Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
DNAzyme-based biosensor as a rapid and accurate verification tool to complement simultaneous enzyme-based media for E. coli detection†
Noor
Zaouri
a,
Zhengfang
Cui
b,
Ana Sol
Peinetti
b,
Yi
Lu
b and
Pei-Ying
Hong
 *a
*a
aKing Abdullah University of Science and Technology (KAUST), Water Desalination and Reuse Center, Division of Biological and Environmental Science and Engineering, Thuwal 239455-6900, Saudi Arabia. E-mail: peiying.hong@kaust.edu.sa; Tel: +966 12 808 2218
bDepartment of Chemistry, University of Illinois at Urbana-Champaign, Urbana, IL 61801, USA
First published on 22nd October 2019
Abstract
Simultaneous enzyme-based media can be used to identify both coliforms and E. coli faster than lactose-based fermentation protocols. However, these media can be prone to false-positive results and would require verification tests to confirm the presence of E. coli. These verification tests (e.g. indole or oxidase tests) work based on identifying specific phenotypic traits in bacteria, and hence are also prone to inaccuracies. This study aimed to achieve rapid verification test of E. coli using DNAzyme-based biosensor. The biosensor was first tested for four E. coli strains against more than 90 non-E. coli strains routinely causing false positive results and showed high specificity towards E. coli strains. The biosensor was then determined for its detection sensitivity. It was observed that a single E. coli colony would generate a robust fluorescence signal after 2 h of sub-culturing. With this short incubation time, non-E. coli strains were unable to generate any apparent fluorescence signal, hence further improving the detection specificity of biosensor. Lastly, we further demonstrate that the biosensor can be applied to different commercial brands of simultaneous enzyme-based agar to consistently identify the presence of E. coli. The biosensor was able to rapidly denote presence of discrete E. coli colonies by binding on to protein targets possibly of DNA replication or motility functions that were predominantly specific to E. coli.
Water impactE. coli detection is an important parameter to monitor when defining water quality. Although enzyme-based agar media require a shorter time to denote if E. coli is present or not, false positive rates associated with these types of media result in unnecessary false alarms for both utility operators and the general public, and incur an additional operational burden for the utilities. We demonstrated that DNAzyme biosensor can be used as a rapid and accurate verification test for colonies growing on agar media. This tool would aid in effective decision-making process to better protect consumers. |
1. Introduction
Determining the presence of Escherichia coli (E. coli) and total coliforms (TC) in drinking water and treated wastewaters is required to denote the water quality and efficiency of a treatment process. Conventional standard protocol determines TC first prior to fecal coliforms and then E. coli.1,2 To achieve a faster decision-making process, in recent years, simultaneous enzyme-based tests are widely used to monitor and enumerate both TC and E. coli simultaneously.3,4 Simultaneous enzyme-based methods (e.g. Colilert or Colilert-18, MI, m-FC, Chromocult) are designed to detect beta-galactosidase and beta-glucuronidase, both of which are enzymes assumed to be uniquely associated with TC and E. coli, respectively.5However, beta-galactosidase and beta-glucuronidase can also be detected in other non-E. coli species that do not fall within the classification of TC (e.g. Aeromonas, Pseudomonas, Sphingomonas, and Flavobacterium).2,5–8 Furthermore, cell viability on media agar can be affected by substances in water matrix that can inhibit the enzymes2,9 and temperature variation.10,11 Therefore, simultaneous enzyme-based methods are susceptible to false positives or false negative results, with varying degrees of false positive rates for E. coli ranging from 0.9 to 37.5% reported.2,9,12–16 Thus, verification tests such as cytochrome oxidase/indole detection,17,18 lactose fermentation,19 and API 20E,20 may be required to verify the identities of colonies growing on the media agar. However, these verification tests still rely on observations of correct phenotypic traits, and expression of these phenotypic traits can be biased depending on the state of bacterial growth and its growth conditions.21–23 The overall time required for some of these verification tests can take up to 48 h, depending on the test type. For instance, indole and cytochrome oxidase tests are quick (less than few minutes) but may not be reliable since both tests can be influenced by dye-containing media. Specifically, dye carried over within the colonies may interfere with the indole color interpretation24 and in the assessment of cytochrome results.25 On the other hand, lactose fermentation test could be reliable but requires more than 48 h to derive the confirmatory results. Inaccuracy and time-consuming verification tests therefore hinder decision-making process necessary to ensure water quality and protect public health.
