Open Access Article
Open Access ArticleMonitoring the integrity of reverse osmosis membranes using novel indigenous freshwater viruses and bacteriophages
Luc M.
Hornstra
 *a,
Tania
Rodrigues da Silva
cd,
Bastiaan
Blankert
be,
Leo
Heijnen
a,
Erwin
Beerendonk
a,
Emile R.
Cornelissen
afg and
Gertjan
Medema
ad
*a,
Tania
Rodrigues da Silva
cd,
Bastiaan
Blankert
be,
Leo
Heijnen
a,
Erwin
Beerendonk
a,
Emile R.
Cornelissen
afg and
Gertjan
Medema
ad
aKWR Watercycle Research Institute, Groningenhaven 7, 3433 PE, Nieuwegein, The Netherlands. E-mail: Luc.Hornstra@kwrwater.nl; Tel: +31306069628
bOasen, Nieuwe Gouwe O.Z. 3, 2801 SB, Gouda, The Netherlands
cNova University of Lisbon – Faculty of Sciences and Technology, Monte da Caparica, Portugal
dDelft University of Technology, Stevinweg 1, 2628 CN, Delft, The Netherlands
eKing Abdullah University of Science and Technology (KAUST), Water Desalination and Reuse Center (WDRC), Biological and Environmental Science and Engineering Division (BESE), Thuwal 23955-6900, Saudi Arabia
fSingapore Membrane Technology Centre, Nanyang Environment and Water Research Institute, Nanyang Technological University, Singapore 637141, Singapore
gParticle and Interfacial Technology Group, Ghent University, Coupure Links 653, B-9000 Ghent, Belgium
First published on 22nd July 2019
Abstract
High pressure membranes are increasingly used for the treatment of contaminated water for various purposes including irrigation and drinking water. The lack of a fast and easy to implement membrane integrity test method with a log removal value (LRV) >3 hampers the implementation of these membranes. Current on-line methods include conductivity, TOC (total organic carbon) and turbidity measurements and can monitor a maximum LRV of 3. Furthermore, challenge tests using chemical or bacteriological virus surrogates such as bacteriophage MS2 show that RO and NF systems can reach LRVs of 6–7, but dosing of these surrogates is not feasible and desirable in full scale drinking water plants. This study describes the identification and use of indigenous viruses, naturally present in surface waters to monitor the integrity of RO membranes in a pilot installation. Natural viruses were identified from fresh source water using metagenomics and qPCR primers developed for a selected set of viruses that were present in high numbers in surface water. The qPCR assays were used to determine the number of gene copies of these viruses in the feed and permeate of the pilot RO installation, and the LRV of these natural viruses was compared with the LRV of spiked MS2 and with on-line conductivity. The concentration of the selected natural viruses in the source water was sufficient to demonstrate a LRV of >7 and was comparable to the results of the spiked MS2 bacteriophage. Furthermore, after inflicting damage to the membrane element by drilling small holes of 1 and 4 mm, both MS2 and the natural viruses detected the damage to the membrane with a nearly identical decrease of LRV, while conductivity lacked sensitivity to monitor any integrity loss. This novel method enables monitoring of the RO membrane integrity at a high sensitivity (LRV > 7), without the addition of chemical or biological virus surrogates. Furthermore, the high concentration of viruses in source water simplifies detection without laborious sample concentration procedures. The implementation of this method facilitates monitoring of the integrity of RO membranes in full scale operation with a much higher sensitivity than current methods.
Water impactThis study describes the identification and use of new indigenous fresh water viruses, for monitoring RO membrane integrity. The performance of these viruses is equal to that of “gold standard” MS2, for intact and compromised RO membranes, and they allow integrity verification with high sensitivity of LRV >7. These results aid broader implementation of high pressure membranes for water treatment. |
1. Introduction
High pressure membranes such as nanofiltration (NF) and reverse osmosis (RO) membranes are increasingly applied in the treatment of conventional and unconventional water sources (ground water, surface water, seawater, effluent and wastewater) for the production of irrigation water, process water and drinking water.1 These membranes are capable of removing particles (inorganics, bacteria, viruses) and dissolved compounds (salts, natural organic matter, compounds of emerging concern) very effectively. One of the key features of high pressure membrane filtration is disinfection, the effective removal of viruses and bacteria. Considering micro-organisms, viruses are the most critical because of their small size, typically between 20 and 400 nm.2–4 High pressure membrane systems should be capable of removing viruses completely due to size exclusion, since the pore sizes or molecular cut-off values of these membranes are much smaller than the dimensions of a typical virus. However, this very effective removal makes operational monitoring of membrane integrity critically important because minor imperfections in membrane modules may result in virus passage and a subsequent human health risk. Membrane element systems may fail due to different mechanisms, such as broken O-seals, leaking glue lines or impaired membranes caused by oxidant damage (due to chemical cleaning), back pressure damage from the permeate side, extreme operational conditions or damage caused by abrasive components in the feed water (particles, chemicals).5–9 As a consequence of membrane element integrity problems, passage of even a small number of pathogenic viruses may lead to a health risk to consumers. Unfortunately, a direct routine measurement of pathogenic viruses in source and permeate water is generally not feasible because of the low concentration of pathogenic viruses in source water, which is even lower in treated water. This would therefore require the sampling and concentration of very large (>1000 l) water volumes in order to detect sufficient levels of pathogenic viruses.10,11 Therefore, membrane integrity monitoring requires a different approach, and regulations have been established to monitor the system integrity. For example, the USEPA membrane filtration