 110 |
| Reverse | CATTTTGCCTTCCGTACACCT | ||
| PGC-1α | Forward | TCATCACCTACCGTTACACCT | 168 |
| Reverse | AAGCAGGGTCAAAATCGTCT | ||
| MyHC1 | Forward | CTCAGCCATGCCAACCGTA | 128 |
| Reverse | ATGTTCTCTTTCAGGTCGTCAT | ||
| MyHC2a | Forward | GGCTTCAGGATTTGGTGGATAAAC | 110 |
| Reverse | GATCTTGCGGAACTTGGATAGATT | ||
| MyHC2x | Forward | CGCTAGTAACATGGAGTGCATCT | 211 |
| Reverse | CCTCGGCTTGAACAGAAGTACC | ||
| MyHC2b | Forward | GGTGAGGGAGCTTGAAAACGAG | 191 |
| Reverse | CCTGCTCTTGTAGGCTTTCACT | ||
| 18srRNA | Forward | CCCACGGAATCGAGAAAGAG | 122 |
| Reverse | TTGACGGAAGGGCACCA |
2.7 Western blot analysis
We used RIPA buffer to isolate the total protein from mice tissues or C2C12 cells. The protein concentration was determined using a Bio-Rad protein assay kit from Bio-Rad Laboratories (Bio-Rad Laboratories, Benicia, California, USA). Sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE) was run for separating the protein. The proteins were transferred to a polyvinylidene difluoride membrane, blocked with 5% non-fat dry milk in 0.05% Tris-buffered saline-Tween 20 for 1 hour at room temperature, and then incubated with primary antibodies in 5% milk overnight at 4 °C. The AdipoQ, AdipoR1, AdipoR2, AMPK, PGC-1α, GAPDH antibodies were obtained from Santa Cruz Biotechnology (Santa Cruz Biotechnology, USA). The membranes were incubated with a horseradish peroxidase-conjugated secondary antibody (Santa Cruz Biotechnology, USA) for 1 h at room temperature. An enhanced chemiluminescence western blot substrate (Pierce Biotechnology) was used to perform the immunodetection, which were detected with the ChemiDoc™ MP Imaging System (Bio-Rad, USA). The bands were analyzed by density analysis with the Image J software. The results for the western blot shown in figures are representative results from at least three independent experiments.2.8 Data analysis
All experimental data are presented as a means ± standard error of the mean (SEM). The Student's t-test was adopted to assess the significance of differences. A P-value of less than 0.05 was considered statistically significant.3 Results
3.1 Effects of RES treatment on mice growth
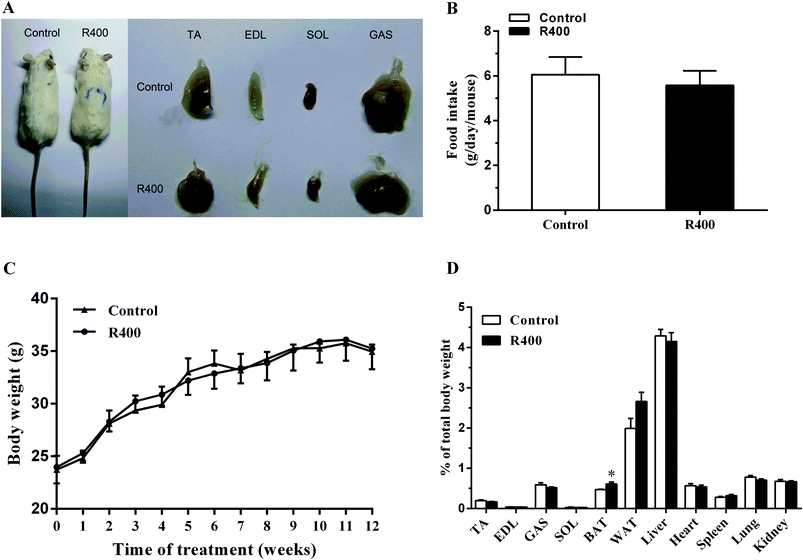
First, we directly examined the effects of the RES treatment on animal growth and tissue/organ weight. We found that the RES treated mice were indistinguishable from their littermates (Fig. 1A). The RES treatment did not affect food intake (Fig. 1B). The RES-treated mice had a similar body weight and growth rate to those of the control mice (Fig. 1C). Moreover, the weight of different tissues, except for brown fat, from the KM mice orally administered with 400 mg kg−1 d−1 RES was unchanged (Fig. 1D). Taken together, the RES treatments did not affect the growth of the mice.3.2 RES treatment affects the fiber type related gene expression in muscle
