Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
The blind men and the elephant: challenges in the analysis of complex natural mixtures
Royston
Goodacre

Department of Biochemistry, Institute of Integrative Biology, University of Liverpool, Biosciences Building, Crown Street, Liverpool L69 7ZB, UK. E-mail: roy.goodacre@liverpool.ac.uk; Web: http://www.twitter.com/roygoodacre
First published on 17th June 2019
Abstract
The identification of molecules from complex mixtures is difficult and full structure determination of the complete chemical milieu is yet to be achieved. Thus the comprehensive analysis of complex natural mixtures continues to challenge physical and analytical chemistry. Over the last 50 years or so, many research laboratories have strived to invent better analytical techniques with complementary physicochemical properties and improved resolving power, and to investigate upfront sample pre-treatments, which are necessary to enhance sample coverage from complex mixtures. The purpose of this Concluding remarks article is to try to capture the recent developments in high-resolution mass spectrometry and nuclear magnetic resonance spectroscopy applied to complex mixtures that were presented and debated, the parallel progress in chemometrics, data processing and machine learning approaches, as well as capturing and highlighting future challenges that still need to be addressed. The summary begins with a brief contextual overview and explains that the title – the blind men and the elephant – reflects that no single method measures everything and that multiple ‘tricorders’ are needed in order to understand complex systems. Next, the meeting highlights are provided, and I hope those that were present are happy that this captures the many diverse areas of research that were discussed and that this article may act as a yardstick to indicate where complex natural mixture analysis stands today.
Introduction
Researchers with highly diverse interests came together on the 13th May 2019 in the John McIntyre Conference Centre at the University of Edinburgh for three days for the 304th Royal Society of Chemistry Faraday Discussion on “Challenges in analysis of complex natural mixtures”. Whilst the sun shone brightly outside the conference venue, with Arthur’s Seat looking very tempting for some outside rambling activity, inside the lecture room science glowed as scientists delivered their latest analytical and data processing developments and findings, with intense discussions and debate warming up the atmosphere.The meeting was chaired by Dušan Uhrin (University of Edinburgh, UK), with excellent help from the conference organisers, including Mark Barrow (University of Warwick, UK), Timothy Ebbels (Imperial College London, UK), Ruth Godfrey (Swansea University, UK), Donald Jones (University of Leicester, UK) and Mathias Nilsson (University of Manchester, UK). There were 93 delegates from some 13 different countries attending the meeting, with a total of 25 oral presentations and 26 posters. One particularly NICE feature was the lightning poster presentations session, which took place in the late afternoon on the first day. These presentations were a mere 45 seconds each with automatic slide advances. Presenters assembled into an orderly queue, and even within just 3/4 of a minute, the audience was treated to NICE summaries that were both Novel and Interesting, and Clearly delivered with great Enthusiasm! The meeting then had its poster session where in depth discussions were held.
The meeting was split into four different sections, which are detailed below, but before getting into this it is worth setting the scene as to why the time was right for a Faraday Discussion to be devoted to the diverse and therefore multidisciplinary sciences behind unraveling complex natural mixtures.
The blind men and the elephant
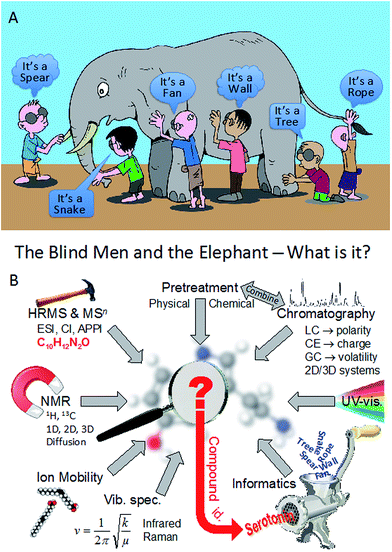
The reader may be familiar with the proverb of the blind men and the elephant, which has its origins in India. This fable is about six blind men who stumble across a strange creature and they try to understand what it is.1 To do this, they each feel a different part of the animal and come to a conclusion based on their limited experience; this is depicted in Fig. 1A. The first blind man feels the elephant’s body and comes to the conclusion that the creature is in fact a wall, while his friend feels the tusk and declares that the elephant is a spear. Another shakes his head after feeling the trunk of the elephant and claims with some anxiety that it’s actually a snake. The fourth blind man, whilst feeling the elephant’s leg, states that they are incorrect and that it’s indeed a tree. The next man has got hold of the elephant’s large ear and announces that it’s a fan, whilst the last blind man, who has hold of its tail, declares that his friends are all wrong and that this elephant is in fact a rope.Of course, all of the men are wrong, and only if they had shared their interactions would they have come to the correct conclusion about the elephant. The meaning of this parable is often used to illustrate that what people perceive as truth or fallacy is based on one’s all too often subjective and narrow experience(s). We can readily extend this to the analysis of complex natural systems.
Fig. 1B illustrates that multiple approaches are used for the analysis of chemical systems, and in this example how multiple physicochemical techniques may be used to identify a specific molecule (e.g.ref. 2). These are based on:
• Sample pretreatment that may involve fractionation or chemical/enzyme reactions.
• Chromatographic separation may then be employed, which uses different physical characteristics to effect separation of molecules in mixtures: viz., polarity, volatility or charge; and combinations of these. This can be combined with pretreatment processes, and would yield some information on a yet to be identified molecule, be it log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P, pKa or volatility.