To overcome the limitations of the above mentioned simultaneous enzyme-based methods, an alternative rapid and accurate verification test is required. To achieve the goal, we propose the use of DNAzyme biosensors to verify the presence of E. coli among colonies obtained from simultaneous enzyme-based media agar. DNAzymes are short single-stranded DNA sequences which can fold into complex tertiary structures to achieve an enzymatic activity.26,27 This activity typically requires the presence of a specific target that acts as a cofactor for proper folding and/or enzymatic reactions. Based on the target, DNAzyme can come in two formats, namely, cis-acting DNAzyme, where the enzyme and substrate strands are linked together through a DNA hairpin, or the trans-acting DNAzyme, where the enzyme and substrate strands are hybridized as individual entities prior to reaction.28 As a result, the enzymatic activity can be used as a basis for signaling output for the cofactor. As the DNAzyme can be easily modified by different signaling agents, such as fluorophores and quencher, many fluorescent sensors have been developed successfully.29–31 Briefly, in the presence of its intended target, specific binding between the target and enzyme strand would cleave the substrate strand, allowing the fluorophore to dissociate from the quencher and generating a turn-on fluorescent signal.
Recently, DNAzymes specific for bacteria target have been developed.32–35 Particularly, Li′s group has developed an E. coli-specific DNAzyme.32 Earlier studies have also designed various biosensors based on this DNAzyme to detect E. coli, and tested for its specificity against other non-E. coli bacteria, for example, Pseudomonas peli, Pediococcus acidilactici, Bacillus subtilis, Yersinia rukeri.28,32,36 However, most of these tested non-E. coli strains were not the ones generating false positives in drinking water utilities.4 Therefore, further studies are needed to test for selectivity of this DNAzyme toward those false-positive isolates recovered by water utilities. This is to ensure the suitability of this DNAzyme biosensor in water quality monitoring. Furthermore, most of the developed DNAzymes were tested for their specificities against E. coli using protocols that involved >7 h incubation to allow propagation till cell density of approximately 1 in terms of OD600 nm.28,32,33 This does not provide competitive edge over other existing verification tests, and DNAzymes should be improved by shortening the time needed to provide verification results. To address these issues, we herein report investigation of the specificity of E. coli DNAzyme sensor against other non-E. coli TC species that were isolated from different water utilities across the U.S.,4 as well as improving the detection sensitivity of DNAzymes by determining the lowest E. coli cell density required to differentiate between target and non-targets. In addition, the nature of the target molecule bound to the E. coli biosensor was further elucidated. These results will facilitate the use of the E. coli biosensor platform as a verification step to improve the accuracy and speed of current simultaneous enzyme-based methods for detecting E. coli.
2. Materials and methods
2.1. Bacterial strains
E. coli and total coliforms were acquired from nine water utilities and laboratories participating in an earlier study.4 The list of bacterial strains tested in this study is provided in Table S1.† To extract the crude intracellular material (CIM), each bacteria was first streaked on LB agar, and incubated for 24 h at 37 °C. Then, a single colony of around 1 mm in diameter was picked from the LB plate and inoculated in 7 mL SOB broth (Hanahan's broth) and incubated for 16 h at 37 °C to reach an OD600 ≈ 1 (OD600 = optical density at 600 nm). 1 mL of the culture broth was then transferred into 2 mL tube, centrifuged for 2 min at 9400g to collect the pellet. The pellet was re-suspended in 1 mL 1× testing buffer (1× testing buffer: 50 mM HEPES, 150 mM NaCl, 15 mM MgCl2, pH 7.5). The pellet suspension was heated at 65 °C to lyse the bacterial cells. After 30 min, the suspension was centrifuged for 2 min at 9400g, and the supernatant was collected to constitute the CIM of that particular bacterium.2.2. E. coli DNAzyme probes