Next, we examined the effects of RES on muscle fiber types. We found that the RES treatment significantly increased the mRNA levels of MyHC1 (36.43%, P < 0.001), MyHC2a (49.63%, P < 0.001) and MyHC2x (109.34%, P < 0.001) in EDL, respectively (Fig. 2A). Likewise, the mRNA levels of MyHC1, MyHC2a, and MyHC2x in SOL were also increased by 145.84% (P < 0.001), 270.48% (P < 0.001) and 181.54% (P < 0.001), respectively (Fig. 2B). These results demonstrated that the RES treatment affects the expression of muscle fiber type related genes.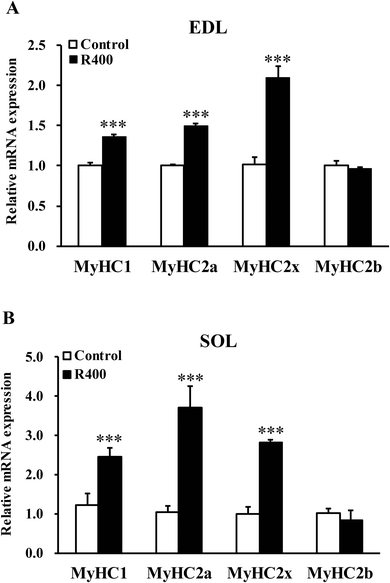 | ||
| Fig. 2 RES treatment affects the expression of MyHC isoform genes in EDL (A) and SOL (B) muscles after RES treatment for 12 weeks. Each value represents the mean ± SEM. ***P < 0.001, n = 10. | ||
3.3 RES treatment affects the myofiber composition in muscle
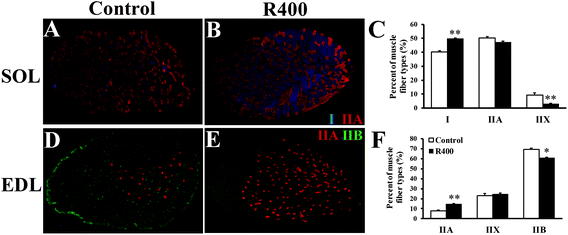
To further explore the effects of RES on skeletal muscle fiber type switching, we examined the myofiber composition in EDL and SOL muscles. Fig. 3A, B, D, and E show that the representative staining of the SOL and EDL muscle sections were labeled with monoclonal antibodies specific for Type I (MyHC1), IIA (MyHC2a), and IIB (MyHC2b) myosin heavy chains. In the slow SOL muscle, the RES treatment increased the percentage of Type I myofibers from 40% to 50% (P < 0.01) (Fig. 3C). Furthermore, the Type IIX myofiber was reduced from 9% to 3% (P < 0.01). In the fast EDL muscle, a significant reduction from 69% to 60% (P < 0.05) in the Type IIB myofiber was observed after the RES treatment. In contrast, an increase of 7% was observed in the Type IIA myofiber (P < 0.01). Taken together, these results indicate that the RES treatment leads to the myofiber type transition from glycolytic to oxidative in the muscles.3.4 RES treatment affects the expression of adiponectin pathway related genes
To determine whether RES could affect the expression of adiponectin pathway related genes, we examined the mRNA and protein levels of AdipoQ, AdipoR1, AdipoR2 in EDL and SOL muscles. We found that the RES treatment drastically increased the expression of AdipoQ, AdipoR1, and AdipoR2 in EDL (Fig. 4A and 5A, B) and SOL (Fig. 4B and 5C, D) muscles. Notably, the RES treatment also significantly increased the expression of AMPK and PGC-1α in the SOL and EDL muscles (Fig. 4A, B and 5A–D). In addition, a significant increase in the AdipoQ concentration of the RES treatment group was detected in comparison with that of the control group (Fig. 6). These findings suggest that the RES treatment may regulate the fiber type via upregulating the expression of AdipoR1, AdipoR2 and their downstream targets, including AMPK and PGC-1α, in the skeletal muscle.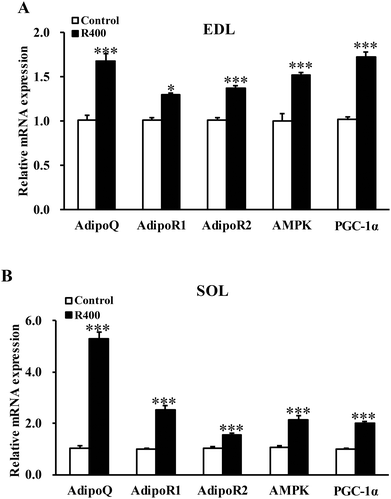 | ||