guidance manual requires membrane system verification by direct and indirect integrity testing.12 Pressure hold and vacuum decay tests are direct integrity verification methods aimed at detecting leaks associated with membrane damage, such as glue line failures or leaks in O-ring seals.6 These direct testing methods require a temporary system shut-down, since these tests are performed off-line.17 Indirect integrity testing methods use typically on-line water quality parameters of the permeate in relation to the feed water. Examples are intrinsic measurements of water parameters like conductivity, total organic carbon (TOC) or turbidity. A major drawback of these methods is that they generally demonstrate a log removal value (LRV) of <3 and are therefore not sensitive enough to detect small breaches in a system that allows a minimal number of viruses to pass.6,13 Instead, challenge tests are developed, using the addition of microbial or non-microbial surrogates to the feed water, which can result in monitoring of a high LRV of intact RO systems.14,15 A good virus surrogate should be similar in size and retention to human pathogenic viruses, detectable with on-line methods, which are inexpensive, do not result in membrane fouling, and are not harmful to humans.16,17 Due to their characteristics, bacteriophages are considered appropriate surrogates, and bacteriophage MS2 is considered the best surrogate and recommended by the USEPA for integrity verification testing of RO membranes.2,18 An overview of current and new membrane verification techniques is presented in Fig. 1, which has been reviewed by Pype et al.4 and Frenkel and Cohen.21 Due to the above mentioned requirements, virus surrogates are frequently used in laboratory and pilot scale installations, but not in full scale utilities, mainly due to costs. Notably, in The Netherlands, addition of surrogates to feed water for the production of drinking water is not permitted by drinking water companies. Therefore there is an urgent need for a membrane integrity testing method that meets the following criteria: (i) able to demonstrate significant log removal values of at least LRV 4,13 (ii) the characteristics (particularly particle size) of the surrogate/indicator virus should be comparable to those of human enteric viruses, (iii) the method must not be expensive or time-consuming and (iv) applicable in full scale (drinking) water treatment plants, indicating that the marker should be indigenously present in the source water.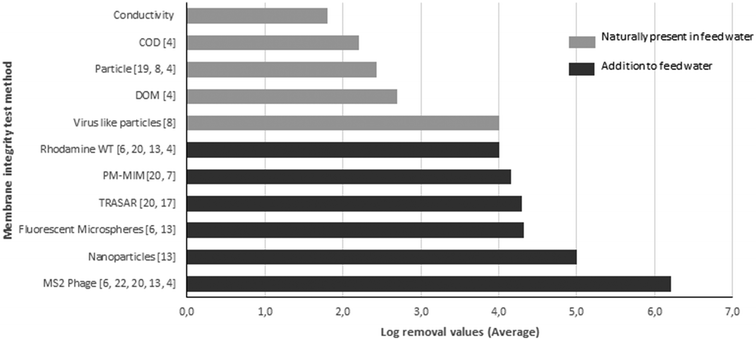 | ||
| Fig. 1 Methods reported in the literature for testing the membrane integrity, the LRVs that can be determined by the described method, and the references for these methods (number between brackets). | ||
The introduction of rapid metagenomics techniques enables the exploration of natural virus populations in surface water samples by identifying the genomes of viruses and bacteriophages present in marine and fresh waters.23–26 Fresh surface water contains an enormous variation of naturally present virus species, and many of these are expected to be present in very high numbers, but information about the population composition of viruses in freshwater is scarce.27 The majority of these viruses have not been identified before, which means that their genomic information is currently not present in the existing genome sequence databases.28 Since viruses are naturally present in freshwater, this offers an opportunity for developing novel water quality indicators for monitoring virus removal, e.g. for membrane integrity monitoring. Previously, due to their presence in surface water, somatic coliphages and F-specific RNA bacteriophages have been proposed and reported as suitable for surveying the performance of UF membranes with LRVs of approximately 3–4.29 Recently, pepper mild mottle virus (PMMoV), a plant RNA virus, but present in human feces and in relatively high concentrations in many water sources, has been evaluated as an indicator virus for monitoring the virus removal by a water treatment process.30,31 The high concentration of PMMoV in river water of 3.0 × 103–1.1 × 106 gene copies per l is highly beneficial since the collection and concentration of large water volumes are not required.32
This study firstly describes a metagenomics approach to identify new natural viruses (NVs) and bacteriophages present in surface water in high concentrations, with the purpose of using these viruses as indicators for virus removal by water treatment processes, specifically for membrane integrity monitoring. Secondly, after identification by metagenomics, a subset of natural viruses is selected to develop standard quantitative PCR (qPCR) assays to easily quantify the selected viruses in freshwater and after subsequent water treatment processes. The utility of the method is shown by integrity monitoring of RO membranes with a LRV of >7, by comparing novel NVs with spiking tests with bacteriophage MS2 and determining integrity loss caused by small drill holes applied to the membrane sheets. We show that by using NVs, it is possible to verify the RO membrane integrity with very high sensitivity and without the need for addition of any bacterial or chemical virus surrogate to the source water.
2. Materials and methods
2.1 Isolation of viruses from surface water and Illumina next generation sequencing
200 l of surface water was collected from the Lek Canal, at Nieuwegein on May 18, 2015. Concentration of the viruses from 200 l to approximately 0.27 l was achieved by cross-flow ultrafiltration (Hemoflow, Fresenius HF80S polysulfon), with a pore size of 10 nm. This was followed by sequential filtration steps using filters with pore sizes of 0.7 and 0.22 μm to remove algae, bacteria and protozoa while passing the viruses. Further concentration was carried out by ultracentrifugation for 2.5 h at 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm to pelletize the viruses. The viruses were resuspended in 500 μl sterile water. Prior to the isolation of DNA and RNA from the viruses, the virus suspension was treated with 2 U μl−1 DNAse (Invitrogen) at 37 °C for 45 min to remove traces of free DNA which could be present in the water sample.