trans-Acting E. coli DNAzyme probe consists of two oligonucleotides, namely, FS1 that is functionalized with fluorophore and quencher (5′-ACT CTT CCT AGC/iFluorT/rA/iDabdT/GGT TCG ATC AAG A-3′), and EC1T (5′-GAT GTG CGT TGT CGA GAC CTG CGA CCG GAA CAC TAC ACT GTG TGG GGA TGG ATT TCT TTA CAG TTG TGT G-3′). In the presence of CIM from the target bacterium, binding between target and EC1T results in catalytic activity that would cleave FS1. Then, the fluorophore and quencher will dissociate to generate a turn-on fluorescent signal, due to a change in the melting temperature after the cleavage. The increase in fluorescence intensity with time correlates with the extent of hybridization. Both oligonucleotides were synthesized by Integrated DNA Technologies (Coralville, IA). 50 μL of E. coli liquid sensor was prepared by mixing 2.5 μL of E. coli FS1 (5 μM), 5 μL of E. coli EC1T (10 μM), 5 μL of 10× testing buffer, and 37.5 μL of sterile water. 1× testing buffer comprised of 50 mM HEPES, 150 mM NaCl, 15 mM MgCl2, adjusted to pH 7.5. Next, the liquid sensor was annealed at 70 °C for 5 minutes and then placed at room temperature before use within 30 min. The specificity of the E. coli DNAzyme in the presence of E. coli CIM and total coliforms CIM was investigated by following the incremental slope rate of the fluorescence signal intensity for 5 minutes. SpectraMax M3 microplate reader was used to record the florescence signal at excitation 485 nm and emission 538 nm. The measurement was done by mixing 200 μL CIM with 2 μL E. coli DNAzyme, and the florescence kinetics exhibited during the first 5 min of reaction was measured.2.3. Determination of fluorescence slope rates with bacterial growth
The slope of the florescence response of E. coli DNAzyme against E. coli and non-E. coli CIM extracted during growth was investigated in triplicates. One colony of E. coli (OH 11 and OH 18) and non-E. coli coliforms, namely Klebsiella (EPA 193), Morganella (MA-35-Ready-2), and Serratia (EPA 74) was individually sub-cultured in SOB broth and incubated at 37 °C. Each colonies picked are of approximately 1 mm in diameter. The OD600 was measured at different time points during the bacterial growth. In parallel, 1 mL samples was collected for CIM extraction as described in section 2.1, and hybridized to liquid sensor as described in section 2.2 to evaluate the fluorescence slope rates.2.4. Verification of simultaneous enzymatic base media method with E. coli liquid sensor
Three types of simultaneous enzymatic media were used to enumerate total and fecal coliforms. The media includes Chromocult (Millipore, Billerica, US), m-FC (Sigma Aldrich, Darmstadt, Germany), and MI (Thermo Fisher Scientific, Suwanee, US) media. Two types of samples were used against the simultaneous enzymatic base media. The first was PBS buffer spiked with known bacteria. To prepare a spiked PBS mixture of known bacterial cells, 1× phosphate buffer saline (PBS buffer) was prepared in sterile 400 mL glass bottle and then spiked with approximately 102E. coli (OH 1) cells. In other separate bottles, 1× PBS was spiked with E. coli and one of the non-E. coli coliforms, namely Klebsiella (EPA 193), Morganella (MA-35-Ready-2) or Serratia (EPA 74) in equal cell densities as that of E. coli. 100 mL of each spiked mixture was filtered through 0.45 μm filter, and the filters were placed on three simultaneous enzyme-based media tested in this study. The media with the respective filters were incubated at temperatures recommended by manufacturers for 24 h (Table 1). The second type of water to be tested against the media was post-MBR effluent collected from KAUST wastewater treatment plant. The effluent was also filtered through 0.45 μm filter and placed on individual media to enumerate coliform numbers. The simultaneous enzyme-based media generates colonies that differed in phenotypic traits depending on whether the colony is derived from E. coli or from other total coliforms (Table 1). After incubation, single colonies, each of approximately 1 mm in diameter, were picked based on their different phenotypic traits and sub-cultured in 200 μL SOB broth in 96-well plate before CIM extraction. After 2 h incubation at 37 °C, CIM was obtained directly on the 96-well plate by first centrifuging plate at 1000g for 20 min, and ca. 180 μL of supernatant from each individual well was transferred to a corresponding well in a clean 96-well plate. The CIM was tested against the E. coli DNAzyme using procedure as described in section 2.2.| Incubation temp. °C | E. coli | Serratia | Morganella | Klebsiella | |