| Fig. 4 RES treatment affects the mRNA level of adiponectin signal genes in EDL (A) and SOL muscles (B). Each value represents the mean ± SEM. *P < 0.05, ***P < 0.001, n = 10. | ||
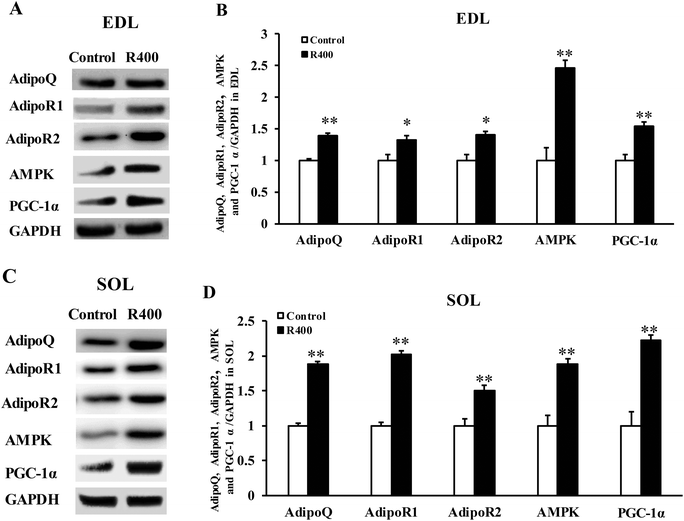 | ||
| Fig. 5 RES treatment affects the protein level of adiponectin signal genes in EDL (A, B) and SOL muscles (C, D). Each value represents the mean ± SEM. *P < 0.05, **P < 0.01, n = 10. | ||
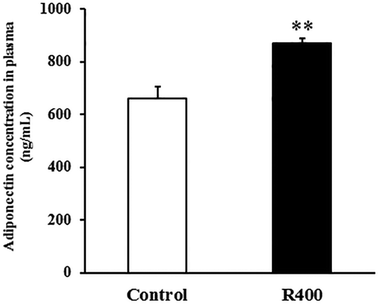 | ||
| Fig. 6 RES treatment affects the plasma AdipoQ concentration. Each value represents the mean ± SEM. **P < 0.01, n = 10. | ||
3.5 RES upregulates MyHC1 expression through the AdipoR1-AMPK-PGC-1α pathway
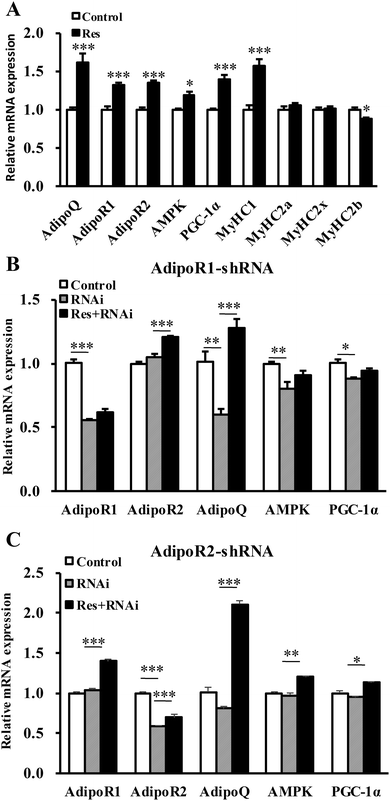
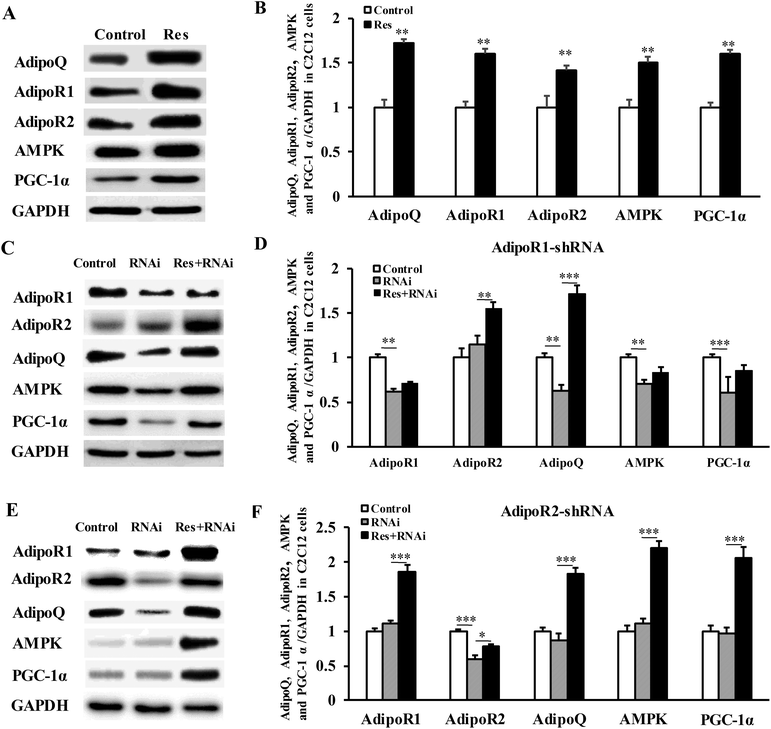
To further confirm that RES could regulate MyHC expression, we treated C2C12 cells with RES. We found that the RES treatment increased the mRNA levels of MyHC1 by 56.61% (P < 0.001), while it decreased the MyHC2b mRNA levels by 11.30% (P < 0.05), compared to the control group (Fig. 7A), indicating a glycolytic to oxidative fiber switching. Notably, the mRNA expression of AdipoQ, AdipoR1, AdipoR2, AMPK and PGC-1α was also increased by 62.18% (p < 0.001), 32.11% (p < 0.001), 35.06% (p < 0.001), 19.37% (p < 0.05) and 40.21% (p < 0.001), respectively (Fig. 7A). The protein expression of AdipoQ, AdipoR1, AdipoR2, AMPK and PGC-1α was also increased by 72.05% (p < 0.01), 60.02% (p < 0.01), 41.05% (p < 0.01), 50.00% (p < 0.01) and 60.04% (p < 0.01), respectively (Fig. 8A). These results suggest that the RES treatment causes muscle fiber switching from glycolytic to oxidative, and that the adiponectin signaling pathway may be involved in this process.To determine whether the RES treatment affected MyHC expression through the adiponectin pathway, we used RNA interference to knockdown the expression of AdipoR1 and AdipoR2 in C2C12 cells.