000 rpm to pelletize the viruses. The viruses were resuspended in 500 μl sterile water. Prior to the isolation of DNA and RNA from the viruses, the virus suspension was treated with 2 U μl−1 DNAse (Invitrogen) at 37 °C for 45 min to remove traces of free DNA which could be present in the water sample.
200 μl of the concentrated virus suspension was used for the isolation of RNA and DNA using a Purelink™ Viral RNA/DNA kit (Invitrogen). To obtain DNA and RNA virus sequences, DNA and RNA sequencing was performed separately using an Illumina HiSeq 2500 system, operated by BaseClear (Leiden, The Netherlands). Data analysis of the raw sequence output was done by BaseClear, and consisted of quality control (based on Illumina Chastity filtering) followed by quality assessment based on the remaining reads using the FASTQC quality control tool version 0.10.0. Subsequently, single sequence reads were assembled to contigs, using the de novo assembly option of the GLC Genomics Workbench version 8.0 program. The contigs were combined to make larger sequences, called scaffolds, using SSpace premium Scaffolder Version 2.3. On the final gap-closed scaffold sequences, a BLAST was performed with NCBI-BLAST (version 2.2.29+) in the NCBI nt database.
2.2 Quantitative polymerase chain reaction (qPCR) assays of the new virus sequences and enumeration of bacteriophage MS2
Based on sequencing coverage, 4 scaffolds were selected from DNA virus sequences which were suspected to be present at high concentration in surface water. These sequences were used to develop qPCR primers for the detection of these viruses in water samples. The primers for the qPCR assays are listed in Table 1.| Forward primer | Reverse primer | Fragment length bp | |
|---|---|---|---|
| NV2247 | AAGCCTGAACGTGTTCCGAT | CTGCCCGCAGGATTGTTAGA | 104 |
| NV2303 | GCCATAATTGGCTTCAGCGG | TCGCGCACTTGGTCAAAAAG | 100 |
| NV2310 | GCATCTTCGTCAATGCGTCC | GAGGTCGTGGTGTGGCTATC | 91 |
| NV2314 | ACCAGGGGCGGTGTATATTG | GACGCCGTTGAAATGTCAGG | 102 |
Samples of 500 ml of surface water for qPCR were taken four times, and the viruses were concentrated by centrifuging at 3000g for 10 min using a Centricon Plus-70 (Merck UFC700018) to a volume of 300 μl. DNA was isolated using a DNA isolation kit (PowerBiofilm TM Qiagen 24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000-50) according to the manufacturer's protocol and eluted in 200 μl elution buffer. Before DNA isolation, an internal standard was added to the sample to determine the DNA isolation efficiency from the water sample. For the qPCR procedure, 5 μl of DNA, 12.5 μl of SYBR-Green mix (Biorad), and forward and reverse primers to a final concentration of 10 μM and 6.5 μl of water were mixed and the following PCR conditions were applied: 3 min, 95 °C; followed by 39 cycles, 10 s, 95 °C; 45 s, 60 °C. After the PCR, a melting curve was examined to verify whether the PCR had resulted in the amplification of a single PCR fragment with the expected melting temperature. Negative controls (sterile ultrapure water) were included for all selected viruses. MS2 was enumerated by culturing using the double agar layer technique as described in ISO 10705-1.
000-50) according to the manufacturer's protocol and eluted in 200 μl elution buffer. Before DNA isolation, an internal standard was added to the sample to determine the DNA isolation efficiency from the water sample. For the qPCR procedure, 5 μl of DNA, 12.5 μl of SYBR-Green mix (Biorad), and forward and reverse primers to a final concentration of 10 μM and 6.5 μl of water were mixed and the following PCR conditions were applied: 3 min, 95 °C; followed by 39 cycles, 10 s, 95 °C; 45 s, 60 °C. After the PCR, a melting curve was examined to verify whether the PCR had resulted in the amplification of a single PCR fragment with the expected melting temperature. Negative controls (sterile ultrapure water) were included for all selected viruses. MS2 was enumerated by culturing using the double agar layer technique as described in ISO 10705-1.
2.3 The reverse osmosis pilot unit
At the Dutch drinking water company Oasen, a RO pilot installation was operated at the drinking water production location Kamerik. The set-up consists of two parallel 8′′ RO membrane elements, and involves a temperature controlled feed water tank with a mixer, kept at 12 °C. The unit was operated in recirculation mode during which the concentrate and permeate are returned to the feed water tank. A cartridge filter (5 μm pore) was installed to retain larger particles from the feed water to prevent feed spacer clogging during the experiment. Control devices such as flow rate meters, pressure meters and conductivity meters placed in the installation provide real-time information about the operation and the status of the installation. The unit contains two parallel pressure vessels (8′′) that each accommodate a single standard spiral wound 8′′ reverse osmosis element (ESPA2 Hydranautics Nitto Group Company) with an active area of 40.9 m2 which can be used independently. The RO unit was operated at a constant permeate flux rate of 25 l m−2 h−1 at a feed flow rate of 6.8 m3 h−1 and a recovery of 15%.2.4 Operation of the unit using challenge tests with MS2 and natural viruses with intact and damaged membranes
Prior to the actual filtration experiment, the feed tank and RO system were flushed with drinking water for 1–2 days at a flow rate of 6.8 m3 h−1 and a recovery of 15%. After this period, the feed tank was filled with surface water from the Grecht Canal located in Kamerik and the experimental run was started. The RO membrane was operated with a feed pressure of 9.5 bar and the permeate conductivity was measured during the experiments. After stabilization of the setup for 30 min, four samples were taken from the feed (after the cartridge filter) and permeate for the NV measurements. Subsequently, MS2 bacteriophages were spiked into the feed water tank at a concentration of 2 × 107 PFU ml−1 and mixed for 15 minutes. Control samples confirmed that the background concentration of F-specific coliphages in the Grecht Canal was insignificant (<20 PFU ml−1) and that MS2 removal by the cartridge filter was negligible (<0.1 LRV). Four samples were taken 30 and 60 minutes after stabilization from the feed and permeate for the MS2 assay. The LRV was calculated according to the following equation:| LRV = log10[Cin/Cout] |
The experiments were conducted with an intact membrane, and the conductivity, the number of MS2 and the number of NVs were determined in the feed and the permeate. Subsequently the membrane was damaged using an electric drill and again these three parameters were determined. After that, the membrane module was replaced by a new module, and the experiment was repeated as above, but with another type of damage. The three categories of inflicted damage included the drilling of a hole with a diameter of 4 mm and 1 mm and four times 1 mm. The holes were drilled with a depth of approximately 2 mm into the external shell of the different membrane elements (Fig. 2). All experiments with intact and damaged membranes were performed in duplicate with new membrane modules, called experiment 1 and experiment 2, and the second experiment was carried out one week after the first experiment. After each run, the system without the membrane element was disinfected with sodium hypochlorite for a minimum of four hours. Blank samples confirmed the complete inactivation of MS2 in the system after disinfection.
