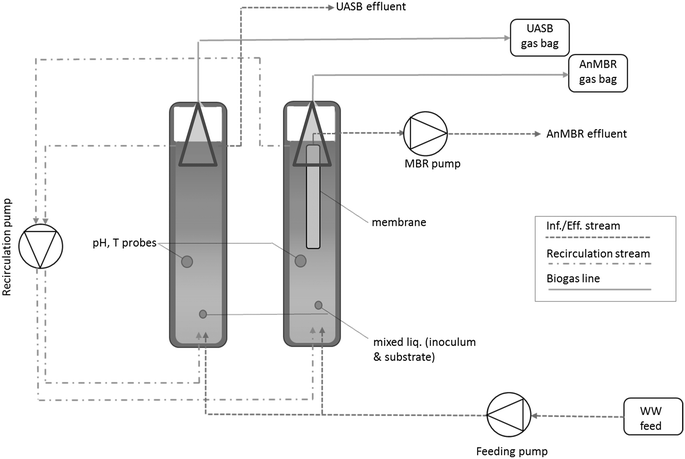
High rate domestic wastewater treatment at 15 °C using anaerobic reactors inoculated with cold-adapted sediments/soils – shaping robust methanogenic communities†
Evangelos
Petropoulos
 *a,
Yongjie
Yu
b,
Shamas
Tabraiz
a,
Aminu
Yakubu
c,
Thomas P.
Curtis
a and
Jan
Dolfing
*a,
Yongjie
Yu
b,
Shamas
Tabraiz
a,
Aminu
Yakubu
c,
Thomas P.
Curtis
a and
Jan
Dolfing
 a
a
aSchool of Engineering, Newcastle University, Newcastle NE17RU, England, UK. E-mail: vagpetrop@gmail.com
bState Key Laboratory of Soil and Sustainable Agriculture, Institute of Soil Science, Chinese Academy of Sciences, Nanjing, 210008, P.R. China
cFaculty of Engineering and the Environment, University of Southampton, UK
First published on 31st October 2018
Abstract
To choose the reactor format in which to employ a low temperature adapted seed for wastewater treatment, we compared a UASB and an AnMBRUASB (UF) reactor at low HRT and temperature (15 °C). The reactors were inoculated with biomass from reactors seeded with soils and sediments from low temperature environments, and fed real municipal wastewater. Both systems met the UWWT Directive (91/271/EEC) COD effluent standard (<125.0 mg L−1) with the AnMBR COD removal efficiency being slightly higher (86.2 ± 1.5%) than that of the UASB (79.3 ± 2.0%). Methane production rates were also higher for the AnMBR than for the UASB, resulting in a COD![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CH4 of 26.1 ± 3.0 and 18.2 ± 2.1%, respectively. Higher methanogenic cell abundance was observed in the AnMBR (3בUASB’). The low conversion of COD to methane was attributed to (i) the presence of SO4 in the influent (120.4 ± 17.4 mg L−1), which scavenged up to 50% of the COD, and (ii) accumulation of un-hydrolysed lipids in the mixed liquor especially in the case of the AnMBR. The UASB was energy positive (0.041 ± 0.013 kW h m−3) whilst the AnMBR was energy negative (−0.221 ± 0.016 kW h m−3). The reactor design appeared to have a profound effect on the numbers and diversity of the methanogens: the hydrogenotrophic activity being favoured in the UASB. But both reactors had comparatively high cell specific rates of methanogenesis. We concluded that the slightly better performance of the AnMBR was not sufficient to offset its higher running cost and the complexity of its design.
CH4 of 26.1 ± 3.0 and 18.2 ± 2.1%, respectively. Higher methanogenic cell abundance was observed in the AnMBR (3בUASB’). The low conversion of COD to methane was attributed to (i) the presence of SO4 in the influent (120.4 ± 17.4 mg L−1), which scavenged up to 50% of the COD, and (ii) accumulation of un-hydrolysed lipids in the mixed liquor especially in the case of the AnMBR. The UASB was energy positive (0.041 ± 0.013 kW h m−3) whilst the AnMBR was energy negative (−0.221 ± 0.016 kW h m−3). The reactor design appeared to have a profound effect on the numbers and diversity of the methanogens: the hydrogenotrophic activity being favoured in the UASB. But both reactors had comparatively high cell specific rates of methanogenesis. We concluded that the slightly better performance of the AnMBR was not sufficient to offset its higher running cost and the complexity of its design.
Water impactWastewater is an energy resource and its recovery using traditional means (anaerobic digestion) is challenging at ambient temperatures (<15 °C). Microbial communities from soils/sediments may realize carbon-positive wastewater treatment. This study investigates how such communities evolve in two mainstream reactor setups (UASB and MBR), fed with actual wastewater. The UASB showed aptitude to further ‘screen’ the community, developing hydrogenotrophic methanogens able to perform at higher rates than the methanogens that developed in the MBR. |
1. Introduction
Wastewater (WW) is a resource. The conversion of polluted effluents to energy (as CH4)1 or other intermediates via anaerobic treatment systems is a way to ‘harvest’ pollution.2,3At present, domestic wastewater is mainly treated aerobically using technologies that not only increase energy usage (0.21 kW h m−3 in Northumbrian Water Ltd, UK) but also tend to increase the greenhouse gas (GHG) footprint (1% of the total annual UK GHG).4 This problem could be tackled via the use of anaerobic treatment systems,5 which have a reduced carbon footprint and allow the recovery of the chemical energy6 that is contained in domestic wastewater (2.1 kW h m−3).7 Temperature is the “Achilles heel” of these biological systems: operation at temperatures <15 °C often limits both methanogenesis and hydrolysis.8
The upflow anaerobic sludge bed (UASB) reactor was a breakthrough in the early 80s and is often used to treat wastewater from various sources.9 This reactor format (and the more recent version EGSB (expanded granular sludge bed)) has often been tried out at low temperatures (4–20 °C).10,15 However, the effluent chemical oxygen demand (COD) could not always meet the E.C. (European Commission) 1991 (UWWTD 91/271/EEC) standard (≤125.0 mg L−1). More recently, the introduction of membrane systems and their application in the wastewater treatment field has led to the development of AnMBRs (anaerobic membrane bio-reactors) for anaerobic treatment at low temperature, with impressive results.16–22 However, this type of reactor incurs increased energy and running costs due to the pumping and cleansing requirements. This energy demand usually varies between 0.3–0.6 kW h m−3.2,16,23–25
Numerous studies have shown that cold adapted microbial communities from soils and sediments can potentially be used in engineering, including low temperature wastewater treatment.26–28 Our most recent work in this area has established the use of psychrophilic seeds under relatively unrealistic conditions: batch reactors fed with sterilised domestic wastewater.28
In this study, we sought to use communities adapted to low temperatures to explore whether moderately low temperature (15 °C), carbon neutral/positive, high rate anaerobic treatment of domestic wastewater is feasible using more realistic reactor formats: a conventional UASB and a ‘modern’ AnMBR. We also wanted to investigate the effect of the reactor setup on the microbial community: whether adaptation is the course that shapes a microbial community, and how shaping occurs in these two different reactor systems. Additionally, many studies in this field use artificial wastewater – typically a mixture of volatile fatty acids (VFAs), long chain fatty acids (LCFAs), starch, and glucose12 – ignoring the true complexity of wastewater, as previously pointed out by Angenent et al. and by Hahn and Figueroa.29,30 In this study, actual, non-sterile primary settled municipal wastewater was used to evaluate the treatment capacity of this low temperature specialized biomass. Thus the adapted biomass would be challenged by autochthonous bacteria in the wastewater in a reactor that retains all biomass (AnMBR) and in a reactor in which some bacteria are washed out (UASB).