|---|---|---|---|---|---|
| Color/fluorescence | Color/fluorescence | ||||
| MI | 37 | Blue/positive | Creamy/negative | Creamy/negative | Creamy/positive |
| m-FC | 37 | Blue | Creamy | Silver | Silver |
| Chromocult | 37 | Violet | Colorless | Colorless | Red |
2.5. Identification of target molecule
3. Results and discussion
3.1. Specificity of E. coli DNAzyme sensor
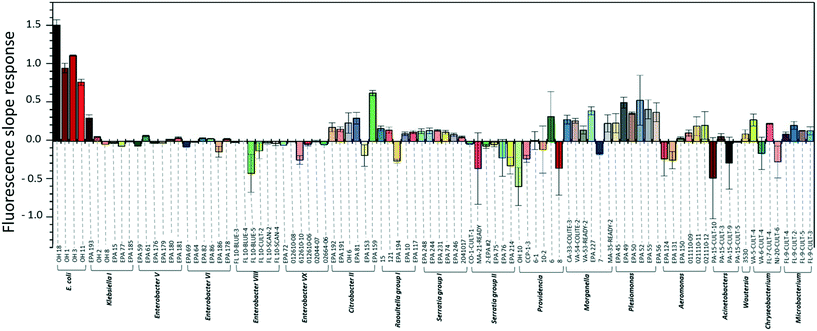
To investigate the specificity of the E. coli DNAzyme sensor, four different E. coli strains and about 90 non-E. coli strains were tested as described in section 2.2. Fig. 1 shows the fluorescence increment slopes in response to the different strains studied. The slopes ranging from 0.7 to 1.6 min−1 were observed for the tested E. coli strains, and the average of these slope values was 55 times higher than those observed for the other non-E. coli bacteria. Even though some non-E. coli bacteria, such as Citrobacter, Klebsiella, Morganella, and Plesiomonas, showed relatively high slope values ranging from 0.2 to 0.6 min−1, these slopes were still lower than those generated by E. coli strains, allowing clear discrimination between E. coli strains and non-E. coli strains. The E. coli DNAzyme sensor showed high specificity to four different target strains of E. coli CIM (OH 1, 3, 11, and 18) but not to other non-target bacteria. Some of the non-E. coli bacteria generated negative slopes, indicating a decrease in fluorescence signal with time (Fig. 1). The exact reasons to account for this observation are not known. However, we postulate that in the presence of some non-target bacteria samples, there may be a potential non-specific quenching of the fluorophores by elements known to quench the fluorophore, such as iron and copper in those samples.3.2. Detection sensitivity of the E. coli DNAzyme sensor
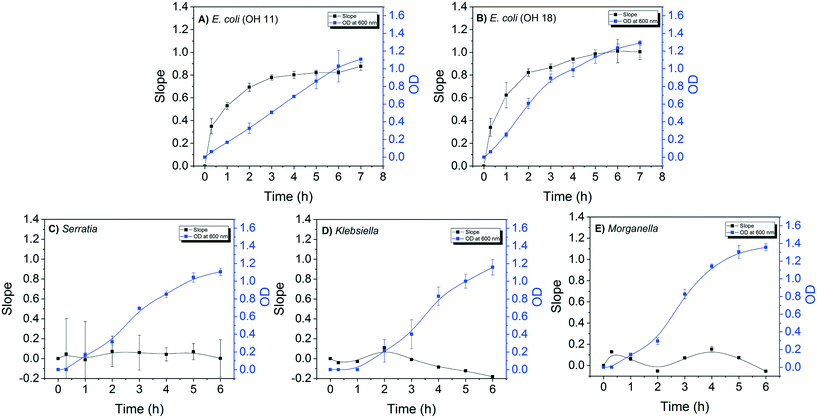
The detection sensitivity of the E. coli DNAzyme sensor was investigated in order to determine the minimum cell density required to distinguish between E. coli and non E. coli strains. The minimal time required to grow the strains for specific detection was also evaluated. For this, the fluorescence signal of the E. coli sensor against E. coli and non-E. coli CIM extracted at different points during the bacterial growth was evaluated. A positive correlation between the slopes of fluorescence increase and OD600 nm values for both E. coli OH11 and OH 18 was observed (Fig. 2A and B). The slope values for E. coli OH11 and OH18 plateau at ca. 3 to 4 h of exponential growth. In contrast, no positive correlation between slope and OD600 nm values were observed for Serratia MA-35-ready-2, Morganella EPA 74 and Klebsiella EPA 193. The highest observed slope values were 0.07 ± 0.15, 0.09 ± 0.01, and 0.21 ± 0.01 for Serratia, Morganella, and Klebsiella, respectively. The average slope values for E. coli OH11 and OH18 were therefore ca. 10 times higher than that observed for the other non-E. coli bacteria after 2 h incubation, suggesting that an improved differentiation can be obtained by applying CIM obtained during early exponential growth to E. coli liquid sensor. Furthermore, 2 h of incubation from a starting colony of 1 mm in diameter was enough to obtain a sufficient fluorescence slope response of E. coli CIM (0.69 ± 0.04, and 0.82 ± 0.33 for OH11 and OH18, respectively), where the OD was ≈0.4 (equivalent to 104–105 cells per mL).Therefore, besides demonstrating high specificity towards E. coli, the developed DNAzyme biosensor also shows high detection sensitivity as it was able to detect the E. coli targets against the non-targets within a short incubation time of 2 h. Based on the growth curve of E. coli (Fig. 2A and B), this result suggests that the biosensor requires minimal cell number of 104–105E. coli cells per mL to result in a significant fluorescence slope increase. In this study, the required cell density was obtained from CIM extraction, and was comparable to that reported by earlier studies.28,32,37 Overall, our developed DNAzyme biosensor was able to differentiate discrete targets against non-targets within 2 h, suggesting that it can be used as a verification method for simultaneous enzyme test by providing results in a faster manner than conventional verification tests.