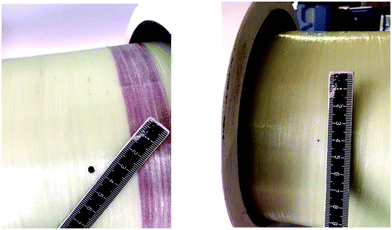 | ||
| Fig. 2 4 mm (left) and 1 mm (right) drilled holes to damage the membrane module and the membrane sheets. | ||
3. Results and discussion
3.1 Identification of potential natural viruses for monitoring of the membrane integrity
After concentration of the viruses from 200 liters of surface water, transmission electron microscopy confirmed the presence of large numbers of viruses, while no bacteria were observed in the samples (Fig. 3).Next generation sequencing of viruses and bacteriophages resulted in 0.6 million RNA sequence reads and 1.2 million DNA sequence reads, leading to 5855 RNA and 7288 DNA scaffolds. BLAST search using these scaffolds revealed 261 similarities (4.5%) for the RNA sequences and 95 similarities (1.3%) for DNA sequences with the BLAST nucleotide database. This low number of similarities suggests that the majority of the sequences obtained are from unknown viruses from fresh surface water and have not been identified before.28
From the DNA scaffolds that showed similarities to the BLAST nucleotide database, four scaffolds were selected primarily based on the sequencing coverage, assuming that a virus that represents a large fraction of the population is reflected by a high coverage in the sequencing library (Table 2). From scaffolds representing these virus primers, four qPCR assays were designed for quantitative detection of these virus genomes in the water samples. The expectation is that the selected viruses are universally present in fresh surface waters, hindering a full metagenomics analysis to define location specific NVs for each water sample of different locations.
| Scaffold nr | Average cov | Homology in BLAST |
|---|---|---|
| 2247 | 33 | Dorcoceras hygrometricum |
| 2303 | 10,9 | Bacteriophage tail assembly protein-like protein [Akkermansia sp. CAG:344] |
| 2310 | 8,5 | Hypothetical protein [uncultured Mediterranean phage] |
| 2314 | 27 | Tail tubular protein B [Phormidium phage Pf-WMP3] |
3.2 Detection of natural viruses in fresh surface water
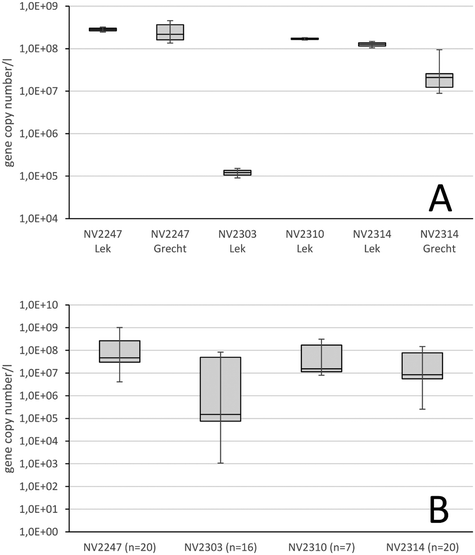
First, the qPCR assays were conducted on water from the Lek Canal which originally was collected for the isolation of the viruses, and to determine whether the four viruses could be detected. The developed qPCR assays quantified the four viruses in the Lek Canal water, of which natural viruses NV2247, NV2310 and NV2314 were present at high concentrations above 1.2 × 108 gene copies per l. Virus NV2303 was present in a lower concentration of 1.1 × 105 gene copies per l (Fig. 4A). Next, it was determined whether the selected viruses were generally present in major surface water locations, used for the production of drinking water. 6 samples from the river Meuse (locations in The Netherlands), 6 samples from the river Rhine (locations in Germany and The Netherlands), 2 samples from the river Schelde (location in Belgium) and 6 samples from the Ijssel lake were analyzed for the number of natural viruses (Fig. 4). The four viruses were detected in all surface water samples. Viruses NV2247 and NV2310 were on average present in the highest concentration of 1.8 × 108 gene copies per l and 1.0 × 108 gene copies per l, respectively. NV2303 and NV2314, with a concentration of 2.0 × 107 gene copies per l and 3.8 × 107 gene copies per l, respectively, were still present in very high concentrations, although lower than the other two. For comparison, the concentration of PMMoV in fresh surface water has been reported to vary between of 3.0 × 103 and 2.9 × 106 gene copies per l approximately.32–34 An expected variation in the number of viruses exists between locations, and in time. This is reflected by the minimum and maximum values that were observed after examining the concentration of the four viruses in various surface waters. The concentration of NV2310 showed, with a log factor of 1.6 between the minimal and maximal gene copy numbers per l, the smallest variation within the various locations, while NV2303 showed a larger variation of 4.9 log, and NV2247 and NV2314 were in between with a log value of 2.4 and 2.8, respectively. Virus removal by water treatment processes is assessed by comparing the number of viruses in water before treatment with the number of viruses in the treated water. Variation of natural virus numbers in source water therefore does not influence the determination of the removal of viruses by water treatments, but it is beneficial when a virus is consistently present in surface water without considerable temporal and spatial variation.3.3 LRV of intact RO membranes using conductivity, model virus MS2 and naturally present viruses