We thus sought to not only evaluate the advantages and disadvantages of the UASB and AnMBR approaches for the anaerobic treatment of domestic wastewater at low temperature, but to also pay special attention to the nature and quality of the biomass relative to the reactor design and hydraulics.
2. Materials and methods
Reactor setup
Two 1 L UASB reactors (Fig. 1) (height![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) diameter ratio: 1
diameter ratio: 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6; height: 600 mm; upflow velocity: 0.6 m h−1) were seeded (17.6 ± 4.0 gTSS L1 and 2.32 ± 0.37 gVSS L−1 mixed liquor; the low VSS
6; height: 600 mm; upflow velocity: 0.6 m h−1) were seeded (17.6 ± 4.0 gTSS L1 and 2.32 ± 0.37 gVSS L−1 mixed liquor; the low VSS![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) TSS reflects the origin of the biomass (soils/sediments rich in silt and gravel)) with an inoculum that had been acclimated (see below) to treat UV-sterilized ‘authentic’ primary settled domestic wastewater at 15 °C under methanogenic conditions. The reactor height was 600 mm with two sampling ports, one at the bottom of the reactor and one at the top (at 0 + 10 cm and at 60–10 cm). One of the two UASBs was equipped with a polyvinylidene fluoride (PVDF) hollow fibre membrane (hydrophobic, pore size 0.1 μm) unit. Both reactors were equipped with a gasbag (Sigma Aldrich, UK) for gas collection.
TSS reflects the origin of the biomass (soils/sediments rich in silt and gravel)) with an inoculum that had been acclimated (see below) to treat UV-sterilized ‘authentic’ primary settled domestic wastewater at 15 °C under methanogenic conditions. The reactor height was 600 mm with two sampling ports, one at the bottom of the reactor and one at the top (at 0 + 10 cm and at 60–10 cm). One of the two UASBs was equipped with a polyvinylidene fluoride (PVDF) hollow fibre membrane (hydrophobic, pore size 0.1 μm) unit. Both reactors were equipped with a gasbag (Sigma Aldrich, UK) for gas collection.
Inoculum
The inoculum was a mixture of cold-adapted sediments from Lake Geneva N 46°23′04′′, E 6°25′07′′ (average temperature −11–17 °C) and soils from Svalbard in the high Arctic at various sampling points situated at N 78° and E 11, 15, and 16° (average temperature −16–6 °C). This inoculum had been adapted for anaerobic wastewater treatment previously as described by Petropoulos et al., 2017 and Petropoulos, 2015.28,31Wastewater
Primary settled domestic wastewater was collected from the Tudhoe Mill (County Durham, UK) wastewater treatment plant (WWTP). The COD concentrations varied considerably (100–800 mg L−1), and the particulate fraction was on average composed of 70% lipids, 20% carbohydrates and 10% proteins. The soluble organic fraction (sCOD) ranged between 8–510 mg L−1. The substrate's total suspended (TSS) and volatile (VSS) solids ranged from 244 ± 26 mg L−1 and 182 ± 20 mg L−1, respectively.32Operation
During the batch operation hydrolysis–fermentation, fermentation (only), and methanogenesis (based on methane production) activities were estimated. The difference between ‘hydrolysis–fermentation’ and ‘fermentation’ is that the first takes into account all the COD that became sCOD, whilst the second only considers the VFA generation excluding soluble longer chain acids (both steps include the fraction of carbon that was methanized). The batch period lasted 14 days, after which feeding was switched to continuous, with the flow adjusted to deliver the required hydraulic retention time (HRT). The HRT was subsequently stepwise reduced, as indicated below. The upflow velocity was kept at 0.8 m h−1. In the AnMBR, the membrane flux (LMH) was adjusted to account for the increased influent flux and clogging (Table 1). The addition of a membrane surface to the existing membrane module was carried out after transferring the (bio)film that had developed on the membrane back into the mixed liquor, cleansing the surface (0.3% HNO3 overnight), by adding membrane fibres to the existing setup.| Phase | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10b | 11 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| a LMH refers to the AnMBR. b Phase 10 is similar to 9; the membrane at the end of P9 was backwashed prior to initiation of the new phase. | |||||||||||
| Day | 0–24 | 25–41 | 42–45 | 46–48 | 53–58 | 61–168 | 187–241 | 249–259 | 291–322 | 336–349 | 350–375 |
| LMHa | 2.02 | 4.04 | 14.15 | 6.96 | 4.04 | 3.58 | 2.68 | 3.59 | 4.05 | 4.05 | 2.18 |
| HRT | 3.5 | 1.75 | 0.50 | 1.00 | 1.75 | 0.83 | 0.58 | 0.43 | 0.32 | 0.32 | 0.32 |
| OLR | 0.07 ± 0.02 | 0.15 ± 0.02 | 0.82 ± 0.08 | 0.41 ± 0.05 | 0.36 ± 0.01 | 0.38 ± 0.03 | 0.45 ± 0.04 | 0.79 ± 0.22 | 1.51 ± 0.10 | 1.60 ± 0.28 | 1.20 ± 0.17 |
Daily operation included a daily 2 h relaxation and a 2 h backwash period (for phases: 2–5, 7, 8, and 11; Table 1). The starting sludge loading rate (SLR) was 0.023 kgCOD kgVSS−1 per day; however since the inocula were initially soils and sediments rich in plant materials not all this VSS encompasses bacteria. From previous enumerations (Petropoulos, 2015)31 we expect a population of ≈5 × 107 cells per ml inoculum. Using a bacterial mass of 10−12 gVSS per cell this would correspond to a start up at an excessive SLR of 46 ± 1.5 kgCOD kg VSbacterial−1 per day.33