P, pKa or volatility.
• For some analyses, additional separation can be performed using ion mobility where molecules are separated on the basis of their size, shape, and charge.
• Detection of molecules can involve many different approaches. These are also highlighted in Fig. 1B and include:
○ Simple UV that may provide information on specific absorbing species in the UV part of the electromagnetic spectrum.
○ Mass spectrometry (MS) may be used to infer chemical formulae (in the example here, accurate mass would suggest: C10H12N2O), and with MS-MS or MSn one can narrow down potential arrangements of atoms within a molecule.
○ Nuclear magnetic resonance (NMR) spectroscopy is accepted as the tool for structural assignments, and provided there is enough sample with sufficiently high purity, it is the tool of choice.
○ Infrared (IR) or Raman spectroscopy3 can also be used, though rarely, and they can provide information about functional groups due to their vibrational fingerprints.
This Faraday Discussion meeting therefore illustrates that systems chemical analysis for understanding complex systems is only really achieved by combining many different methods as they supply complementary information needed to identify an unknown substance. In addition to human interpretation, integration of multiple analytical approaches with appropriate informatics is needed for molecular identification. This is depicted by the ‘mincer’ in Fig. 1B and the informatics used may include statistics, chemometrics or some machine learning approach, in order to reveal in an objective fashion what the molecules may be within a mixture of diverse chemicals.
Thus we can see that chemical analysis is a multidisciplinary subject practiced by many scientists with diverse interdisciplinary skills. All of these disciplines were represented at this Faraday Discussion, and only with cooperation and integration can the whole molecular picture be ‘seen’: in Fig. 1B, this would be for the identification of serotonin. In reality, complex systems are much more complicated!
Chicken tikka masala
In this Faraday Discussion meeting, many different complex systems were analysed; these included plants, plant products (traditional medicines and essential oils), soils, coal, soot and petroleum fractions (petroleomics) as well as human derived samples such as urine.Particularly complex systems are the food that we eat.4 If we take a plate of chicken tikka masala and maybe have side accompaniments of raita and roti, then the meal we eat is very complex. This meal would (for example; other recipes do exist!) contain chicken marinated in tikka masala paste containing oil or butter, onion, ginger, garlic, cumin, turmeric, coriander, paprika, chilli powder, tomato, cream and coriander. This would then be skewered on bamboo or wood and cooked (which may release chemicals in the wood into the food) and then this chicken tikka added to a curry gravy containing yogurt, lemon juice, garlic, ginger, salt, cumin, garam masala and paprika. The raita may contain (e.g.) yoghurt, cucumber and mint, and the roti (e.g.) flour, salt and oil. With the exception (perhaps) of salt, each of these individual ingredients are highly complex mixtures and so the ensemble on the plate is an incredibly diverse mixture of chemicals. The analysis of this concoction would be very detailed and multifaceted.
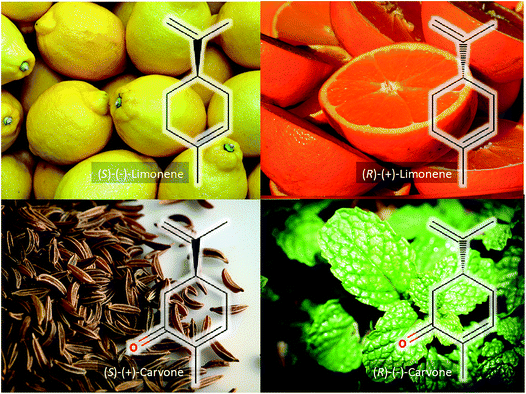
If we consider just two of the ingredients – lemon and mint – we recognise these as having distinct aromas and flavours, yet the chemicals that give rise to these characteristics are very simple. As illustrated in Fig. 2, the distinct lemony flavour comes from (S)-(−)-limonene, whilst its enantiomer (R)-(+)-limonene is found in oranges and is responsible for their aroma.5 The simple addition of a carbonyl group to the benzene ring of limonene gives rise to either the mint or caraway aroma and the flavour from (R)-(−)-carvone or (S)-(+)-carvone, respectively.6 These four very simple monoterpenes give highly diverse flavours and this highlights the importance of chirality in molecules, and in particular the interaction of such molecules with our taste receptors.
Whilst clearly important, the chiral nature of analysis was not really explored within this Faraday Discussion, and neither were positional isomers and their importance. By way of further example, if the food prepared above had used olive oil, then the major component would be (9Z)-octadec-9-enoic acid (oleic acid). This cis-isomer is considered healthy, while the trans-isomer (E)-octadec-9-enoic acid is not.7 The point being made here is that the analysis of complex systems requires careful analysis and the analyst needs to decide which chemical resolution is sufficient to report.