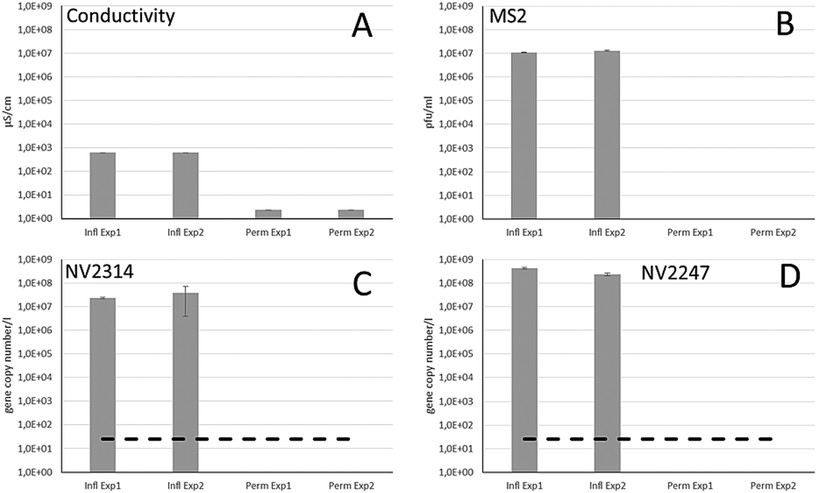
Different integrity testing methods including on-line conductivity measurements, spiked bacteriophage MS2 and the indigenously present natural viruses were compared in a RO pilot system, using feed water from the Grecht Canal. First, it was evaluated whether NVs were present in the source water of the Grecht Canal (Fig. 4A). Because of high abundance, NV2247 and NV2314 were selected for evaluation of the RO membrane system, and these NVs were present in high concentration. The integrity of intact RO membranes was determined in two successive experimental runs, by monitoring the conductivity and the number of bacteriophage MS2, NV2247 and NV2314 in the feed and the permeate stream (Fig. 5). The conductivity in the source water was 620 μS cm−1, and in the permeate 2.3 μS cm−1, which is the background value of the conductivity measurement, resulting in an average LRV of 2.43 in this system. To determine the removal of MS2 bacteriophage using the system, MS2 was added to the feed water in a concentration of approx. 1.0 × 107 pfu ml−1 in the feed, and after passage of the RO element, MS2 was not detected in the permeate, herewith demonstrating that RO membranes are capable of completely blocking viruses. With MS2 bacteriophages, a LRV of >7 log was demonstrated. Before dosing of MS2, the feed and permeate were sampled to determine the concentration of NV2314 and NV2247 by qPCR. The concentration of NV2314 in the RO feed water was 2.3 × 107 and 3.9 × 107 gene copies per l for experiments 1 and 2, respectively. qPCR analysis of the RO permeate showed that NV2314 was not present in the permeate in both experiments (Fig. 5) above the detection limit of 25 gene copies per l. This shows that NV2314 is, similar to MS2, fully retained by the RO membrane and can be monitored with this virus with a LRV of more than 6. The concentration of NV2247 in the feed was 4.2 × 108 and 2.4 × 108 gene copies per l for experiments 1 and 2, respectively, which was present in even higher numbers in surface water than NV2314. Also this virus was not detected in the permeate, and therefore retained by the membrane, and because of the very high concentration in the source water, a LRV of 7.1 could be demonstrated.3.4 The use of MS2 and natural viruses NV2247 and NV2314 to monitor membrane damage
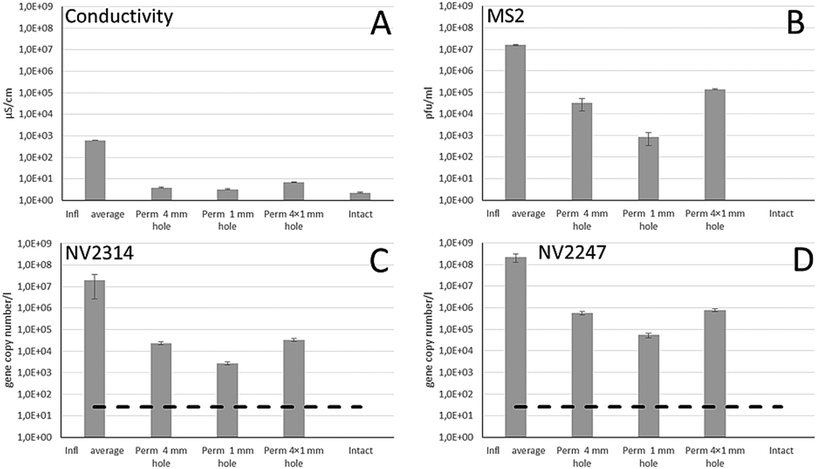
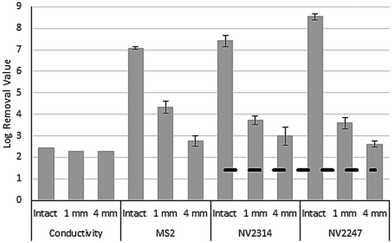
The membrane integrity of the damaged membrane inflicted by drill holes to membrane modules was assessed by measuring the conductivity and concentrations of MS2, NV2314 and NV2247 in the feed and the permeate (Fig. 6). Due to operational limitations, one experiment was carried out per week for each inflicted damage. For the first week, the effect of a single 4 mm drill hole was determined, followed by 1 mm and 4 × 1 mm drill holes. Within this time period the concentration of NV2314 and NV2247 showed minor variation in the feed water (Fig. 4A).Fig. 6 shows the result of the membrane integrity testing results. The damage inflicted to the membrane by the drill holes has only a minor effect on the measured conductivity. The conductivity in the feed water during the experiments was on average 640 μS cm−1, and in the permeate 3.9, 3.3 and 6.9 μS cm−1 for the 4 mm, the 1 mm and the 4 × 1 mm drill holes, respectively, The impairments result in a minimal increase of the conductivity in the permeate compared to the conductivity measurement of the intact system. The MS2 dosing experiments, however, show that this 1 mm drill hole results in loss of virus integrity of 2.8 LRV (Fig. 6B). Therefore, it was concluded that although conductivity can be measured on-line, it is not sensitive enough to determine a small but significant breach of a membrane module. The inflicted damage had a clear effect on both MS2 and NV2314 and NV2247 removal, and these effects were more severe for the 4 mm and 4 × 1 mm holes, where obviously more viruses can pass the membrane. The LRVs are significantly reduced by the presence of the inflicted damage to the membrane modules (Fig. 7). Notably, both MS2 and NV2314 and NV2247 are capable of monitoring this failure in membrane integrity, and show a similar reduction in LRVs caused by the inflicted damage on the membranes. Hence, these natural viruses have the capacity to monitor the integrity of the RO membranes, similar to the spiked MS2 marker. The fact that NV2314 and NV2247 are indigenous viruses and therefore both naturally present in the source water enables monitoring of the membrane integrity in a full scale RO installation with a LRV of >7 log units without the addition of chemical or biological surrogates, a prerequisite