Gas Analysis
CH4 in the headspace (gasbag) was monitored as % by volume using gas chromatography (GC). Gas samples (50 μl) withdrawn from the bag using a gas-tight syringe (SGE-Europe) were injected to a Carlo Erba HRGC S160 GC fitted with an FID detector and a HP-PLOTQ column (0.32 mm diameter, 30 m length and 20 μm film). The dissolved methane in both the mixed liquor and the effluent was also measured (from day 90 onwards) by quantifying (%) the formed methane from a 20 ml sample in a closed Wheaton vial (60 ml) after vigorous shaking at 25 °C.VSS
The VSS content of the biomass was estimated gravimetrically.32VFA
Samples from the liquid phase were removed from reactors using sterile syringes and transferred to sterile 2 ml micro-centrifuge tubes and then centrifuged (3 min at 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g) to obtain a supernatant for analysis. The supernatant was analysed by ion exchange chromatography.34
000 × g) to obtain a supernatant for analysis. The supernatant was analysed by ion exchange chromatography.34
Anions
SO42− and NO3− were measured after filtration (0.45 μm) in a Dionex, ICS-1000 ion chromatograph fitted with an AS40 automated sampler.Flux
The membrane flux was estimated from the volume of the effluent that passed through the membrane in a 24 h period. The excess aqueous volume that remained in the AnMBR due to reduced flux was collected and centrifuged (4000 RPM for 20 minutes); the solid part was returned to the reactor.Organic content
COD and soluble COD (sCOD) in the influent, effluent and mixed liquor were measured based on APHA, 2006.32Proteins
The protein content of the mixed liquor and the biomass (after lysis) was spectrophotometrically (Merck, UK) measured (Bradford, 1976), using a Protein Quantification Kit – Rapid (Sigma Aldrich, Fluka, UK). The conversion to COD was carried out using (C4H6.1O1.2N)x as a point of reference.35Lipids
The lipid content of the mixed liquor was measured gravimetrically using the Bligh and Dyer (1959)36 extraction protocol (10 ml sample). The conversion to COD was carried out stoichiometrically using palmitate (C36H60O2) as a point of reference.Carbohydrates
The carbohydrate content was estimated using the anthrone method.37 The concentration was spectrophotometrically estimated (Merck, UK), standardizing a curve with known amounts of glucose. The conversion to COD was implemented stoichiometrically using glucose (C6H12O6) as a point of reference.DNA extraction
Biomass samples were obtained from pellets generated by centrifugation (3 minutes, 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm) of 1 ml of mixed liquor sample. Total genomic DNA was extracted using a modified Griffiths et al. (2000)38 protocol based on CTAB and C6H6O:CHCl3:C5H12O in which the addition of CHCl3:C5H12O was carried out twice to minimize the presence of C6H6O in the sample.37 The use of phase lock light 2 ml Eppendorf tubes with phase lock gel® (VWR, UK) was also incorporated to further preclude potential C6H6O interference. The DNA extractions were carried out on samples collected on days 30, 102, 166, 242 and 375. The quality of the DNA was examined with a Nanodrop (ThermoFisher, UK) to ensure that the ratios 260
000 rpm) of 1 ml of mixed liquor sample. Total genomic DNA was extracted using a modified Griffiths et al. (2000)38 protocol based on CTAB and C6H6O:CHCl3:C5H12O in which the addition of CHCl3:C5H12O was carried out twice to minimize the presence of C6H6O in the sample.37 The use of phase lock light 2 ml Eppendorf tubes with phase lock gel® (VWR, UK) was also incorporated to further preclude potential C6H6O interference. The DNA extractions were carried out on samples collected on days 30, 102, 166, 242 and 375. The quality of the DNA was examined with a Nanodrop (ThermoFisher, UK) to ensure that the ratios 260![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 280 and 230
280 and 230![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 260 were between 1.8 and 2.1, to guarantee the absence of PCR inhibitors.
260 were between 1.8 and 2.1, to guarantee the absence of PCR inhibitors.
Sequencing and bioinformatics
Sequencing was carried out by the Earlham Institute (Norwich, UK) using an Illumina Hi-Seq.39 The 16 s rRNA gene data was processed using the Quantitative Insights Into Microbial Ecology (QIIME) 1.9.1 pipeline.40 We obtained >150![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 sequences per sample, covering ≥90% of the diversity.41 The sequencing data was pruned (top 15
000 sequences per sample, covering ≥90% of the diversity.41 The sequencing data was pruned (top 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000) per sample for downstream analysis.42 The differences of the bacterial communities between reactors were statistically verified using the STAMP® platform. A heatmap was generated to show a data matrix where colouring gives an indication of the numeric differences at the genus level.
000) per sample for downstream analysis.42 The differences of the bacterial communities between reactors were statistically verified using the STAMP® platform. A heatmap was generated to show a data matrix where colouring gives an indication of the numeric differences at the genus level.