Compared with the control group, the gene mRNA expression level of AdipoR1 was clearly decreased by 56% by transfecting the AdipoR1 shRNA (Fig. 7B). Compared with the control group, the gene mRNA expression level of AdipoR2 was notably decreased by 59% by transfecting the AdipoR2 shRNA (Fig. 7C). The knockdown of AdipoR1 significantly decreased the mRNA and protein levels of AMPK and PGC-1α (Fig. 7B and 8C, D). Furthermore, the knockdown of AdipoR1 abolished the effects of RES on AMPK and PGC-1α expression (Fig. 7B and 8C, D). However, the knockdown of AdipoR2 did not affect the mRNA and protein levels of the expression of AMPK and PGC-1α (Fig. 7C and 8E, F). In addition, the RES treatment could still upregulate the mRNA and protein levels of AMPK and PGC-1α expression in AdipoR2-silencing C2C12 cells (Fig. 7C and 8E, F). Taken together, these results demonstrate that RES affects MyHC1 expression through activating the AdipoR1-AMPK-PGC-1α pathway.
4 Discussion
In this study, we showed that the RES treatment could upregulate MyHC1 expression in vivo and in vitro. Furthermore, we found that the RES treatment increased the plasma adiponectin content and the expression of AdipoR1, AMPK and PGC-1α. Mechanistically, we demonstrated that RES upregulates MyHC1 expression through the AdipoR1-AMPK-PGC-1α pathway.RES, a natural polyphenol, can be extracted from different plant species such as peanuts and grapes. In this study, we observed that the treatment with RES at the dose of 400 mg kg−1 d−1 for 12 weeks had no significant effect on food intake or body weight, which is consistent with previous studies.24,25 In mammals, there are two adipose tissue types: white adipose tissue (WAT) and brown adipose tissue (BAT). WAT is considered to be the primary site of triglyceride storage, while BAT is specialized in energy expenditure and thermogenesis, dissipating energy as heat.26–28 Andrade et al. (2014) found that Resveratrol promotes BAT thermogenic gene expression and energy metabolism in mice fed a standard diet.29 Wang et al. (2016) found that RES enhances brown adipocyte formation and function in mice that were fed a high-fat diet through activating AMPKα1.30 In agreement with the previous findings, we found that RES increased the BAT expression. In our study, RES effectively increased the AdipoQ protein content in the serum of mice, which led to a significant effect on the target tissue, including the adipose tissue and muscle through blood circulation. AdipoQ can also target its receptors, including AdipoR1 and AdipoR2, in the peripheral tissues through blood circulation. Our previous results reveal that the AdipoQ gene transfer by hydrodynamic based-gene delivery can enhance lipid catabolism in epididymal white fat, gastrocnemius, and extensor digitorum longus of mice through activating the AMPK, SIRT1, and PGC-1α.31 Therefore, the effects of RES on brown adipocyte formation may be through the AdipoQ signaling pathway. Thus, we will focus on the effect of RES on BAT in future studies.