of many drinking water companies. The maximum LRV of natural viruses is determined by its concentration in the source water. The fact that both viruses but specifically NV2247 are present in very high numbers can facilitate the development of fast and easy membrane monitoring, because laborious pretreatment (concentration) of the sample can be avoided to a large extent due to the high concentrations. Furthermore, both viruses are DNA viruses, in contrast to PMMoV which is a RNA virus and therefore requires an additional cDNA synthesis step before it can be used as target for the qPCR. For DNA viruses, this step is not required, and the DNA is directly available for qPCR after the DNA isolation.
3.5 Natural viruses: Promising process indicators to verify membrane integrity
The current methods for monitoring the membrane integrity have limitations, predominantly due to the lack of sensitivity to monitor virus passage caused by minor but significant imperfections of the membrane system. Current methods that use intrinsic parameters of the source water, like conductivity, TOC or turbidity, cannot determine a LRV of >3, while intact RO systems can achieve complete virus removal. Application of RO or UF for water reuse, treating impaired water to produce safe drinking water or water for food crop irrigation, would benefit from integrity methods to securely maintain a LRV of >6, particularly when membrane filtration is the only disinfection step in the water treatment process. The use of specific natural indigenous viruses as process indicators to determine RO membrane integrity, as demonstrated in this work, has two major advantages for routinely determining virus removal log credits in RO units. 1: The presence of high numbers of NVs in source water enables the measurement of the viruses before and after RO in sufficiently high numbers to be able to determine a LRV of >6, and achieve this without extensive concentration of the permeate samples. 2: No addition of chemical or biological virus surrogates to the source water is required. This is a major advantage, as addition of any virus surrogate or tracer material to a full scale water treatment RO unit for drinking water production is not permitted in The Netherlands. Moreover, the addition of surrogates or tracers is costly, can potentially result in membrane fouling, and is required to be harmless for humans, which limits possible solutions. Although the proposed method requires grab sampling, implementation of NVs allows frequent and inexpensive monitoring of full scale RO installations on a routine basis within the required LRV range of 6 or more. Obviously, the metagenomics approach as demonstrated in this study can be used to identify new viruses different from the ones identified in this study, to extend the set of viruses for monitoring virus removal by water treatment processes.Natural viruses in freshwater are a greatly unexplored area, and are referred to as viral dark matter due to the fact that most of the viral sequences identified by metagenomics approaches do not align to any viral reference sequence.28 This is what we encountered in this study; only a very small percentage of the scaffolds showed similarity to genome fragments in the NCBI nucleotide libraries. Indigenous viruses are a previously unexplored source of new indicator organisms for integrity monitoring of water treatment processes. Here, based on their natural presence in source water in high concentrations, NV2314 and NV2247 were selected for verification of RO membrane integrity. These or similar viruses could serve to determine the LRV for other (physical) water treatment processes, such as UF, but also conventional treatment processes such as (slow) sand filtration, dune infiltration and riverbank filtration.
NV2314 shows homology in the nucleotide database with tail tubular protein B from Phormidium phage Pf-WMP3, infecting cyanobacterium Phormidium foveolarum.35Phormidium phages belong to Podoviridae, a group of dsDNA, tailed phages that are characterized by having a very short non-contractile tail. NV2247 shows homology with Dorcoceras hygrometricum, a plant species, and might therefore be a plant infecting virus. Homology with plant genomes can be explained by the presence of viral sequences in plant genomes. The BLAST homology search in this study has been performed with a section of the virus genome, and the number of virus sequences in the BLAST nucleotide database is at this moment very limited. Therefore, future BLAST comparisons, when more virus sequences are expected to be present in the database, might be more precise, with results differing from our comparison.