qPCR standards
Methanosarcina barkeri cultures were employed for the preparation of the qPCR standards. DNA was extracted with an MP-bio ‘for soil DNA’ extraction kit (UK) following the manufacturer's instructions. The mcrA gene was amplified with the mlas-f primer according to Steinberg and Regan (2008).43 PCR products were used to transform Escherichia coli cells according to the manufacturer's instructions (TA cloning kit; Invitrogen, UK). Positive clones were grown in LB broth containing ampicillin at 37 °C. Plasmids were extracted and purified with a plasmid purification kit (ROCHE, UK) and quantified based on Quant-It (Invitrogen, UK). Plasmid DNA from each clone was diluted in PCR-grade distilled water to create the dilution series that was used for the qPCR standards.qPCR
Quantitative PCR (qPCR) was used for the quantitation of methanogens and total bacteria in the reactors. The methanogenic groups were quantified using functional gene primers (mlas-f, mcrA-rev) for methanogens by using a previously described method by Steinberg and Regan (2008).43 The qPCR took place on a CFX96 real-time PCR system (Biorad, UK) using 39 cycles. Reaction conditions included an initial denaturation step at 98 °C for 3 min, followed by 39 cycles of denaturation at 95 °C for 5 s, annealing at 66 °C for 10 s, and extension at 65 °C for 5 s with a 0.5 °C increment until a final extension step at 95 °C for 0.5 min according to the manufacturer (BIORAD, UK for Ssofast Evagreen® Supermix). The reaction mixture consisted of 3 μl of DNA template, 1 μl of sterile de-ionized water, 0.5 μl each of the forward and reverse primers (concentration of 10 pmol ul) and 5 μl of Ssofast EvaGreen Supermix (Biorad, UK). The analysis incorporated a 5-point calibration curve using DNA standards of known concentration; no-template controls were prepared from filter-sterilized de-ionized water. All qPCR reactions were performed in triplicate and efficiency values were calculated based on standards. Starting quantity (SQ) values from the qPCR in gene copies per ml were converted to cells per ml.26Activity test
At the end of the experiment the methanogenic activity of the biomass in the two reactors was tested with acetate and formate as test substrates. Both substrates were supplied at final concentrations equivalent to 1000 mgCOD L−1, and biomass was added to achieve an F![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) M of approx. 0.55. The assays took place at 15 and 37 °C (incubators). Controls with un-amended biomass were also prepared (fed with distilled water) in 80 ml Wheaton vials. All treatments were prepared in duplicate. Prior to incubation, the pH was set to 7.0 ± 0.1. The activity was initially expressed as ‘total activity’ (including the methane produced from both the intermediates provided and from the carbon trapped at the inoculum), then as ‘net’ methanogenic activity, subtracting the methane formed from the un-amended controls from the total to evaluate the gas that was produced strictly from the intermediates. This leads to a very conservative and putative approach for quantifying the minimum capacity of the seed. Methane was measured twice per day at 12 h intervals.
M of approx. 0.55. The assays took place at 15 and 37 °C (incubators). Controls with un-amended biomass were also prepared (fed with distilled water) in 80 ml Wheaton vials. All treatments were prepared in duplicate. Prior to incubation, the pH was set to 7.0 ± 0.1. The activity was initially expressed as ‘total activity’ (including the methane produced from both the intermediates provided and from the carbon trapped at the inoculum), then as ‘net’ methanogenic activity, subtracting the methane formed from the un-amended controls from the total to evaluate the gas that was produced strictly from the intermediates. This leads to a very conservative and putative approach for quantifying the minimum capacity of the seed. Methane was measured twice per day at 12 h intervals.
Energy usage
For the estimation of the energy requirement during the operation of the reactors, the following assumptions were made: a) all pumping requirements as 0.02 kW h m−3 (ref. 44) (including wastewater pumping to the anaerobic biozone, pumping for mixing (via up-flow velocity), and fouling mitigation via pumping the effluent (backwash16)); b) membrane operation as 0.3 kW h m−3;16,44 this is generally a non-conservative approach: the energy used for fouling mitigation and membrane operation varies considerably, and these values are at the low end of the range.2,24,25 The energy demand for the recovery of dissolved methane was also taken into consideration (0.05 kW h m−3).2,25 For the energy production from the methane produced, the average methane volume (in mmol) from the most efficient periods (9, 10, and 11) was converted to energy (kW h m−3), assuming a methane energy content of 10 kW h per m3 of CH4 produced. The efficiency of a combined heat power (CHP) engine was taken into consideration as 61.8% as per the optimized operation suggested by Li et al. (2011)45 (an analytical table for the estimation methodology is given in the ESI,† Table S1). The potential energy production in the absence of sulphate was estimated stoichiometrically, assuming that all organic carbon (as acetate) that was utilized to achieve SO4 reduction remained available to form methane instead.3. Results and discussion
3.1 The batch period
3.2 The continuous period
| Phase | 1 | 2 | 3 | 4a | 5a | 6 | 7 | 8 | 9 | 10 | 11 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| a Phases 4 and 5 are combined as they both refer to the overloading period and recovery from the overloading period; SE stands for standard error (n = 2). | |||||||||||
| AnMBR removal efficiency (%) | 35.8 ± 11.8 | 52.1 ± 7.5 | 85.6 ± 2.4 | 86.2 ± 5.9 | 70.9 ± 7.1 | 79.3 ± 1.3 | 71.3 ± 14.5 | 78.7 ± 2.3 | 68.7 ± 12.8 | 86.2 ± 1.5 | |
| UASB removal efficiency (%) | 45.9 ± 30.1 | 50.1 ± 14.8 | 53.2 ± 1.3 | 82.5 ± 5.6 | 68.8 ± 4.3 | 63.7 ± 3.4 | 70.3 ± 13.8 | 68.1 ± 3.0 | 68.5 ± 10.1 | 79.3 ± 2.0 | |
UASB systems have been used for the anaerobic treatment of domestic WW at ∼15 °C since the 80s.14,15 Many of these studies employed synthetic wastewaters, which are not necessarily representative of real world conditions. Our study is one of the few that used real wastewater. Table 3 summarizes the performance of the current study for the UASB and AnMBR as well as previous studies with similar objectives. The COD removal and effluent quality achieved at 15 °C and a low HRT of 0.32 days are superior to previous studies using real wastewater. This suggests, but does not prove, that using our biomass facilitated improved wastewater treatment at 15 °C.