Many studies in recent years have shown that RES plays a crucial role in improving mitochondrial function15,32 and regulating muscle fiber type transformation.15,16 In a previous study, dietary RES supplementation was shown to play a positive role in increasing the transcription level of MyHC2a and reducing that of MyHC2b, along with the decrease of myofiber cross-sectional area.17 In this study, we also observed that RES could significantly upregulate MyHC1, MyHC2a, and MyHC2x gene expression levels in skeletal muscle (EDL and SOL) in KM mice. Furthermore, the muscle immunostaining of the results showed that RES treatment led to the myofiber type transition from glycolytic to oxidative in muscles, which was the direct evidence of the effects of RES on skeletal muscle fiber type switching.
The AMPK/PGC-1α signaling pathway has been found to play a key role in the energy metabolism of muscle tissues and be closely associated with the muscle fiber type transformation. Exercise training activates AMPK ascribed to the increase of the AMP/ATP ratio and Ca2+ flux during muscle contraction, enhancing the activity of PGC-1α, which is a vital regulatory factor of fiber transformation from type 2 to type 1 in the skeletal muscle.33–35 The skeletal muscle-specific PGC-1α knockout mice exhibited a muscle fiber switch from oxidative type 1 and 2a to type 2x and 2b muscle fibers.36 Adiponectin, one of the adipocytokines, could regulate AMPK/PGC-1α transcription in energy metabolism by Hydrodynamics-based gene delivery.31 Also of note, the incubation with gAcrp30 (2.5 μg ml−1) for 30 min on EDL led to an increase in the AMPK expression level and glucose uptake.37 The serum adiponectin levels positively correlated with type 1 muscle fiber and negatively correlated with type 2b muscle fiber.38 In a previous study, the adiponectin gene has been reported to increase the oxidative type I myofiber by regulating the AMPK and PGC-1α expression.20 In our study, we found that RES effectively increased the AdipoQ protein content in the serum of mice, as well as the gene expression of AdipoQ in EDL and SOL. In addition, the muscle fiber type results showed that RES could regulate the transcriptional activity of type 1 fiber by raising the transcription level of AMPK and PGC-1α in vivo and in vitro. Therefore, these results indicate that RES may regulate the skeletal muscle fiber switching through the adiponectin signaling pathway.
In the adiponectin signaling pathway, there are two different receptors for adiponectin: AdipoR1 and AdipoR2, which are abundant in skeletal muscle and liver, respectively. They are distinct from the G-protein-coupled receptors in structure and function, although both receptors contain seven-transmembrane domains.39,40 Moreover, it has been shown that AdipoR1 activates the AMPK pathway; in contrast, AdipoR2 activates the PPARα pathway in the liver.41 The muscle-specific disruption of AdipoR1 can decrease the activation of AMPK and PGC-1α by adiponectin, oxidative type I muscle fiber, and exercise endurance. However, the suppression of AdipoR2 expression by specific short interfering RNA (siRNA) failed to reduce the mitochondrial biogenesis that has been induced by adiponectin.20 To ascertain whether RES regulates muscle fiber types by AdipoR1 or AdipoR2 in the adiponectin signal pathway, an experiment was performed to determine the effects of resveratrol on the expression of adiponectin signal pathway genes and AMPK/PGC-1α pathway genes in C2C12 cells by transfecting them with specific short hairpin RNA (shRNA) of AdipoRs. Here, our research showed that the expression of AMPK and PGC-1α significantly decreased after silencing AdiopR1, but no change was observed after silencing AdiopR2. The knockdown of AdipoR1 expression could abolish the effect of RES on AMPK and PGC-1α expression. However, RES significantly increased the AMPK and PGC-1α expression in the AdipoR2-silencing C2C12 cells. The results indicate that AdipoR1 is involved in the regulatory process of RES on muscle fiber type switching.
5 Conclusions
In conclusion, the RES treatment results in muscle fiber switching from glycolytic to oxidative through the AdipoR1-AMPK-PGC-1α pathway. These results enhance our understanding of the regulation of RES on muscle fiber type switching and further suggest that RES may be used as a muscle fiber regulator for controlling the proportion of type I muscle fiber. This work may provide a new strategy for enhancing endurance and relieving muscle diseases caused by oxidative muscle fiber deficiency.Conflicts of interest
The authors have no competing interests to declare.Acknowledgements
This work was supported by the grants from National Natural Science Foundation of China (31460606 and 31760672) and by Guangxi Natural Science Foundation (2014GXNSFAA118131).References
- D. Pette and R. S. Staron, Myosin isoforms, muscle fiber types, and transitions, Microsc. Res. Tech., 2000, 50, 500–509 CrossRef CAS PubMed.
- D. Pette and R. S. Staron, Transitions of muscle fiber phenotypic profiles, Histochem. Cell Biol., 2001, 115, 359–372 CAS.