Dealing with complexity
The first session discussed the latest advances in high resolution mass spectrometry (HRMS) and chromatography, and their hyphenation. Philippe Schmitt-Kopplin gave a fascinating Introductory lecture that set the scene perfectly. He discussed complexity and diversity and remarked that the former is subjective and often hard to define. In terms of the diversity of chemicals, Philippe used small molecule chemistry (DOI: 10.1039/c9fd00078j) and highlighted that there are currently 97.3 × 106 different molecules in PubChem (https://pubchem.ncbi.nlm.nih.gov), of which 113![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 are detailed in the Human Metabolome Database (http://www.hmdb.ca) and if we confine this to lipids there are around 43
000 are detailed in the Human Metabolome Database (http://www.hmdb.ca) and if we confine this to lipids there are around 43![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 entries in Lipid Maps (https://lipidmaps.org). He also horrified the audience by explaining that for linear molecules of 700 amu containing just carbon, hydrogen, and oxygen, there were 1046 possible isomers!
000 entries in Lipid Maps (https://lipidmaps.org). He also horrified the audience by explaining that for linear molecules of 700 amu containing just carbon, hydrogen, and oxygen, there were 1046 possible isomers!
My own calculations for linear peptides show even more degrees of freedom. A simple peptide containing a mere 20 amino acids has 2020 = 1026 possible amino acid sequences. If we extend this to the average protein in archaea, bacteria or eukaryotes, which contain 283, 311 and 438 residues, respectively,8 then the complexity by numbers becomes astronomical with 10368 archaeal proteins, 10404 bacterial ones and 10569 eukaryotic proteins! Whilst during the discussion, we learnt that 21 Tesla FT-ICR-MS has enough resolving power to resolve two analytes that differ by the mere mass of an electron (DOI: 10.1039/c9fd00005d) and with 7 dimensional NMR spectroscopy 1018 analytes can be resolved (ref. 9, DOI: 10.1039/c8fd00213d), we are likely to run out of time before all of these proteins are measured as the lifetime of the universe is 1017 s (ref. 10), and you’ve probably used several of those reading this far!
For the analysis of any system, what is needed is an ideal detector, along with upfront sample separation or preparative chromatography (DOI: 10.1039/c8fd00234g), and of course like Father Christmas, the Easter Bunny or an honest politician, there is no such thing; there is no magic ‘tricorder’! If it were to exist, then the ideal detector (DOI: 10.1039/c8fd00233a) would be fast, have good orthogonality, provide uniform ionization, allow simplified data analysis and have improved (perhaps absolute) quantification.
For MS, electrospray ionisation (ESI) dominates most LC-MS and direct infusion-MS analyses, the latter mainly employing FT-ICR-MS. To a degree, this is a rather crude ionisation technique as ions are generated by squirting a conducting liquid through a needle to which a high voltage is then applied. As compound identification requires two orthogonal features,11 MS-MS or MSn is needed. This is however also rather crude and uncontrolled as the ions are often bombarded with an inert gas and the resulting fragmentation is akin to hitting a nut with a hammer and working out which nut was destroyed in the experiment. Thus library matching with standards is the key to compound identification along with more orthogonal techniques like NMR spectroscopy, which provides detailed structural analysis.
Other ionisation techniques are thus needed and applied, and within this session and elsewhere in the meeting, electron ionisation (EI) and various chemical ionisation (CI) methods as well as atmospheric pressure photoionization (APPI) were discussed. Each ionisation method has a bias to specific chemical classes and thus only part of the chemical milieu is ionized. The discussion of MS and fragmentation highlighted that any MS detector will be compromised in terms of having enough scanning speed, high enough mass resolution and enough duty cycle time to perform MS-MS or higher. Thus, for analyses with MS, there are always some concessions to be made.
All in all, this first session was interesting and perfectly set the scene. However, the most memorable thing that was elegantly and rather terrifyingly illustrated by Ryan Rodgers (DOI: 10.1039/c9fd00005d), was that all analyses were only scraping the top of the iceberg in terms of the comprehensiveness of analysis. For the analysis of petroleum fractions, the routine approach was to perform ESI FT-ICR-MS of aminopropyl silica (APS) extracts of bitumen. However, this revealed only a very small fraction of peaks compared to the same MS approach on six different modified aminopropyl silica (MAPS) fractions (Fig. 3). This relatively simple pre-fractionation revealed that the standard analysis had failed to ionise so many of the components within bitumen, and advocates for the use of prior separation and increased ionisation methods. Up front separation was also used by Jeffrey Hawkes who used exclusion chromatography coupled to MS in order to reveal dark matter that could be detected by UV but was seemingly invisible to MS (DOI: 10.1039/c8fd00222c).
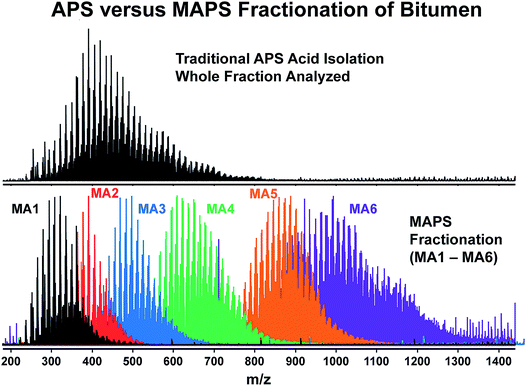 | ||
| Fig. 3 This figure illustrates that many analytes are not ionised in ESI and so a lot of information is missing. The top spectrum is a broadband negative ion mode ESI FT-ICR mass spectrum of the aminopropyl silica combined acids extract obtained from MacKay bitumen. When modified aminopropyl silica extracts from the same material are analysed in 6 fractions (MA1 (black)–MA6 (purple)), the broadband ESI FT-ICR mass spectra shown in the bottom spectra contain considerably more information, and this ‘dark matter’ missing from the first analysis is revealed. Reproduced from DOI: 10.1039/c9fd00005d with permission from the Royal Society of Chemistry. | ||
Within this context, the words ‘brutal’ and ‘depressing’ were used with reference to the above analysis and to the realisation that so much ‘dark matter’ was missing. Whilst da Silva and colleagues refer to dark matter in metabolomics as instances where there are MS data but no reference structure,12 we consider here dark matter to also include small and large molecules that are not even measured by the analytical method and so go undetected.13 The worrying thing is that there is no real way to estimate the level of dark matter when a complex sample is analysed.