| Reference | Reactor type | Substrate | Temperature (°C) | OLR (kgCOD m−3 per day) | HRT (d) | COD effluent (mg L−1) | COD removal efficiency (%) | CH4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) COD (LCH4 gCOD−1) COD (LCH4 gCOD−1) |
|---|---|---|---|---|---|---|---|---|
| P.S.WW stands for primary settled wastewater; actual wastewater stands for the substrate that is not well defined whether it is raw or primary settled. a Expressed per gBOD5. | ||||||||
| Current study | AnMBR | Actual P.S.WW | 15 | 1.60 ± 0.28 | 0.32 | 39.6 ± 4.1 | 86.2 ± 1.5 | 0.09 |
| Current study | UASB | Actual P.S.WW | 15 | 1.60 ± 0.29 | 0.32 | 83.9 ± 8.3 | 79.3 ± 2.0 | 0.06 |
| Lew et al., 2009 (ref. 22) | AnMBR | Actual P.S.WW | 25 | 2.16 | 0.25 | 65 | 88 | — |
| Rizvi et al., 2015 (ref. 10) | UASB | Actual WW | 17 | ≈1.3 | 0.375 | 180–203 | 57–62 | — |
| Mahmoud et al., 2004 (ref. 13) | UASB | Actual WW | 15 | 2.88 | 0.25 | 218.4 | 44 | 0.47–0.36 |
| Angenent et al., 2001 (ref. 29) | AMBR | Actual WW | 20–15 | 3.5 | 0.17 | 246 | 59 | — |
| Van der Last and Lettinga, 1992 (ref. 53) | EGSB | Actual WW | 13–19 | 2.7–4.7 | 0.14–0.04 | 200 | 16–34 | — |
| Hahn and Figueroa, 2015 (ref. 30) | ABR | Raw WW | 12–23 | 1.3 | 0.125 | 433.2 | 43 ± 15 | 0.28a |
| Watanabe et al., 2017 (ref. 17) | AnMBR | Synthetic WW | 25–10 | 0.15–1.2 | 2–0.25 | <100 | <75 | — |
| Smith et al., 2015 (ref. 18) | AnMBR | Synthetic WW | 15–3 | 3–1.2 | 1.2–0.7 | <70 | 86–96 | — |
| Ozgun et al., 2015 (ref. 19) | AnMBR | Synthetic WW | 25 | 2 | 0.25 | 42 ± 4.4 | ≈92 | — |
| Bialek et al., 2013 (ref. 11) | EGSB | Synthetic WW | 10 | 0.5–2 | 2–0.5 | <140 | <85 | |
| Smith et al., 2013 (ref. 20) | AnMBR | Synthetic WW | 15 | 0.44–0.66 | 1–0.66 | 36 ± 21 | 92 ± 5 | — |
| Martinez-Sosa et al., 2011 (ref. 21) | AnSMBR | Synthetic WW | 20 | 0.4–0.9 | 1.5–0.67 | 50 | 88 | 0.23 |
| Lin et al., 2011 (ref. 54) | SAnMBR | Synthetic WW | 30 | 1 | 0.42 | 40 | 90 | 0.26 |
| McKeown et al., 2009 (ref. 12) | EGSB-AF | Synthetic WW | 15–4 | 3.75–10 | 0.5–1 | 900 | 82 | — |
After 375 operational days, the AnMBR and UASB mixed liquor and the AnMBR biofilm harboured different populations of both bacteria and methanogenic archaea. The archaeal communities of the UASB and AnMBR mixed liquor-AnMBR biofilm were ≥75% different (P = 0.05), whilst the community of the AnMBR mixed liquor and the biofilm was <10% (P = 0.05) different. Heatmaps (Fig. 2a and b) show that acetoclastic methanogens of the genus Methanosaeta (order Methanosarcinales) dominated the archaeal populations in the AnMBR reactor, growing especially as a biofilm on the surface of the membrane and, to a lesser extent, in the mixed liquor. Acetoclastic methanogens were less prominent in the UASB reactor, where hydrogenotrophic methanogens (Methanobrevibacter and Methanoculleus genera, belonging to the order Methanobacteriales and Methanomicrobiales, respectively) were proportionally more abundant. Hydrogenotrophs in the AnMBR were mainly from the genus Methanospirillum.
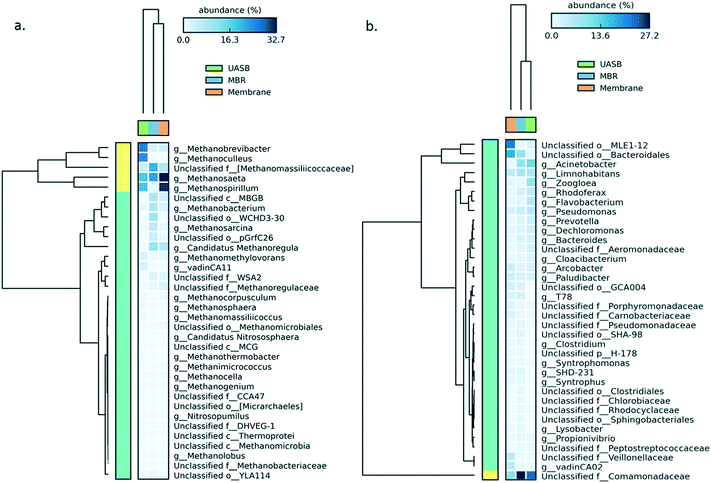 | ||
| Fig. 2 Heatmap of the community developed in the UASB and AnMBR mixed liquor as well as at the AnMBR biofilm for (left to right): a) archaea and b) bacteria genera; the sample was taken on day 375. | ||
For the bacterial kingdom the differences were less substantial, with the UASB and AnMBR mixed liquor being similar (35%; P = 0.05), while the biofilm was more different (75%, P = 0.05). The majority of the microorganisms in the mixed liquor belonged to the family Comamonadaceae, whilst in the biofilm Bacteroidales and MLEI1-12 dominated.
| Units/conditions | AnMBRbiom. Acetate 15 °C | UASBbiom. Acetate 15 °C | AnMBRbiom. Formate 15 °C | UASBbiom. Formate 15 °C | AnMBRbiom. Acetate 37 °C | UASBbiom. Acetate 37 °C | AnMBRbiom. Formate 37 °C | UASBbiom. Formate 37 °C |
|---|---|---|---|---|---|---|---|---|
| a The activity from un-amended controls was subtracted from the results presented in the table to solely quantify the activity from the direct intermediates and preclude the activity that might appear from later hydrolysis of previously un-hydrolysed organic materials. b For the expression of the activity per cell, the qPCRmcrA enumeration was used. c For the expression of the activity per reactor, the activity per cell per ml was multiplied by the reactor volume. d Further details/plot are given in ESI S3. | ||||||||
| Total mmolCH4 gVS−1 h−1 | 0.066 ± 0.013 | 0.065 ± 0.01 | 0.038 ± 0.013 | 0.148 ± 0.029 | 0.199 ± 0.054 | 0.156 ± 0.035 | 0.103 ± 0.054 | 0.154 ± 0.035 |
| Net mmolCH4 gVS−1 h−1 | 0.03 ± 0.01 | 0.02 ± 0.01 | 0.01 ± 0.01 | 0.10 ± 0.01 | 0.10 ± 0.03 | 0.08 ± 0.02 | 0.00 ± 0.00 | 0.08 ± 0.02 |
| Net mgCOD gVS−1 d−1 | 50.9 ± 11.4 | 26.1 ± 11.3 | 7.9 ± 3.9 | 156.3 ± 15.0 | 147.5 ± 41.6 | 121.0 ± 32.2 | 0.6 ± 0.3 | 118.0 ± 23.3 |
| Net fmolCH4 per cell per dayb | 662.0 ± 148.0 | 339.7 ± 140.2 | 102.5 ± 56.3 | 2032.5 ± 201.5 | 1917.3 ± 504.3 | 1573.7 ± 418.8 | 7.5 ± 3.8 | 1534.4 ± 302.0 |
| Net gCOD reactorcell−1 d−1c | 0.72 ± 0.16 | 0.13 ± 0.25 | 0.11 ± 0.06 | 0.76 ± 0.08 | 2.10 ± 0.59 | 0.59 ± 0.16 | 0.01 ± 0.00 | 0.58 ± 0.11 |
A likely scenario for the differentiation between the methanogenic communities is that a less well adapted (to low temperature) community (mainly acetoclastic methanogens) remained in the AnMBR and acclimated, whilst in the UASB these cells were washed out allowing only the more tolerant hydrogenotrophs to predominate (Fig. 2a). The difference between methanogenic cell abundances in the two systems was assessed on the final experimental day where the methanogenic population in the AnMBR reached 1.71 × 107 ± 3.12 × 106, three times higher than that in the UASB (5.88 × 106 ± 2.46 × 106) (Fig. 3).