3.3. Coupling the DNAzyme sensors for verification of colonies on simultaneous enzymatic media
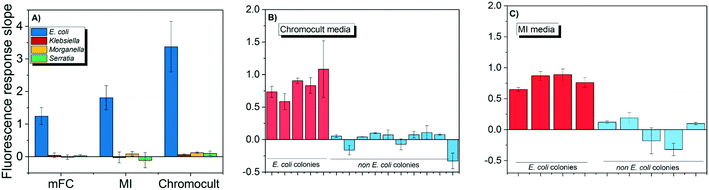
Both E. coli OH1 and non-E. coli strains spiked into the buffer matrix displayed the correct phenotypic traits when grown on Chromocult, m-FC, and MI simultaneous enzyme media. Individual colonies were picked and further incubated for 2 h to obtain their respective CIM. As demonstrated in Fig. 3A, E. coli OH 1 colonies can be clearly differentiated from the other non-E. coli strains based on the fluorescence intensity slope values.When wastewater effluent was filtered and placed on Chromocult and MI, interference effect of the wastewater effluent resulted in some difficulties differentiating between E. coli and non-E. coli colonies using both simultaneous-enzymatic media. Although there were colonies that showed the distinct phenotypic color representative of E. coli, there were also other colonies that displayed atypical phenotypic traits and would require further verification. Thus, the E. coli DNAzyme sensor was tested against individual colonies and was again able to distinguish between the E. coli and non-E. coli colonies based on fluorescence intensity slope value (Fig. 3B and C). The specificity of the liquid sensor towards E. coli was not altered by the type of simultaneous enzyme-based medium used or the complex matrix of the water samples (e.g., effluent wastewater).
The conventional verification methods (e.g. oxidase and indole test) are particularly vulnerable to compounds like glucose, tryptophan or indicator dyes that could cause aberrant results,38 and would hence require sub-culturing on non-selective media in order to accurately denote the correct phenotypic traits. In contrast, our findings suggest that the DNAzyme biosensors were not subjected to the same limitations. The DNAzyme biosensor was able to correctly discern E. coli despite being grown on different brands of commercially available simultaneous enzymatic media (Fig. 3), including colored media like m-FC and MI agar. This result indicates that the dyes and the fluorescence compounds (which originated from chromogens or fluorogen) formed in individual colonies during bacteria growth on enzymatic media, do not interfere with the fluorescence signal of the DNAzyme biosensor (Fig. 3). Therefore, the identification of E. coli with the DNAzyme biosensor is not affected by the different media composition and complexity of different water matrix like treated wastewater. Our observations are in agreement with earlier studies demonstrating the ability of DNA-based biosensor to identify its intended target present in a range of complex samples, e.g., apple juice, milk, and blood serums.36,39
3.4. Molecular nature of target hybridized to DNAzyme sensor
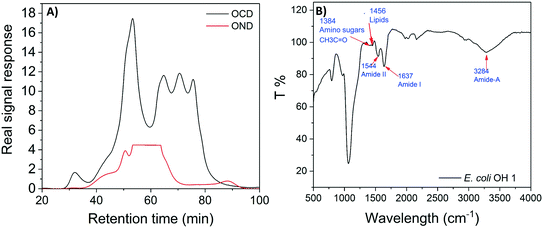
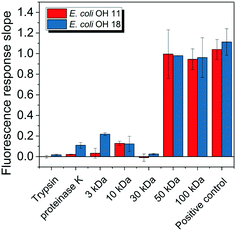
E. coli CIM consists of organic fractions that showed variation in molecular weights as observed from LC-OCD profile (Fig. 4A). Those organic fractions showed high nitrogen signal response, which is an indicator of the presence of proteins. Specifically, these organic fractions are functionalized with amide-A (3294 cm−1), amide-B (3090 cm−1), amide-I (1650 cm−1), amide-II (1555 cm−1), and amide-III (1410 cm−1) groups (Fig. 4B), further indicating the presence of proteins.40 These observations were in agreement with the study that conducted by Ali et al. 2011 with E. coli DNAzyme sensor, in which they reported on the proteinaceous nature of the target molecule.32 To further verify that the target of E. coli DNAzyme sensors was protein, E. coli CIM from OH11 and OH18 strains were individually treated with enzymes (trypsin and proteinase K) prior to mixing with liquid sensor. Negligible fluorescence response was detected after treatment with both enzymes compared to the non-treated E. coli CIM control (Fig. 5). Furthermore, the estimated molecular weight of the target proteins in OH 11 and OH 18 CIM is >30 kDa (Fig. 5), typical of the protein molecular weight.3.5. Identities of the protein targets
A total of 44 and 61 different proteins were identified by LC-MS/MS to be present in E. coli OH11 and OH18 CIM, respectively. Tables S2 and S3† list the ubiquitous proteins found in both OH11 and OH18 with their corresponding peptide sequences, suggesting that these are the possible proteins candidate that consistently bind onto the DNAzyme biosensor. These target proteins can be classified into two categories. First, proteins that interact with nucleic acids (e.g. P0A8T7, P46889, P06612, P0AES4, P0A7Z4, P36659, and P46850). DNAzyme probe is a single-stranded DNA that binds to protein/proteins molecule for activation. Hence the target protein is inferred to mimic the feature of interacting with DNA molecule.33 Second, protein associated with flagellum functions (P04949). High score value of these proteins was observed when analyzing the LC-MS/MS data with DNAzyme/OH11 and DNAzyme/OH18 complex, respectively (Tables S2 and S3†), suggesting their high predominance in the sample.A further analysis was made on those pulled down protein sequences of relatively longer length by matching for their identities against protein databases (Table S4†). Although most of the pulled-down protein sequence identified by LC-MS/MS showed high similarity with that present in E. coli (e.g. P04949, P0AE4S4, P0A6Y8, P36659, P75937, P33235), two proteins (e.g. P0A8T7, P06612, P0A7Z4) demonstrated a reasonably high similarity percentage to other non-targets like Klebsiella spp., Enterobacter spp., Citrobacter spp. and Pseudomonas spp. (Table S4†), suggesting that a small portion of the proteins that bound on to the DNAzyme biosensor may originate from non-targets like Klebsiella spp., Enterobacter spp., Citrobacter spp. and Pseudomonas spp. (Table S4†).