High resolution techniques
In this session, high resolution NMR spectroscopy was highlighted as a complementary technique to high resolution MS. NMR spectroscopy is very powerful, as in contrast to MS it has the capability to solve structures, but often struggles with complexity in systems where there are many analytes present that vary in their concentration over large dynamic ranges. It was thus highly appropriate to hear about the latest developments and challenges with high resolution NMR and MS.In this session, much consideration was also given to upfront sample preparation and separation. Supercritical fluid extraction (SFE) coupled to both MS (DOI: 10.1039/c9fd00011a) and in-line sample concentration for NMR (DOI: 10.1039/c8fd00237a) were illustrated and discussed in terms of the selection of analytes extracted in this manner. The audience learnt that modifications of the supercritical fluid can readily allow for the analysis of both non-polar14 and polar analytes with NMR spectroscopy (DOI: 10.1039/c8fd00237a): by simply adding methanol to CO2, one can shift the mobile phase from non-polar to be more polar. A different approach highlighted for NMR analysis was to use viscous materials such as sucrose or 1% agarose gels to enable spin-diffusion during NMR acquisitions (DOI: 10.1039/c8fd00226f), an alternative to the popular DOSY-NMR approach.15,16
We were to learn later in the meeting that 7D NMR is possible, but even with modest 3D NMR, time is a limiting factor. In Nicholle Bell’s paper and presentation (DOI: 10.1039/c9fd00008a), (3,2)D NMR was introduced as a method of reducing the dimensionality of hyphenated NMR whilst still keeping the information content of 3D spectra, but offering the speed advantages of 2D NMR measurements.
Combining different methods was also highlighted in this session in terms of high resolution NMR and MS, but also combining these along with bioassays to decide which fractions from Chinese medicinal plants contained pharmacologically active substances, and hence which fractions to concentrate on for structural elucidation (DOI: 10.1039/c8fd00223a). This is an essential component in the analysis of highly complex mixtures, such as those derived from plant sources.
Finally in this session, the use of hydrogen–deuterium exchange (HDX), which in ambient conditions normally only occurs on exchangeable protons such as –OH and –NH, was extended to labeling protons on aromatic rings and –CH side chains (from substances found in coal) for their identification with FT-ICR-MS (DOI: 10.1039/c9fd00002j). The conditions used for this HDX were somewhat harsh as they involved treatments with 4 M NaOD or 16% DCl and heating to 120 °C for 40 h, so would only be useful for non-labile chemical species such as the components found within lignin.
Optimisation of sample pretreatment prior to high resolution analyses featured heavily in the discussion and it was suggested that this process needed to be done for each individual scenario – there was ‘no free lunch’. It would seem that most of this optimisation was done by brute force and with tongues maybe firmly in cheek by armies of PhD students. It is possible that this could be performed better by improved design of experiments and this would feature in the next session.
Data mining and visualisation
Collecting data on complex mixtures is only the start of the journey, and the next session of discussion was on chemometrics along with data mining, multivariate calibration or multi-way analysis, and how best these can be applied to different types of complex mixture.We were reminded by Johan Trygg (DOI: 10.1039/c8fd00243f) that:
“The challenge is not in data collection but in maximising information in data and transforming data into information, knowledge and wisdom.”
Johan Trygg, Faraday Discussion on Challenges in analysis of complex natural mixtures, 2019
This had perhaps been borrowed from an early quote by Henry Nix who was discussing national geographic information systems:
“Data does not equal information; information does not equal knowledge; and, most importantly of all, knowledge does not equal wisdom. We have oceans of data, rivers of information, small puddles of knowledge, and the odd drop of wisdom.”
Henry Nix, Keynote address, AURISA, 1990
Of course, this processing is important, but Johan also reminded us that the design of the experiment was vital in order to maximise the extraction of knowledge about a complex natural system, and hence become a wiser person after the data have been collected and analysed.
As had already been discussed, the analysis of chemical systems using more than one tool is important but the challenge is then what to do with such data. Multiblock analysis was suggested as one potential approach (DOI: 10.1039/c8fd00243f) and with JUMBA (Joint and Unique MultiBlock Analysis) this would allow for the extraction of variation in the systems under analysis at three different levels: (i) globally joint level that would provide information on common features across all data sets; (ii) locally joint information that would provide knowledge within one particular block (analytical technique); and (iii) unique features that may be specific to (e.g.) lipidomics rather than metabolomics or oxylipin analyses; in the example given for differentiation between people with mild or severe malaria from control populations.