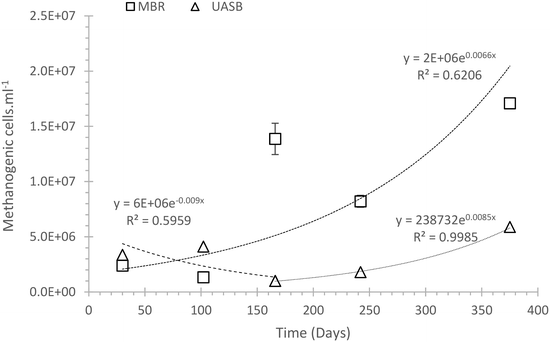 | ||
| Fig. 3 Estimation of the exponential growth and decay of the methanogenic population in a UASB and an AnMBR reactor setup. Error bars indicate standard error (n = 3). | ||
Fig. 3 suggests that initially (day 1–150) the methanogenic cells in the UASB that poorly adapted to the system were washed out. After day 150 the population was dominated by cells that were able to grow in this reactor and at this temperature, and their number gradually increased. In the AnMBR, on the other hand, washout was precluded by the membrane and the number of cells increased from day zero. We speculate that the bacteria in the AnMBR may thus have become relatively less well adapted than those in the UASB. The change in the number of total phyla also supports this interpretation, showing that one phylum disappeared in the UASB every 30 days (Nphyla −0.031tdaysR2 = 0.56), whilst for the AnMBR this number did not change significantly (Nphyla 0.023tdays; R2 = 0.21). The more dynamic character of the UASB compared to the AnMBR is also visible in the relative abundance plots (ESI† S3 and S4).
The activity of the UASB cells was similar at 15 °C and 37 °C (Table 4), revealing not only the importance of hydrogen at low temperatures but also the tolerance of these cells to low temperature (rates cell−1). For the AnMBR both the activity from formate and the presence of hydrogenotrophs were minimal. Interestingly the cell specific rates of the hydrogenotrophs were considerably higher than those previously estimated by Petropoulos et al., 2017 using this inoculum, fed with sterile raw WW as the feed during the first experimental trials.28 The maximum cell specific methanogenic rates at 15 °C were in the same range or higher (with formate) than the rates estimated by Dolfing and Mulder (1985) at 37 °C; meanwhile the cell specific rates from both systems at 37 °C exceeded those reported in the 1985 study.46
The cell specific activities reported are conservative (possibly underestimates) because a substantial amount of methane was also produced by the un-amended controls (Table 4; ESI† S5), and this “background” was subtracted prior to the calculation of the cell specific rates. The expression of the activity as ‘total activity’ has not changed the pattern which highlights the importance of hydrogenotrophy in the UASB reactors (this is further visualized in ESI† S3). The amount of methane that was generated from the un-amended controls indirectly shows some kind of affinity of the communities to some substrates, since previously-accumulated carbon was utilized in the absence of these other substrates.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
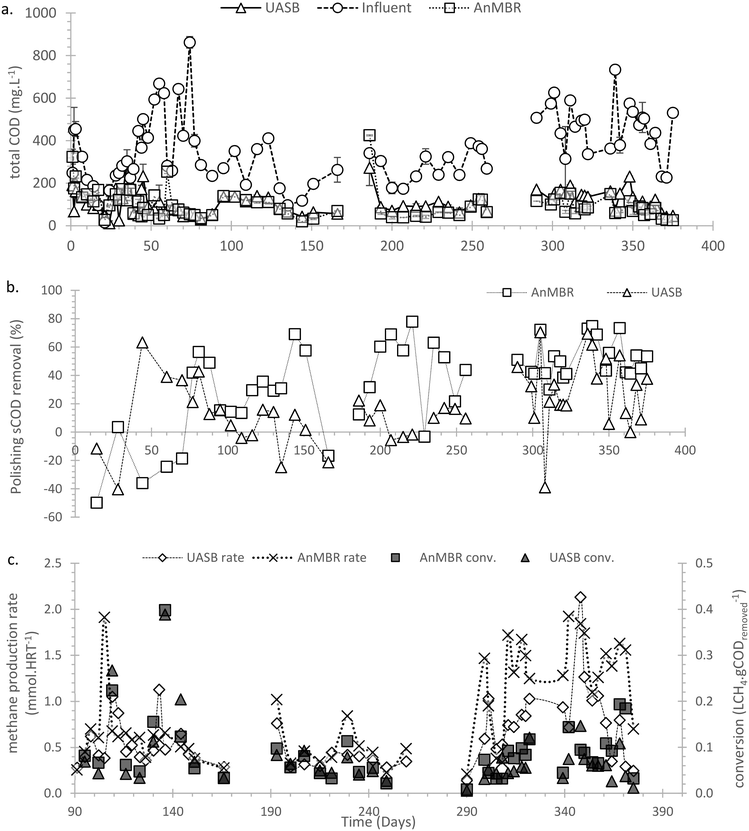
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CH4) but increased after day 60 (>10%). Before day 60 sulphate reduction was (in the absence of gaseous CH4 production) presumed to be the main mechanism of COD removal. From day 90 (Fig. 4c) the methane production rate was quantified as the sum of the methane in the gaseous phase and that dissolved in the aqueous phase (mixed liquor and effluent); before day 90 only the methane in the headspace gas was quantified. After day 90 the CH4 conversion rates became more stable, with the AnMBR performing better than the UASB, leading to an average COD
CH4) but increased after day 60 (>10%). Before day 60 sulphate reduction was (in the absence of gaseous CH4 production) presumed to be the main mechanism of COD removal. From day 90 (Fig. 4c) the methane production rate was quantified as the sum of the methane in the gaseous phase and that dissolved in the aqueous phase (mixed liquor and effluent); before day 90 only the methane in the headspace gas was quantified. After day 90 the CH4 conversion rates became more stable, with the AnMBR performing better than the UASB, leading to an average COD![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CH4 conversion of 0.09 ± 0.01 and 0.06 ± 0.01 LCH4 gCODremoved−1 in the AnMBR and UASB, respectively, corresponding to 26.1 ± 3.0 and 18.2 ± 2.1% conversion. In phases 9–11 the COD
CH4 conversion of 0.09 ± 0.01 and 0.06 ± 0.01 LCH4 gCODremoved−1 in the AnMBR and UASB, respectively, corresponding to 26.1 ± 3.0 and 18.2 ± 2.1% conversion. In phases 9–11 the COD![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CH4 reached the maximum observed for the UASB with an average of 21.5 ± 2.1%; for the AnMBR no significant further increase was observed.