Since fluorescence slope increment arising from interaction between DNAzyme biosensor and Klebsiella, Enterobacter, Citrobacter and Pseudomonas spp. were lower than that observed in E. coli (Fig. 1), these results suggest a likewise poor binding efficiency between these non-target proteins with the DNAzyme biosensor. However, this non-specific binding might create an interference effect that would not allow a conclusive identification of E. coli based on fluorescence slope increment when the target and non-target bacteria are present together in a mixed culture form. Since the non-targets would be reversibly bound to the biosensor on an intermittent basis to generate a slope value, it might be difficult to confidently denote which slope values would benchmark to a definitive presence of E. coli and not to the presence of non-targets in a mixed culture. Hence, the DNAzyme biosensor in its current form would only suffice as a verification tool to differentiate discrete colonies, and would require further development to ensure 100% specificity to only E. coli if it is to be developed into a tool for mixed culture identification.
4. Conclusions
In this study, we adapted a DNAzyme-based biosensor that was able to rapid identify discrete E. coli colonies and differentiate them against non-E. coli colonies reported to cause false positives in other tests using simultaneous enzyme-based agar media. The DNAzyme biosensor demonstrates a high detection sensitivity and requires 2 h of cultivation from a single colony (of about 1 mm in diameter) to obtain sufficient cell numbers that generate a robust fluorescence signal. When demonstrated on different types of commercially available simultaneous enzyme-based agar, the DNAzyme-based biosensor was not subjected to any interference effect arising from colorants, media composition or water matrices. Our findings suggest the E. coli DNAzyme biosensor remains competitive to serve as a rapid and specific verification test to be complemented with the use of simultaneous enzyme media. The current regulatory process require laboratories to report suspected E. coli colonies grown on agar plate as positive. We envision the use of this biosensor as a verification step for E. coli first, prior to reporting their data. Specifically, regular sampling to denote for the presence of total coliforms and E. coli using simultaneous enzyme media can still proceed according to the standard protocol. Single colonies observed on the agar can then be isolated for 2 h sub-culturing in SOB medium before lysing for the intracellular matrix and reacting with the biosensor for fluorescence measurement. False positive rates associated with simultaneous enzyme-based agar results in unnecessary false alarms for both utility operators and public, and incur an additional operational burden for utilities. By providing a rapid and specific verification test result, we believe that this DNAzyme biosensor would aid in effective decision-making process to protect general public.Conflicts of interest
Y. L. is a co-founder of ANDalyze, Inc. Zhengfang Cui was an employer of ANDalyze, Inc. ANDalyze, Inc. had no involvement in the experimental design, data interpretation and writing of this manuscript. All other authors declare no conflict of interest.Acknowledgements
The work was supported by KAUST Competitive Research Grant URF/1/3407-01-01 awarded to P.-Y. H. and Y. L. The authors would like to acknowledge Professor Wen-Tso Liu for gifting the E. coli and non-E. coli strains that were used in this study. The authors would also like to acknowledge Dr Ya Zhang's effort in curating and preparing the strains for shipment to KAUST.References
- APHA, Standard methods for the examination of water and wastewater, American Public Health Association, American Water Works Association, Water Environment Foundation, Washington, DC, 22nd edn, 2012, pp. 9–65 Search PubMed.
- K. P. Brenner, C. C. Rankin, Y. R. Roybal, G. N. Stelma Jr., P. V. Scarpino and A. P. Dufour, New medium for the simultaneous detection of total coliforms and Escherichia coli in water, Appl. Environ. Microbiol., 1993, 59(11), 3534–3544 CAS.
- APHA, Standard methods for the examination of water and wastewater, American Public Health Association, American Water Works Association, Water Environment Federation, 22nd edn, 2012, pp. 9–93 Search PubMed.
- Y. Zhang, P.-Y. Hong, M. W. LeChevallier and W.-T. Liu, Phenotypic and Phylogenetic Identification of Coliform Bacteria Obtained Using 12 Coliform Methods Approved by the U.S. Environmental Protection Agency, Appl. Environ. Microbiol., 2015, 81(17), 6012–6023 CrossRef CAS PubMed.