The need for comparison of multiple data analysis algorithms on the same set of data also featured in this session, and this is always necessary when a new algorithmic approach is proposed. This was exemplified in one paper (DOI: 10.1039/c9fd00004f) where immunological markers from cells were measured using flow cytometry and the conclusion was again that there was ‘no free lunch’ as the performance of the algorithm depended on the nuances of the multivariate approach used.17
Structural analysis of molecules that have not already been measured and thus do not feature in databases of known substances is a challenge. This was addressed in two papers (DOI: 10.1039/c8fd00235e and DOI: 10.1039/c8fd00227d) that used GNPS libraries and data sets (https://gnps.ucsd.edu/, ref. 18). A series of algorithms was developed that allowed in a semi-automated fashion for sub-structural analysis and annotation of MSn data (DOI: 10.1039/c8fd00235e). In the future, more molecules will be identified using in silico predictions,19 as it will not be possible to acquire or make all possible standards for confirmatory MSn testing.
I reflected on this significant challenge that Justin van der Hooft was addressing and was reminded of the quote by a famous French philosopher:
“Science is built up with facts, as a house is with stones. But a collection of facts is no more a science than a heap of stones is a house”
Jules Henri Poincaré, La Science et l’hypothèse, 1854–1912
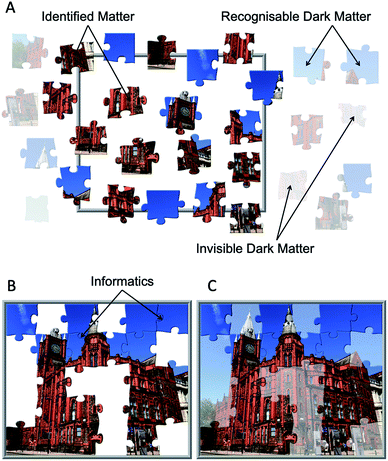
If we are to measure a complex mixture of molecules in order to understand the whole system, then currently this is like doing a jigsaw puzzle with only some of the pieces (or for Jules he can only find some of the bricks). This process is illustrated in Fig. 4. We can ask: what does the system do? Who interacts with whom? And in this example: what is in the picture? This is especially complex when we have three types of jigsaw piece with no idea as to whether we have a large or small proportion of the identified components (identified matter), and what we need to do to uncover the dark matter. Informatics is key to fitting the jigsaw pieces together. These computational approaches must infer what goes where, where and what the gaps are, and how to fill them; a central feature in any systems chemistry or biology analysis.20,21 In Fig. 4 some of the pieces can be put together and after gap filling we can see that the picture is revealed as the Victoria Gallery & Museum at the University of Liverpool; the VGM resides within the Victoria Building, which was constructed in 1892 and was the inspiration for the term ‘red brick university’.22
In this session, the discussion was directed towards having transparency in the data analysis process, standardisation in data collection and reporting the full informatics analysis pipeline. It was agreed that this could be enabled by suitable training activities where the ambition is to ingrain objectivity into the whole process. In order to enable this transparency, this community agreed that it is desirable to have more people make their data and code freely available. For discussions on this process within the metabolomics community, the interested reader is directed here.23–25
Future challenges and new approaches
The final day of the meeting was spent discussing new sample processing and instrumental advantages and what these may offer in the future, along with how data from different sources can be fused to understand more of the whole (DOI: 10.1039/c8fd00242h). Novel approaches to feature extraction from high-resolution data were also discussed (DOI: 10.1039/c9fd00014c).Over the last 5 years or so, ion mobility spectrometry (IMS) has been coupled with mass spectrometry.26,27 IMS is potentially useful as this technique adds a new dimension to analyte separation as ionised molecules are separated in the gas phase based on their mobility within a carrier gas; the orthogonal characteristics are therefore based on drift time in the carrier gas and these can be represented as collision cross sections (CCS), which can be computationally predicted.28 Two papers detailed IMS coupled with MS: the first with a 12 T FT-ICR-MS system for the analysis of heavy oil (DOI: 10.1039/c8fd00239h), and a further study exploited trapped IMS (TIMS) for the elucidation of isomeric species from dissolved organic matter (DOM) from aquatic systems (DOI: 10.1039/c8fd00221e). It was clear from both studies that this extra dimension of separation offered by IMS was highly useful for resolving components within complex mixtures.
As shown in other sessions, complex natural systems analysis must embrace sample pretreatment and separation. This was shown for human urine where solid phase extraction (SPE) was assessed using diverse types of column chemistry (DOI: 10.1039/c8fd00220g). This allowed for enrichment of specific molecular fractions and, perhaps as expected, the matching of the SPE type to the polarity or anion/cation enrichment is predictable and thus enhances the molecular content of the analyses.
Finally, a particularly elegant approach highlighted in one paper (DOI: 10.1039/c8fd00213d) was to use substrates with stable isotopes to label organisms prior to NMR spectroscopy. As was mentioned in the discussion session after this paper, “you are what you eat” and the novel aspect of feeding Daphnia magna with 13C labeled algae (Chlamydomonas reinhardtii) enhanced the ability to investigate metabolism as 13C–12C bond formation could be selectively observed. With further work, this could lead to accurate quantification of in vivo processes as these measurements could be made inside the NMR instrument. A mind boggling memory of the discussion on this paper was when the presenting author Ronald Soong admitted to spinning whole shrimp inside the instrument; I’m sure the poor creature also had its mind boggled!