CH4 reached the maximum observed for the UASB with an average of 21.5 ± 2.1%; for the AnMBR no significant further increase was observed.
The sulphate concentration of the influent WW (average influent concentration SO4: 120.4 ± 17.4 mg L−1) partly accounts for the low conversion rates. The AnMBR and UASB lost 0.89 ± 0.08 and 0.60 ± 0.06 mmolCH4 HRT−1 due to SO4 reduction, respectively. In the absence of sulphate this would have led to a COD![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CH4 of 0.18 ± 0.02 and 0.13 ± 0.01 LCH4 gCODremoved (or conversions of 51.7 ± 5.0 and 37.0 ± 3.5%). COD losses due to oxygen ingress and aerobic oxidation were unlikely as the redox was maintained ≤150 mV for both setups (≤200 during the last 60 days).
CH4 of 0.18 ± 0.02 and 0.13 ± 0.01 LCH4 gCODremoved (or conversions of 51.7 ± 5.0 and 37.0 ± 3.5%). COD losses due to oxygen ingress and aerobic oxidation were unlikely as the redox was maintained ≤150 mV for both setups (≤200 during the last 60 days).
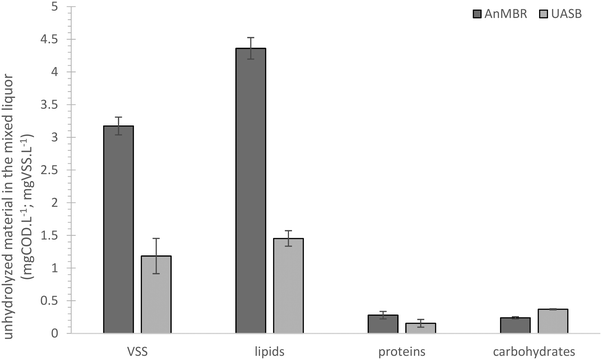
Although the presence and reduction of sulphate reduce the energy efficiency, SRB contributes to treatment with advantages previously discussed.47 The sulphate concentration in domestic wastewater influents varies depending on numerous factors but generally, the concentration in the current study is relatively high (compared to a common composition proposed by Tchobanoglous et al., 2002).48 The slightly higher SO4 removal in the AnMBR compared to the UASB might be a result of electron donors being supplied from hydrolysis of the material that was retained in the mixed liquor (the AnMBR sCODmixedliquor was higher than the UASB sCODmixedliquor (average of 157.3 ± 15.6 and 107.8 ± 11.3 mg L−1 for the AnMBR and UASB, respectively)).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) M was presumably kept high (low cell abundance and high organic material in the early and final stage, respectively).
M was presumably kept high (low cell abundance and high organic material in the early and final stage, respectively).
The membrane also assisted in stripping dissolved methane from the liquid phase (8.9 ± 1.6 and 16.3 ± 1.6% of the total methane was found in the effluent of the AnMBR and UASB, respectively), an amount that was higher than what would stoichiometrically be expected.46 It is unknown under what mechanism the membrane degasification functions; possibly this is an effect of the turbulence generated by the passage of the aqueous body through the membrane biofilm.
Their benefits notwithstanding, it is common knowledge that membranes tend to foul. This usually results in a decrease in the operational flux (as LMH). We used the rate of the LMH loss to monitor fouling (Table 5). The variable LMH (flux in L m−2 h−1) indicated that for a specific treatment system (anaerobic, at low temperature – high solids, OLR ∼0.4 kg m−3 per day or less) the optimal flux should be kept < 7.0 L m−2 h−1 (phase 3, 4); in the case of similar operational conditions but at higher OLR the LMH should be <4.0 L m−2 h−1 (including consistent backwash). The average-fouling rate with and without backwash was 0.091 and 0.017 L m−2 HRT−1, respectively. For more conservative LMH (<3.0 L m−2 h−1, phase 6) backwashing is not essential (fouling rate −9.4 ml m−2 d−1, 30% faster than without backwashing phase 7). At lower HRTs even fluxes equal to 3.0 L m−2 h−1 are relatively high due to the sludge increase and SRT reduction is obligatory. For the last phase (11) the flux was set as previously (2.8 L m−2 h−1) with backwash every 3 HRT. This regime maintained the membrane functionality almost at 100% until the end of the experimentation.
| Phase | 1 | 2 | 3 | 4 | 5 | 6 | 7 | 8 | 9 | 10 | 11 |
|---|---|---|---|---|---|---|---|---|---|---|---|
| a Y stands for yes; N stands for no; ‘—‘ stands for the fouling that occurred in an already fouled membrane and was excluded from further analysis. | |||||||||||
| Backwasha | N | Y | Y | Y | Y | N | Y | Y | N | N | Y |
| J loss (L m−2 d−1) | 0.009 | 0.006 | 2.397 | 0.744 | 0.009 | 0.009 | 0.006 | 0.049 | 0.066 | 0.038 | 0.002 |
| J loss (L m−2 HRT−1) | 0.032 | 0.021 | — | — | 0.032 | 0.011 | 0.011 | — | 0.206 | 0.119 | 0.007 |
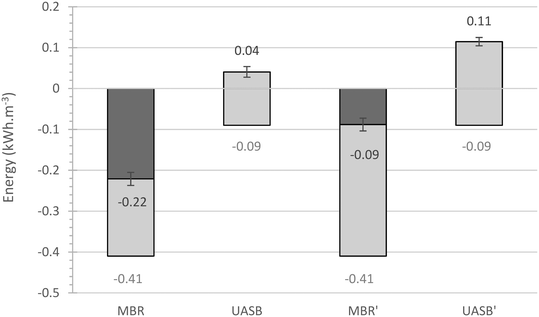
The energy theoretically available from the produced methane implies that the UASB can be energy positive (0.041 ± 0.013 kW h m−3), while the addition of the membrane renders the treatment system slightly energy negative (−0.221 ± 0.016 kW h m−3) (Fig. 5; detailed calculation in ESI† Table S1). The energy costs for the UASB are considerably lower than what is usually required for aerobic treatment processes (0.23 kW h m−3 (Northumbrian Water Ltd, UK)); for the AnMBR this is not the case though. The methane energy produced by the AnMBR yields only 0.06 kW h m−3 more than the UASB. More energy could have been produced in the absence of sulphate, rendering both systems more sustainable but not necessarily carbon neutral in the case of the AnMBR (from −0.088 ± 0.016 to 0.145 ± 0.01 kW h m−3 for the AnMBR and the UASB, respectively). In reality, all real wastewaters contain sulphate and some dissolved methane could escape in the effluent.