The described metagenomics approach allows the detection and identification of many more indigenous viruses for the verification of water treatment processes. Currently, the knowledge about natural viruses is extremely limited and we lack structural information such as size and hydrophobicity and ecological information like host organism. It is expected that the amount of information about natural viruses will rapidly increase in the coming years, and the characteristics of natural viruses will be gradually revealed. When structural information can be derived, it will be possible to use a specific set of natural viruses for different purposes, e.g. a set of natural viruses with different sizes for membrane characterization based on size exclusion or virus charge. Also, natural viruses with characteristics that resemble the human pathogenic viruses are obviously good candidates as indicators for the removal of human pathogenic viruses. Furthermore, ecological information is beneficial to understanding the background of the viruses in natural water. This helps to understand whether the natural viruses are expected to be present in all natural water systems or geographically limited to specific areas or temporally restrained to various seasons. However, the current lack of this knowledge is not an obstacle to the implementation of natural viruses for integrity monitoring of RO membranes specifically, or water purification processes in general, as has been shown in this study.
4. Conclusions
It is possible to identify indigenous viruses from water sources by using a metagenomics approach, with the aim of using these viruses for the verification of virus removal by water treatment processes.Indigenous freshwater viruses are capable of monitoring intact RO membrane integrity by showing a LRV of more than 7 log units, due to their presence in high concentrations in the source water.
The use of specific indigenous freshwater viruses enables sensitive detection of small inflicted damage to a standard membrane module, with similar results to “gold standard” bacteriophage MS2 (that has to be dosed and is therefore only usable in lab/pilot) and in contrast to conductivity that lacks sensitivity (LRV <3) for detection of small breaches.
The use of these viruses for membrane integrity monitoring does not require the addition of chemical or biological tracers or surrogates, which greatly reduces costs and fouling of the membranes and allows monitoring of full scale treatment plants.
Natural viruses make routine monitoring of virus removal in full scale RO operations possible, without extensive additional concentration procedures.
The use of natural viruses is not limited to RO membranes, but can be implemented in many water treatment procedures where the verification of virus removal is of crucial importance.
Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
This study was financed by the Dutch water supply companies as part of the joint research program (BTO). Anita van der Veen is greatly acknowledged for her technical assistance. Cheryl Bertelkamp (KWR) is greatly acknowledged for her assistance in making the literature overview of membrane integrity methods.References
- C. Y. Tang, Z. Yang, H. Guo, J. J. Wen, L. D. Nghiem and E. Cornelissen, Potable Water Reuse through Advanced Membrane Technology, Environ. Sci. Technol., 2018, 52(18), 10215–10223 CrossRef CAS PubMed.
- A. Antony, J. Blackbeard and G. Leslie, Removal Efficiency and Integrity Monitoring Techniques for Virus Removal by Membrane Processes, Crit. Rev. Environ. Sci. Technol., 2012, 42(9), 891–933 CrossRef.
- M. L. Pype, B. C. Donose, L. Martí, D. Patureau, N. Wery and W. Gernjak, Virus removal and integrity in aged RO membranes, Water Res., 2015, 90, 167–175 CrossRef PubMed.
- M. L. Pype, G. M. Lawrence, J. Keller and W. Gernjak, Reverse osmosis integrity monitoring in water reuse: The challenge to verify virus removal - A review, Water Res., 2016, 98, 384–395 CrossRef CAS PubMed.
- M. Kitis, J. C. Lozier, J. H. Kim, B. Mi and B. Mariñas, Microbial removal and integrity monitoring of RO and NF membranes, J. - Am. Water Works Assoc., 2003, 95, 105–119 CrossRef CAS.
- J. Lozier, M. Kitis, C. Colvin, J. H. Kim, B. Mi and B. Mariñas, Microbial Removal and Integrity of High-Pressure Membranes, IWA Publishing, London, UK, 2003, p. 220 Search PubMed.
- S. Surawanvijit, J. Thompson, A. Rahardianto, V. Frenkel and Y. Cohen, Pulsed marker method for real-time detection of reverse osmosis membrane integrity loss, Desalination, 2015, 370, 25–32 CrossRef CAS.
- X. Huang, J. H. Min, W. Lu, K. Jaktar, C. Yu and S. C. Jiang, Evaluation of methods for reverse osmosis membrane integrity monitoring for wastewater reuse, Journal of Water Process Engineering, 2015, 7, 161–168 CrossRef.
- E. R. Ostarcevic, J. Jacangelo, S. R. Gray and M. J. Cran, Current and emerging techniques for high-pressure membrane integrity testing, Membranes, 2018, 8, 60 CrossRef PubMed.
- N. Albinana-Gimenez, M. P. Miagostovich, B. Calgua, J. M. Huguet, L. Matia and R. Girones, Analysis of adenoviruses and polyomaviruses quantified by qPCR as indicators of water quality in source and drinking-water treatment plants, Water Res., 2009, 43, 2011–2019 CrossRef CAS PubMed.
- E. Rames, A. Roiko, H. Stratton and J. Macdonald, Technical aspects of using human adenovirus as a viral water quality indicator, Water Res., 2016, 96, 308–326 CrossRef CAS PubMed.
- USEPA, Membrane filtration guidance manual EPA 815-R-06-009, Office of Water, Washington, DC, 2005 Search PubMed.
- J. G. Jacangelo and S. Gray, in Assessment of Selected Methodologies for Monitoring the Integrity of Reverse Osmosis Membranes for Water Recycling, ed. WateReuse, The WateReuse Research Foundation, 2015, https://watereuse.org/watereuse-webcast/assessment-of-selected-methodologies-for-monitoring-the-integrity-of-reverse-osmosis-membranes-for-water-recycling/ Search PubMed.
- M. Kumar, S. Adham and J. DeCarolis, Reverse osmosis integrity monitoring, Desalination, 2007, 214(1–3), 138–149 CrossRef CAS.