Conclusions and prospects
This Faraday Discussion certainly showcased the current state-of-the-art for the analysis of complex natural systems. These analyses are in themselves complex and require diverse analytical techniques in combination with robust data analysis, interpretation and visualisation. I hope those that were present would agree that the above captures the main outcomes of the meeting and what was discussed.Within any research area there is always room for improvement. In addition to improvements in high resolution instrument hardware and informatics software, there were three main areas that would make the analysis of complex natural mixtures more complete. Each of these is readily achievable and these are based on sound analytical chemistry.
The first is that in many studies presented at this Faraday Discussion meeting, there seemed to be a lack of ownership and suitable level of background knowledge of the sample under interrogation. Some fantastic analyses were performed using HRMS and NMR, but little was mentioned in terms of where the sample had come from and whether the sample was relevant to answer the question under examination. Sampling and experimental design should be considered as an essential aspect of the analysis of complex systems29 and this was discussed at some length throughout the meeting. Collecting material may be achieved by a grab sample, though this single sample may under-represent the chemical diversity. It was considered that passive sampling over many days would generate a more comprehensive specimen from a heterogeneous environment, as well as the ability to take miniaturised analytical equipment into the field for on-site point and shoot analyses.30–32
The second was that there was in general a lack of figures of merit presented. With the exception of a few posters presented by early career researchers, no one discussed the precision in the amount of material measured, nor the accuracy of these levels. Here I am referring to the ordinate (MS ion count or NMR intensity) and not the abscissa, where for HRMS and NMR there certainly was excellent precision in m/z or δ (ppm); hence the high-resolution terminology. Similarly, where analytes were identified in complex mixtures, very few data were presented in terms of limits of detection (LOD) and quantification (LOQ), or limits of linearity (LOL). All of these are useful metrics and vital to appreciate the robustness of an analytical technique.33
The final challenge, which brings both of the above together, is the need to have validation in the measurement process. The 4 Rs of any analytical experiment are that it should have excellent reproducibility and robustness, and this is only achieved by resampling and repeating the experiment. Statistics within the sampling process are paramount. One would never perform a highly detailed and intricate analysis of a single grain of sand and then declare that this could be extrapolated to the point where every beach, sand dune or desert on the planet were fully understood!
In the extensive discussion sessions, which are a huge benefit of these Faraday Discussion meetings, it was debated how, as a community, analyses could be improved over the next few years. Two main conclusions arose from these deliberations: the first was that this was a friendly, welcoming community with diverse interests, and that people should talk, be listened to, learn the common language, and collaborate; the second was that one should address the question first and not the technology – no one really thought that the tail should wag the dog.
I would hope that if in 10 years or so I were to read a follow up Faraday Discussion volume on “challenges in analysis of complex natural mixtures”, there would be more thought into the background of the sample and whether it is representative of the problem, along with experimental design being used by many. In addition, I would like to see more confidence in the reproducibility and robustness of the process, with proof, and that suitable statistical figures of merit were presented alongside the data. Only then will we know whether we are addressing the challenge of analysing complex natural mixtures.
As was clear from these three days in Edinburgh, chemical systems analysis is vibrant and has brought together scientists from many different disciplines. I believe the future of this field is sunny and bright, and that the next decade will see further improvements in analytics and data processing that will allow for even more comprehensive analysis of complex natural mixtures. The pinnacle of these analyses is to reach the top of the complexity mountain, just like those that climbed to the peak of Arthur’s Seat after this most memorable Faraday Discussion meeting was over!
Conflicts of interest
There are no conflicts to declare.Acknowledgements
The inspiration for the title – the blind men and the elephant – came from seeing my colleague David Ellis at the University of Manchester use this in a recent talk. I would also like to thank in particular Dušan Uhrin, Tim Ebbels, and the organising committee for their very kind invite to spend three days at this very stimulating Faraday Discussion.References
- E. B. Goldstein, Encyclopedia of Perception, SAGE Publications Inc., 2009 Search PubMed.
- W. B. Dunn, D. Broadhurst, H. J. Atherton, R. Goodacre and J. L. Griffin, Systems level studies of mammalian metabolomes: the roles of mass spectrometry and nuclear magnetic resonance spectroscopy, Chem. Soc. Rev., 2011, 40, 387–426 RSC.
- D. I. Ellis and R. Goodacre, Metabolic fingerprinting in disease diagnosis: biomedical applications of infrared and Raman spectroscopy, Analyst, 2006, 131, 875–885 RSC.
- D. I. Ellis, V. L. Brewster, W. B. Dunn, J. W. Allwood, A. P. Golovanov and R. Goodacre, Fingerprinting food: current technologies for the detection of food adulteration and contamination, Chem. Soc. Rev., 2012, 41, 5706–5727 RSC.
- P. Laszlo, Citrus: A History, The University of Chicago Press, Chicago, 2007 Search PubMed.
- C. C. C. R. De Carvalho and M. M. R. Da Fonseca, Carvone: Why and how should one bother to produce this terpene, Food Chem., 2006, 95, 413–422 CrossRef CAS.
- A. Kiritsakis, Olive Oil, American Oil Chemists Society, Champagin. IL, USA, 1991 Search PubMed.