- I. Tryland and L. Fiksdal, Enzyme characteristics of beta-D-galactosidase- and beta-D-glucuronidase-positive bacteria and their interference in rapid methods for detection of waterborne coliforms and Escherichia coli, Appl. Environ. Microbiol., 1998, 64(3), 1018–1023 CAS.
- P. Kampfer, A. Nienhuser, G. Packroff, F. Wernicke, A. Mehling, K. Nixdorf, S. Fiedler, C. Kolauch and M. Esser, Molecular identification of coliform bacteria isolated from drinking water reservoirs with traditional methods and the Colilert-18 system, Int. J. Hyg. Environ. Health, 2008, 211(3–4), 374–384 CrossRef PubMed.
- J. P. Landre, A. A. Gavriel and A. J. Lamb, False-positive coliform reaction mediated by Aeromonas in the Colilert defined substrate technology system, Lett. Appl. Microbiol., 1998, 26(5), 352–354 CrossRef CAS PubMed.
- H. Leclerc, D. A. Mossel, S. C. Edberg and C. B. Struijk, Advances in the bacteriology of the coliform group: their suitability as markers of microbial water safety, Annu. Rev. Microbiol., 2001, 55, 201–234 CrossRef CAS PubMed.
- J. Olstadt, J. J. Schauer, J. Standridge and S. Kluender, A comparison of ten USEPA approved total coliform/E. coli tests, J. Water Health, 2007, 5(2), 267–282 CrossRef.
- C. M. Davies, S. C. Apte and S. M. Peterson, Possible interference of lactose-fermenting marine vibrios in coliform beta-D-galactosidase assays, J. Appl. Bacteriol., 1995, 78(4), 387–393 CrossRef CAS PubMed.
- F. M. Schets, P. J. Nobel, S. Strating, K. A. Mooijman, G. B. Engels and A. Brouwer, EU Drinking Water Directive reference methods for enumeration of total coliforms and Escherichia coli compared with alternative methods, Lett. Appl. Microbiol., 2002, 34(3), 227–231 CrossRef CAS PubMed.
- C. Bernasconi, G. Volponi and L. Bonadonna, Comparison of three different media for the detection of E. coli and coliforms in water, Water Sci. Technol., 2006, 54(3), 141–145 CrossRef CAS PubMed.
- W. L. Chao, Evaluation of Colilert-18 for the detection of coliforms and Escherichia coli in tropical fresh water, Lett. Appl. Microbiol., 2006, 42(2), 115–120 CrossRef CAS PubMed.
- T. C. Covert, L. C. Shadix, E. W. Rice, J. R. Haines and R. W. Freyberg, Evaluation of the Autoanalysis Colilert test for detection and enumeration of total coliforms, Appl. Environ. Microbiol., 1989, 55(10), 2443–2447 CAS.
- A. F. Maheux, V. Huppe, M. Boissinot, F. J. Picard, L. Bissonnette, J. L. Bernier and M. G. Bergeron, Analytical limits of four beta-glucuronidase and beta-galactosidase-based commercial culture methods used to detect Escherichia coli and total coliforms, J. Microbiol. Methods, 2008, 75(3), 506–514 CrossRef CAS PubMed.
- C. R. Fricker, P. S. Warden and B. J. Eldred, Understanding the cause of false negative beta-D-glucuronidase reactions in culture media containing fermentable carbohydrate, Lett. Appl. Microbiol., 2010, 50(6), 547–551 CrossRef CAS PubMed.
- R. Maarit Niemi, J. Mentu, A. Siitonen and S. I. Niemelä, Confirmation of Escherichia coli and its distinction from Klebsiella species by gas and indole formation at 44 and 44.5°C, J. Appl. Microbiol., 2003, 95(6), 1242–1249 CrossRef PubMed.
- S. Frankel, S. Reitman and A. C. Sonnenwirth, Gradwohl's clinical laboratory methods and diagnosis, C.V. Mosby Co., St. Louis., vol. 2, p. 1117 Search PubMed.
- J. Bartram, R. Ballance, W. H. Organization and U. N. E. Programme, Water Quality Monitoring - A Practical Guide to the Design and Implementation of Freshwater Quality Studies and Monitoring Programmes, 1996 Search PubMed.
- C. M. O'Hara, D. L. Rhoden and J. M. Miller, Reevaluation of the API 20E identification system versus conventional biochemicals for identification of members of the family Enterobacteriaceae: a new look at an old product, J. Clin. Microbiol., 1992, 30(1), 123–125 Search PubMed.
- F. Ben Abdallah, K. Chaieb, M. Snoussi, A. Bakhrouf and K. Gaddour, Phenotypic variations and molecular identification of Salmonella enterica serovar Typhimurium cells under starvation in seawater, Curr. Microbiol., 2007, 55(6), 485–491 CrossRef CAS PubMed.