- U. Bergh, A. Thorstensson, B. Sjodin, B. Hulten, K. Piehl and J. Karlsson, Maximal oxygen uptake and muscle fiber types in trained and untrained humans, Med. Sci. Sports, 1978, 10, 151–154 CAS.
- P. G. Schantz and G. K. Dhoot, Coexistence of slow and fast isoforms of contractile and regulatory proteins in human skeletal muscle fibres induced by endurance training, Acta Physiol. Scand., 1987, 131, 147–154 CrossRef CAS PubMed.
- V. Ljubicic, M. Burt, J. A. Lunde and B. J. Jasmin, Resveratrol induces expression of the slow, oxidative phenotype in mdxmouse muscle together with enhanced activity of the SIRT1-PGC-1α axis, Am. J. Physiol.: Cell Physiol., 2014, 307, C66–C82 CrossRef CAS PubMed.
- J. R. Zierath and J. A. Hawley, Skeletal Muscle Fiber Type: Influence on Contractile and Metabolic Properties, PLoS Biol., 2004, 2, e348 CrossRef PubMed.
- K. Leisson, U. Jaakma and T. Seene, Adaptation of equine locomotor muscle fiber types to endurance and intensive high speed training, J. Equine Vet. Sci., 2008, 28, 395–401 CrossRef.
- L. E. Donnelly, R. Newton, G. E. Kennedy, P. S. Fenwick, R. H. Leung, K. Ito, R. E. Russell and P. J. Barnes, Anti-inflammatory effects of resveratrol in lung epithelial cells: molecular mechanisms, Am. J. Physiol.: Lung Cell. Mol. Physiol., 2004, 287, 774–783 CrossRef PubMed.
- W. Zhu, S. Chen, Z. Li, X. Zhao, W. Li, Y. Sun, Z. Zhang, W. Ling and X. Feng, Effects and mechanisms of resveratrol on the amelioration of oxidative stress and hepatic steatosis in KKAy mice, Nutr. Metab., 2014, 11, 35 CrossRef PubMed.
- S. Jarolim, J. Millen, G. Heeren, P. Laun, D. S. Goldfarb and M. Breitenbach, A novel assay for replicative lifespan in saccharomyces cerevisiae, FEMS Yeast Res., 2004, 5, 169–177 CrossRef CAS PubMed.
- M. Viswanathan, S. K. Kim and A. Berdichevsky, A role for SIR-2.1 regulation of ER stress response genes in determining C. elegans life span, Dev. Cell, 2005, 9, 605–615 CrossRef CAS PubMed.
- M. Azorín-Ortuño, M. J. Yañéz-Gascón, F. J. Pallarés, J. Rivera, A. González-Sarrías, M. Larrosa, F. Vallejo, M. T. García-Conesa, F. Tomás-Barberán and J. C. Espín, A dietary resveratrol-rich grape extract prevents the developing of atherosclerotic lesions in the aorta of pigs fed an atherogenic diet, J. Agric. Food Chem., 2012, 60, 5609–5620 CrossRef PubMed.
- M. Lagouge, M. C. Argmann, Z. Gerharthines, H. Meziane, C. Lerin, F. Daussin, N. Messadeq, J. Milne, P. Lambert, P. Elliott, B. Geny, M. Laakso, P. Puiqserver and J. Auwerx, Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1 alpha, Cell, 2006, 127, 1109–1122 CrossRef CAS PubMed.
- G. Ramadori, L. Gautron, T. Fujikawa, C. R. Vianna, J. K. Elmquist and R. Coppari, Central administration of resveratrol improves diet-induced diabetes, Endocrinology, 2009, 150, 5326–5333 CrossRef CAS PubMed.
- N. L. Price, A. P. Gomes, A. J. Y. Ling, F. V. Duarte, A. Martin-Montalvo, B. J. North, B. Agarwal, L. Ye, G. Ramadori, J. S. Teodoro, B. P. Hubbard, A. T. Varela, J. G. Davis, B. Varamini, A. Hafner, R. Moaddel, A. P. Rolo, R. Coppari, C. M. Palmeira, R. de Cabo, J. A. Baur and D. A. Sinclair, SIRT1 is required for AMPK activation and the beneficial effects of resveratrol on mitochondrial function, Cell Metab., 2012, 15, 675–690 CrossRef CAS PubMed.
- C. Zhang, J. Luo, B. Yu, P. Zheng, Z. Huang, X. Mao, J. He, J. Yu, J. Chen and D. Chen, Dietary resveratrol supplementation improves meat quality of finishing pigs through changing muscle fiber characteristics and antioxidative status, Meat Sci., 2015, 102, 15–21 CrossRef CAS PubMed.