- L. P. Kozlowski, Proteome-pI: proteome isoelectric point database, Nucleic Acids Res., 2017, 45, D1112–D1116 CrossRef CAS PubMed.
- S. Hiller, C. Wasmer, G. Wider and K. Wüthrich, Specific Resonance Assignment of soluble non-globular proteins by 7D APSY-NMR Spectroscopy, J. Am. Chem. Soc., 2007, 129, 10823–10828 CrossRef CAS PubMed.
- J. D. Barrow and J. Silk, The left hand of creation: the origin and evolution of the expanding universe, Penguin, London, 1995 Search PubMed.
- L. W. Sumner, A. Amberg, D. Barrett, R. Beger, M. H. Beale, C. Daykin, T. W.-M. Fan, O. Fiehn, R. Goodacre, J. L. Griffin, N. Hardy, R. Higashi, J. Kopka, J. C. Lindon, A. N. Lane, P. Marriott, A. W. Nicholls, M. D. Reily and M. Viant, Proposed minimum reporting standards for chemical analysis, Metabolomics, 2007, 3, 211–221 CrossRef CAS PubMed.
- R. R. da Silva, P. C. Dorrestein and R. A. Quinn, Illuminating the dark matter in metabolomics, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 12549–12550 CrossRef CAS PubMed.
- O. A. H. Jones, Illuminating the dark metabolome to advance the molecular characterisation of biological systems, Metabolomics, 2018, 14, 101 CrossRef PubMed.
- F. H. M. van Zelst, S. G. J. van Meerten, P. J. M. van Bentum and A. P. M. Kentgens, Hyphenation of supercritical fluid chromatography and NMR with in-line sample concentration, Anal. Chem., 2018, 90, 10457–10464 CrossRef CAS PubMed.
- H. Barjat, G. A. Morris, S. Smart, A. G. Swanson and S. C. R. Williams, High-resolution diffusion-ordered 2D spectroscopy (HR-DOSY) – a new tool for the analysis of complex-mixtures, J. Magn. Reson., Ser. B, 1995, 108, 170–172 CrossRef CAS.
- M. Nilsson, The DOSY Toolbox: A new tool for processing PFG NMR diffusion data, J. Magn. Reson., 2009, 200, 296–302 CrossRef CAS PubMed.
- P. S. Gromski, H. Muhamadali, D. I. Ellis, Y. Xu, E. Correa, M. L. Turner and R. Goodacre, A tutorial review: Metabolomics and partial least squares-discriminant analysis – a marriage of convenience or a shotgun wedding, Anal. Chim. Acta, 2015, 879, 10–23 CrossRef CAS PubMed.
- M. Wang, J. J. Carver, V. V. Phelan, L. M. Sanchez, N. Garg, Y. Peng, D. D. Nguyen, J. Watrous, C. A. Kapono, T. Luzzatto-Knaan, C. Porto, A. Bouslimani, A. V. Melnik, M. J. Meehan, W.-T. Liu, M. Crüsemann, P. D. Boudreau, E. Esquenazi, M. Sandoval-Calderón, R. D. Kersten, L. A. Pace, R. A. Quinn, K. R. Duncan, C.-C. Hsu, D. J. Floros, R. G. Gavilan, K. Kleigrewe, T. Northen, R. J. Dutton, D. Parrot, E. E. Carlson, B. Aigle, C. F. Michelsen, L. Jelsbak, C. Sohlenkamp, P. Pevzner, A. Edlund, J. McLean, J. Piel, B. T. Murphy, L. Gerwick, C.-C. Liaw, Y.-L. Yang, H.-U. Humpf, M. Maansson, R. A. Keyzers, A. C. Sims, A. R. Johnson, A. M. Sidebottom, B. E. Sedio, A. Klitgaard, C. B. Larson, C. A. P. Boya, D. Torres-Mendoza, D. J. Gonzalez, D. B. Silva, L. M. Marques, D. P. Demarque, E. Pociute, E. C. O’Neill, E. Briand, E. J. N. Helfrich, E. A. Granatosky, E. Glukhov, F. Ryffel, H. Houson, H. Mohimani, J. J. Kharbush, Y. Zeng, J. A. Vorholt, K. L. Kurita, P. Charusanti, K. L. McPhail, K. Fog Nielsen, L. Vuong, M. Elfeki, M. F. Traxler, N. Engene, N. Koyama, O. B. Vining, R. Baric, R. R. Silva, S. J. Mascuch, S. Tomasi, S. Jenkins, V. Macherla, T. Hoffman, V. Agarwal, P. G. Williams, J. Dai, R. Neupane, J. Gurr, A. M. C. Rodríguez, A. Lamsa, C. Zhang, K. Dorrestein, B. M. Duggan, J. Almaliti, P.-M. Allard, P. Phapale, L.-F. Nothias, T. Alexandrov, M. Litaudon, J.-L. Wolfender, J. E. Kyle, T. O. Metz, T. Peryea, D.-T. Nguyen, D. VanLeer, P. Shinn, A. Jadhav, R. Müller, K. M. Waters, W. Shi, X. Liu, L. Zhang, R. Knight, P. R. Jensen, B. Ø. Palsson, K. Pogliano, R. G. Linington, M. Gutiérrez, N. P. Lopes, W. H. Gerwick, B. S. Moore, P. C. Dorrestein and N. Bandeira, Sharing and community curation of mass spectrometry data with Global Natural Products Social Molecular Networking, Nat. Biotechnol., 2016, 34, 828–837 CrossRef CAS PubMed.