- B. W. Ciebin, M. H. Brodsky, R. Eddington, G. Horsnell, A. Choney, G. Palmateer, A. Ley, R. Joshi and G. Shears, Comparative evaluation of modified m-FC and m-TEC media for membrane filter enumeration of Escherichia coli in water, Appl. Environ. Microbiol., 1995, 61(11), 3940–3942 CAS.
- S. R. Monday, T. S. Whittam and P. C. Feng, Genetic and evolutionary analysis of mutations in the gusA gene that cause the absence of beta-glucuronidase activity in Escherichia coli O157:H7, J. Infect. Dis., 2001, 184(7), 918–921 CrossRef CAS PubMed.
- Himedia Kovac's Indole Reagent, 2018.
- Bactident® Oxidase, Merck Microbiology Manual 12th edn.
- J. Liu, Z. Cao and Y. Lu, Functional Nucleic Acid Sensors, Chem. Rev., 2009, 109(5), 1948–1998 CrossRef CAS PubMed.
- C. E. McGhee, K. Y. Loh and Y. Lu, DNAzyme sensors for detection of metal ions in the environment and imaging them in living cells, Curr. Opin. Biotechnol., 2017, 45, 191–201 CrossRef CAS PubMed.
- S. D. Aguirre, M. M. Ali, B. J. Salena and Y. Li, A sensitive DNA enzyme-based fluorescent assay for bacterial detection, Biomolecules, 2013, 3(3), 563–577 CrossRef PubMed.
- J. Li and Y. Lu, A highly sensitive and selective catalytic DNA biosensor for lead ions, J. Am. Chem. Soc., 2000, 122(42), 10466–10467 CrossRef CAS.
- J. Liu, A. K. Brown, X. Meng, D. M. Cropek, J. D. Istok, D. B. Watson and Y. Lu, A catalytic beacon sensor for uranium with parts-per-trillion sensitivity and millionfold selectivity, Proc. Natl. Acad. Sci. U. S. A., 2007, 104(7), 2056–2061 CrossRef CAS PubMed.
- W. Chiuman and Y. Li, Efficient signaling platforms built from a small catalytic DNA and doubly labeled fluorogenic substrates, Nucleic Acids Res., 2006, 35(2), 401–405 CrossRef PubMed.
- M. M. Ali, S. D. Aguirre, H. Lazim and Y. Li, Fluorogenic DNAzyme probes as bacterial indicators, Angew. Chem., Int. Ed., 2011, 50(16), 3751–3754 CrossRef CAS PubMed.
- Z. Shen, Z. Wu, D. Chang, W. Zhang, K. Tram, C. Lee, P. Kim, B. J. Salena and Y. Li, A catalytic DNA activated by a specific strain of bacterial pathogen, Angew. Chem., Int. Ed., 2016, 55(7), 2431–2434 CrossRef CAS PubMed.
- M. M. Ali, A. Slepenkin, E. Peterson and W. Zhao, A simple DNAzyme-based fluorescent assay for Klebsiella pneumoniae, ChemBioChem, 2019, 20(7), 906–910 CrossRef CAS PubMed.
- M. M. Ali, M. Wolfe, K. Tram, J. Gu, C. D. M. Filipe, Y. Li and J. D. Brennan, A DNAzyme-based colorimetric paper sensor for Helicobacter pylori, Angew. Chem., Int. Ed., 2019, 58(29), 9907–9911 CrossRef CAS PubMed.
- K. Tram, P. Kanda, B. J. Salena, S. Huan and Y. Li, Translating bacterial detection by DNAzymes into a litmus test, Angew. Chem., Int. Ed., 2014, 53(47), 12799–12802 CrossRef CAS PubMed.
- M. M. Ali, C. L. Brown, S. Jahanshahi-Anbuhi, B. Kannan, Y. Li, C. D. M. Filipe and J. D. Brennan, A printed multicomponent paper sensor for bacterial detection, Sci. Rep., 2017, 7(1), 12335 CrossRef PubMed.
- C. Darkoh, C. Chappell, C. Gonzales and P. Okhuysen, A rapid and specific method for the detection of indole in complex biological samples, Appl. Environ. Microbiol., 2015, 81(23), 8093–8097 CrossRef CAS PubMed.
- M. Liu, Q. Zhang, J. D. Brennan and Y. Li, Graphene-DNAzyme-based fluorescent biosensor for Escherichia coli detection, MRS Commun., 2018, 8(3), 687–694 CrossRef CAS.
- L. Dramas, Ceramic ultrafiltration of marine algal solutions: a comprehensive study, PhD, King Abdullah University of Science and Technology, 2014 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9ew00795d |
| This journal is © The Royal Society of Chemistry 2019 |