- F. Pellme, U. Smith, T. Funashashi, Y. Matsuzawa, H. Brekke, O. Wiklund, M. R. Taskinen and P. A. Jansson, Circulating adiponectin levels are reduced in nonobese but insulin-resistant first-degree relatives of type 2 diabetic patients, Diabetes, 2003, 52, 1182–1186 CrossRef CAS PubMed.
- J. Krakoff, T. Funahashi, C. D. Stehouwer, C. G. Schalkwijk, S. Tanaka, Y. Matsuzawa, S. Kobes, P. A. Tataranni, R. L. Hanson, W. C. Knowler and R. S. Lindsay, Inflammatory markers, adiponectin, and risk of type 2 diabetes in the pima Indian, Diabetes Care, 2003, 26, 1745–1751 CrossRef CAS PubMed.
- M. P. Krause, Y. Liu, V. Vu, L. Chan, A. Xu, M. C. Riddell, G. Sweeney and T. J. Hawke, Adiponectin is expressed by skeletal muscle fibers and influences muscle phenotype and function, Am. J. Physiol.: Cell Physiol., 2008, 295, C203–C212 CrossRef CAS PubMed.
- M. Iwabu, T. Yamauchi, M. Okada-Iwabu, K. Sato, T. Nakagawa, M. Funata, M. Yamaguchi, S. Namiki, R. Nakayama, M. Tabata, H. Ogata, N. Kubota, I. Takamoto, Y. K. Hayashi, N. Yamauchi, H. Waki, M. Fukayama, I. Nishino, K. Tokuyama, K. Ueki, Y. Oike, S. Ishii, K. Irose, T. Shimizu, K. Touhara and T. Kadowaki, Adiponectin and AdipoR1 regulate PGC-1α and mitochondria by Ca2+ and AMPK/SIRT1, Nature, 2010, 464, 1313–1319 CrossRef CAS PubMed.
- J. H. Um, S. J. Park, H. Kang, S. Yang, M. Foretz, M. W. McBurney, M. K. Kim, B. Viollet and J. H. Chung, AMP-activated protein kinase-deficient mice are resistant to the metabolic effects of resveratrol, Diabetes, 2010, 59, 554–563 CrossRef CAS PubMed.
- M. Wang, H. Yu, Y. S. Kim, C. A. Bidwell and S. Kuang, Myostatin facilitates slow and inhibits fast myosin heavy chain expression during myogenic differentiation, Biochem. Biophys. Res. Commun., 2012, 426, 83–88 CrossRef CAS PubMed.
- S. T. Kim, K. Marquard, S. Stephens, E. Louden, J. Allsworth and K. H. Moley, Adiponectin and adiponectin receptors in the mouse preimplantation embryo and uterus, Hum. Reprod., 2011, 26, 82–95 CrossRef CAS PubMed.
- G. Alberdi, V. M. Rodríguez, J. Miranda, M. T. Macarulla, N. Arias, C. Andrés-Lacueva and M. P. Portillo, Changes in with adipose tissue metabolism induced by resveratrol in rats, Nutr. Metab., 2011, 8, 29 CrossRef CAS PubMed.
- M. T. Macarulla, G. Alberdi, S. Gómez, I. Tueros, C. Bald, V. M. Rodríguez, J. A. Martínez and M. P. Portillo, Effects of different doses of resveratrol on body fat and serum parameters in rats fed a hypercaloric diet, J. Physiol. Biochem., 2009, 65, 369–376 CrossRef CAS PubMed.
- S. Gesta, Y. H. Tseng and C. R. Kahn, Developmental origin of fat: tracking obesity to its source, Cell, 2007, 131(2), 242–256 CrossRef CAS PubMed.
- M. Saito, Brown adipose tissue as a regulator of energy expenditure and body fat in humans, Diabetes Metab. J., 2013, 37, 22–29 CrossRef PubMed.
- M. J. Vosselman, W. D. van Marken Lichtenbelt and P. Schrauwen, Energy dissipation in brown adipose tissue: from mice to men, Mol. Cell. Endocrinol., 2013, 379, 43–50 CrossRef CAS PubMed.
- J. M. Andrade, A. C. Frade, J. B. Guimarães, K. M. Freitas, M. T. Lopes, A. L. Guimarães, A. M. de Paula, C. C. Coimbra and S. H. Santos, Resveratrol increases brown adipose tissue thermogenesis markers by increasing SIRT1 and energy expenditure and decreasing fat accumulation in adipose tissue of mice fed a standard diet, Eur. J. Nutr., 2014, 53(7), 1503–1510 CrossRef CAS PubMed.
- S. Wang, X. Liang, Q. Yang, X. Fu, M. Zhu, B. D. Rodgers, Q. Jiang, M. V. Dodson and M. Du, Resveratrol enhances brown adipocyte formation and function by activating AMP-activated protein kinase (AMPK) α1 in mice fed high-fat diet, Mol. Nutr. Food Res., 2017, 61(4), 1600746 CrossRef PubMed.