- T. De Vijlder, D. Valkenborg, F. Lemière, E. P. Romijn, K. Laukens and F. Cuyckens, A tutorial in small molecule identification via electrospray ionization-mass spectrometry: The practical art of structural elucidation, Mass Spectrom. Rev., 2018, 137, 607–629 CrossRef PubMed.
- D. B. Kell, Metabolomics, modelling and machine learning in systems biology towards an understanding of the languages of cells. The 2005 Theodor Bücher lecture, FEBS J., 2006, 273, 873–894 CrossRef CAS PubMed.
- D. B. Kell and J. D. Knowles, The role of modeling in systems biology, in System modeling in cellular biology: from concepts to nuts and bolts, ed. Z. Szallasi, J. Stelling and V. Periwal, MIT Press, Cambridge, 2006, pp. 3–18 Search PubMed.
- A. L. Mackenzie and A. R. Allan, Redbrick University Revisited, Liverpool University Press, 1996 Search PubMed.
- P. Rocca-Serra, R. M. Salek, M. Arita, E. Correa, S. Dayalan, A. Gonzalez-Beltran, T. M. D. Ebbels, R. Goodacre, J. Hastings, K. Haug, A. Koulman, M. Nikolski, M. Oresic, S.-A. Sansone, D. Schober, J. Smith, S. Steinbeck, M. R. Viant and S. Neumann, Data standards can boost metabolomics research, and if there is a will, there is a way, Metabolomics, 2016, 12, 14 CrossRef PubMed.
- R. J. M. Weber, T. N. Lawson, R. M. Salek, T. M. D. Ebbels, R. C. Glen, R. Goodacre, J. L. Griffin, K. Haug, A. Koulman, P. Moreno, M. Ralser, C. Steinbeck, W. B. Dunn and M. R. Viant, Computational tools and workflows in metabolomics: An international survey highlights the opportunity for harmonisation through Galaxy, Metabolomics, 2017, 13, 12 CrossRef PubMed.
- B. Burla, M. Arita, M. Arita, A.K. Bendt, A. Cazenave-Gassiot, E.A. Dennis, K. Ekroos, X. Han, K. Ikeda, G. Liebisch, M.K. Lin, T.P. Loh, P.J. Meikle, M. Orešič, O. Quehenberger, A. Shevchenko, F. Torta, M.J. Wakelam, C.E. Wheelock and M.R. Wenk, MS-based lipidomics of human blood plasma – a community-initiated position paper to develop accepted guidelines, J. Lipid Res., 2018, 59, 2001–2017 CrossRef CAS PubMed.
- A. B. Kanu, P. Dwivedi, M. Tam, L. Matz and H. H. Hill, Ion mobility–mass spectrometry, J. Mass Spectrom., 2008, 43, 1–22 CrossRef CAS PubMed.
- F. Lanucara, S. W. Holman, C. J. Gray and C. E. Eyers, The power of ion mobility-mass spectrometry for structural characterization and the study of conformational dynamics, Nat. Chem., 2014, 6, 281–294 CrossRef CAS PubMed.
- V. Gabelica, A. A. Shvartsburg, C. Afonso, P. Barran, J. L. P. Benesch, C. Bleiholder, M. T. Bowers, A. Bilbao, M. F. Bush, J. L. Campbell, I. D. G. Campuzano, T. Causon, B. H. Clowers, C. S. Creaser, E. De Pauw, J. Far, F. Fernandez-Lima, J. C. Fjeldsted, K. Giles, M. Groessl, C. J. Hogan Jr, S. Hann, H. I. Kim, R. T. Kurulugama, J. C. May, J. A. McLean, K. Pagel, K. Richardson, M. E. Ridgeway, F. Rosu, F. Sobott, K. Thalassinos, S. J. Valentine and T. Wyttenbach, Recommendations for reporting ion mobility Mass Spectrometry measurements, Mass Spectrom. Rev., 2019, 38, 291–320 CrossRef CAS PubMed.
- L. Leardi, Experimental design in chemistry: A tutorial, Anal. Chim. Acta, 2009, 652, 161–172 CrossRef PubMed.
- L. Li, T.-C. Chen, Y. Ren, P. I. Hendricks, R. G. Cooks and Z. Ouyang, Mini 12, Miniature Mass Spectrometer for Clinical and Other Applications—Introduction and Characterization, Anal. Chem., 2014, 86, 2909–2916 CrossRef CAS PubMed.
- X. Meng, X. Zhang, Y. Zhai and W. Xu, Mini 2000: A Robust Miniature Mass Spectrometer with Continuous Atmospheric Pressure Interface, Instruments, 2018, 2, 2 CrossRef.
- D. I. Ellis, H. Muhamadali, S. A. Haughey, C. T. Elliott and R. Goodacre, Point-and-shoot: rapid quantitative detection methods for on-site food fraud analysis – moving out of the laboratory and into the food supply chain, Anal. Methods, 2015, 7, 9401–9414 RSC.
- J. C. Miller and J. N. Miller, Statistics for Analytical Chemistry, Ellis Horwood, London, 1988 Search PubMed.
| This journal is © The Royal Society of Chemistry 2019 |