- USEPA, Guidelines for Water Reuse, Epa/625/R-04/108, 2012 (September), pp. p1–28 Search PubMed.
- H. Guo, Y. Wyart, J. Perot, F. Nauleau and P. Moulin, Low-pressure membrane integrity tests for drinking water treatment: A review, Water Res., 2010, 44(1), 41–57 CrossRef CAS PubMed.
- M. Portillo, Monitoring Reverse Osmosis Membrane Integrity for Direct Potable Reuse Applications, in WateReuse Conference, WateReuse, Texas, USA, 2015 Search PubMed.
- M. Blumenstein, B. Bartley and J. Q. Adams, Environmental Technology Verification Report; Removal of Microbial Contaminants in Drinking Water Dow Chemical Company - Water Solutions SFD-2880 Ultrafiltration Module, U.S. Environmental Protection Agency, Washington, DC, EPA/600/R-11/004, 2011 Search PubMed.
- S. Adham, P. Gagliardo, D. Smith, D. Ross, K. Gramith and R. Trussell, Monitoring the integrity of reverse osmosis membranes, Desalination, 1998, 119(1–3), 143–150 CrossRef CAS.
- Australian Guidelines, Australian guidelines for water recycling: Managing health and environmental risks (Phase 2) Augmentation of drinking water supplies, National Water Quality Management Strategy, 2008 Search PubMed.
- V. Frenkel and Y. Cohen, New Techniques for Real-Time Monitoring of Reverse Osmosis (RO) Membrane Integrity, WateReuse Research Foundation, 2014 Search PubMed.
- B. Mi, C. L. Eaton, J. H. Kim, C. K. Colvin, J. C. Lozier and B. J. Marinas, Removal of biological and non-biological viral surrogates by spiral-wound reverse osmosis membrane elements with intact and compromised integrity, Water Res., 2004, 38(18), 3821–3832 CrossRef CAS PubMed.
- M. Breitbart and F. Rohwer, Here a virus, there a virus, everywhere the same virus?, Trends Microbiol., 2005, 13(6), 278–284 CrossRef CAS PubMed.
- S. J. Williamson, D. B. Rusch, S. Yooseph, A. L. Halpern, K. B. Heidelberg, J. I. Glass, C. Andrews-Pfannkoch, D. Fadrosh, C. S. Miller, G. Sutton, M. Frazier and J. C. Venter, The Sorcerer II Global Ocean Sampling Expedition: metagenomic characterization of viruses within aquatic microbial samples, PLoS One, 2008, 3(1), e1456 CrossRef PubMed.
- K. Rosario, C. Nilsson, Y. W. Lim, Y. Ruan and M. Breitbart, Metagenomic analysis of viruses in reclaimed water, Environ. Microbiol., 2009, 11(11), 2806–2820 CrossRef CAS PubMed.
- I. Hewson, J. G. Barbosa, J. M. Brown, R. P. Donelan, J. B. Eaglesham, E. M. Eggleston and B. A. LaBarre, Temporal dynamics and decay of putatively allochthonous and autochthonous viral genotypes in contrasting freshwater lakes, Appl. Environ. Microbiol., 2012, 78(18), 6583–6591 CrossRef CAS PubMed.
- M. Mohiuddin and H. E. Schellhorn, Spatial and temporal dynamics of virus occurrence in two freshwater lakes captured through metagenomic analysis, Front. Microbiol., 2015, 6, 960 Search PubMed.
- S. R. Krishnamurthy and D. Wang, Origins and challenges of viral dark matter, Virus Res., 2017, 239, 136–142 CrossRef CAS PubMed.
- O. A. Ferrer, S. Casas, C. Galvan, F. Lucena, A. Bosch, B. Galofre, J. Mesa, J. Jofre and X. Bernat, Direct ultrafiltration performance and membrane integrity monitoring by microbiological analysis, Water Res., 2015, 83, 121–131 CrossRef CAS PubMed.
- T. Zhang, M. Breitbart, W. H. Lee, J. Run, C. L. Wei, S. W. L. Soh, M. L. Hibberd, E. T. Liu, F. Rohwer and Y. Ruan, RNA Viral Community in Human Feces: Prevalence of Plant Pathogenic Viruses, PLoS Biol., 2005, 4(1), e3 CrossRef PubMed.
- N. Shirasaki, T. Matsushita, Y. Matsui and R. Yamashita, Evaluation of the suitability of a plant virus, pepper mild mottle virus, as a surrogate of human enteric viruses for assessment of the efficacy of coagulation-rapid sand filtration to remove those viruses, Water Res., 2018, 129, 460–469 CrossRef CAS PubMed.
- I. A. Hamza, L. Jurzik, K. Uberla and M. Wilhelm, Evaluation of pepper mild mottle virus, human picobirna virus and Torque teno virus as indicators of fecal contamination in river water, Water Res., 2011, 45, 1358–1368 CrossRef CAS PubMed.
- E. Eiji Haramoto, M. Kitajima, N. Kishida, Y. Konno, H. Katayama, M. Asami and M. Akibac, Occurrence of Pepper Mild Mottle Virus in Drinking Water Sources in Japan, Appl. Environ. Microbiol., 2013, 79(23), 7413–7418 CrossRef PubMed.
- M. Kitajima, H. P. Sassi and J. R. Torrey, Pepper mild mottle virus as a water quality indicator, npj Clean Water, 2018, 1, 19, DOI:10.1038/s415.
- X. Liu, S. Kong, M. Shi, L. Fu, Y. Gao and C. An, Genomic Analysis of Freshwater Cyanophage Pf-WMP3 Infecting Cyanobacterium Phormidium foveolarum: The Conserved Elements for a Phage, Microb. Ecol., 2008, 56, 671 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2019 |