- Y. N. Huang, J. H. Qi, L. Xiang and Y. Z. Wang, Construction of adiponectin-encoding plasmid DNA and overexpression in mice in vivo, Gene, 2012, 502, 87–93 CrossRef CAS PubMed.
- S. Timmers, E. Konings, L. Bilet, R. H. Houtkooper, T. van de Weijer, G. H. Goossens, J. Hoeks, S. van der Krieken, D. Ryu, S. Kersten, E. Moonen-Kornips, M. K. C. Hesselink, I. Kunz, V. B. Schrauwen-Hinderling, E. Blaak, J. Auwerx and P. Schrauwen, Calorie Restriction-like Effects of 30 Days of Resveratrol Supplementation on Energy Metabolism and Metabolic Profile in Obese Humans, Cell Metab., 2011, 14, 612–622 CrossRef CAS PubMed.
- C. Handschin, Regulation of skeletal muscle cell plasticity by the peroxisome proliferator-activated receptor γ coactivator 1α, J. Recept. Signal Transduction Res., 2010, 30, 376–384 CrossRef CAS PubMed.
- T. Akimoto, S. C. Pohnert, P. Li, M. Zhang, C. Gumbs, P. B. Rosenberg, R. S. Williams and Z. Yan, Exercise Stimulates Pgc-1α Transcription in Skeletal Muscle Through Activation of the p38 MAPK Pathway, J. Biol. Chem., 2005, 280, 19587–19593 CrossRef CAS PubMed.
- J. Lin, H. Wu, P. T. Tarr, C. Y. Zhang, Z. Wu, O. Boss, L. F. Michael, P. Puigserver, E. Isotani, E. N. Olson, B. B. Lowell, R. Bassel-Duby and B. M. Spiegelman, Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres, Nature, 2002, 418, 797–801 CrossRef CAS PubMed.
- C. Handschin, S. Chin, P. Li, F. Liu, E. Maratos-Flier, N. K. Lebrasseur, Z. Yan and B. M. Spiegelman, Skeletal muscle fiber-type switching, exercise intolerance, and myopathy in PGC-1alpha muscle-specific knock-out animals, J. Biol. Chem., 2007, 282, 30014–30021 CrossRef CAS PubMed.
- E. Tomas, T. S. Tsao, A. K. Saha, H. E. Murrey, C. C. Zhang, S. I. Itani, H. F. Lodish and N. B. Ruderman, Enhanced muscle fat oxidation and glucose transport by ACRP30 globular domain: Acetyl-CoA carboxylase inhibition and AMP-activated protein kinase activation, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 16309–16313 CrossRef CAS PubMed.
- A. M. Brandstetter, B. Picard and Y. Geay, Regional variations of muscle fibre characteristic in m. semitendinosus of growing cattle, J. Muscle Res. Cell Motil., 1997, 18, 57–62 CrossRef CAS PubMed.
- J. Wess, G-protein-coupled receptors: molecular mechanisms involved in receptor activation and selectivity of G-protein recognition, FASEB J., 1997, 11, 346–354 CrossRef CAS PubMed.
- T. Yamauchi, J. Kamon, Y. Ito, A. Tsuchida, T. Yokomizo, S. Kita, T. Sugiyama, M. Miyagishi, K. Hara, M. Tsunoda, K. Murakami, T. Ohteki, S. Uchida, S. Takekawa, H. Waki, N. H. Tsuno, Y. Shibata, Y. Terauchi, P. Froguel, K. Tobe, S. Koyasu, K. Taira, T. Kitamura, T. Shimizu, R. Nagai and T. Kadowaki, Cloning of adiponectin receptors that mediate antidiabetic metabolic effects, Nature, 2003, 423, 762–769 CrossRef CAS PubMed.
- T. Yamauchi, Y. Nio, T. Maki, M. Kobayashi, T. Takazawa, M. Iwabu, M. Okada-Iwabu, S. Kawamoto, N. Kubota, T. Kubota, Y. Ito, J. Kamon, A. Tsuchida, K. Kumagai, H. Kozono, Y. Hada, H. Ogato, K. Tokuyama, M. Tsunoda, T. Ide, K. Murakami, M. Awazawa, I. Takamoto, P. Froguel, K. Hara, K. Tobe, R. Nagai, K. Ueki and T. Kadowaki, Targeted disruption of AdipoR1 and AdipoR2 causes abrogation of adiponectin binding and metabolic actions, Nat. Med., 2007, 13, 332–339 CrossRef CAS PubMed.
Footnote |
| † These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2019 |