| Features | COD removal | TSS-VSS removal | HRT | SRT | Community structure | Acidification | Cost | Energy potential | Operation | Self sufficiency |
|---|---|---|---|---|---|---|---|---|---|---|
| AnMBR | Very good | Very good | Low | Short | Stable | Likely | High | Moderate | Challenging | Less likely |
| UASB | Very good | Good | Low | Moderate | Dynamic | Less likely | Low | Good | Simplified | Likely |
4. Conclusions
We have qualitatively compared two reactor formats in this study and on that basis we would choose to scale up our use of the cold-adapted seed in a UASB rather than an AnMBR. The slightly better treatment performance of the latter could not offset its cost, complexity and potential complications. The use of real wastewaters in this study was shown to be pivotal in drawing conclusions about the applicability of putative treatment technologies. It is likely that the reactor design, presumably the membrane, had a profound effect on the microbial ecology, and thus the performance of the system; however, there may be circumstances in which the use of the AnMBR is justified.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was funded by the BBSRC (BB/K003240/1; Engineering synthetic microbial communities for biomethane production). Sequencing was performed at Earlham Institute, Norwich, UK by the Prof. Swarbreck team.References
- I. Angelidaki, L. Ellegaard and B. K. Ahring, A comprehensive model of anaerobic bioconversion of complex substrates to biogas, Biotechnol. Bioeng., 1999, 63, 363–372 CrossRef CAS PubMed.
- P. L. McCarty, J. Bae and J. Kim, Domestic Wastewater Treatment as a Net Energy Producer–Can This be Achieved?, Environ. Sci. Technol., 2011, 45, 7100–7106 CrossRef CAS PubMed.
- P. L. McCarty and D. P. Smith, Anaerobic wastewater treatment, Environ. Sci. Technol., 1986, 20, 1200–1206 CrossRef CAS.
- J. Keller and K. Hartley, Greenhouse gas production in wastewater treatment: process selection is the major factor, Water Sci. Technol., 2003, 47, 43–48 CrossRef CAS PubMed.
- J. L. Campos, D. Valenzuela-Heredia, A. Pedrouso, A. Val del Rio, M. Belmonte and A. Mosquera-Corral, Greenhouse gases emissions from wastewater treatment plants: minimization, treatment, and prevention, J. Chem., 2016, 3796352, 1–12 Search PubMed.
- A. C. Van Haandel and G. Lettinga, Anaerobic sewage treatment: a practical guide for regions with a hot climate, John Wiley & Sons, 1994 Search PubMed.
- E. S. Heidrich, T. P. Curtis and J. Dolfing, Determination of the internal chemical energy of wastewater, Environ. Sci. Technol., 2010, 45, 827–832 CrossRef PubMed.
- E. J. Bowen, J. Dolfing, R. J. Davenport, F. L. Read and T. P. Curtis, Low-temperature limitation of bioreactor sludge in anaerobic treatment of domestic wastewater, Water Sci. Technol., 2014, 69, 1004–1013 CrossRef CAS PubMed.
- L. Seghezzo, G. Zeeman, J. B. van Lier, H. V. M. Hamelers and G. Lettinga, A review: the anaerobic treatment of sewage in UASB and EGSB reactors, Bioresour. Technol., 1998, 65, 175–190 CrossRef CAS.
- H. Rizvi, N. Ahmad, F. Abbas, I. H. Bukhari, A. Yasar, S. Ali, T. Yasmeen and M. Riaz, Start-up of UASB reactors treating municipal wastewater and effect of temperature/sludge age and hydraulic retention time (HRT) on its performance, Arabian J. Chem., 2015, 8, 780–786 CrossRef CAS.
- K. Bialek, J. Kim, C. Lee, G. Collins, T. Mahony and V. O'Flaherty, Quantitative and qualitative analyses of methanogenic community development in high-rate anaerobic bioreactors, Water Res., 2011, 45, 1298–1308 CrossRef CAS PubMed.
- R. M. McKeown, C. Scully, T. Mahony, G. Collins and V. O'Flaherty, Long-term (1243 days), low-temperature (4–15° C), anaerobic biotreatment of acidified wastewaters: bioprocess performance and physiological characteristics, Water Res., 2009, 43, 1611–1620 CrossRef CAS PubMed.
- N. Mahmoud, G. Zeeman, H. Gijzen and G. Lettinga, Anaerobic sewage treatment in a one-stage UASB reactor and a combined UASB-Digester system, Water Res., 2004, 38, 2348–2358 CrossRef CAS PubMed.
- A. W. A. De Man, P. C. Grin, R. E. Roersma, K. C. F. Grolle and G. Lettinga, Anaerobic Treatment of Municipal Wastewater at Low Temperatures Anaerobic Treatment, A Grown-Up Technology, Conference papers, Aquatech '86, 1986, pp. 453–466 Search PubMed.
- G. Lettinga, R. Roersma and P. Grin, Anaerobic treatment of raw domestic sewage at ambient temperatures using a granular bed UASB reactor, Biotechnol. Bioeng., 1983, 25, 1701–1723 CrossRef CAS PubMed.
- S. Judd, The MBR book: principles and applications of membrane bioreactors for water and wastewater treatment, Elsevier, 2010 Search PubMed.
- R. Watanabe, Y. Nie, S. Wakahara, D. Komori and Y.-Y. Li, Investigation on the response of anaerobic membrane bioreactor to temperature decrease from 25° C to 10° C in sewage treatment, Bioresour. Technol., 2017, 243, 747–754 CrossRef CAS PubMed.
- A. L. Smith, S. J. Skerlos and L. Raskin, Anaerobic membrane bioreactor treatment of domestic wastewater at psychrophilic temperatures ranging from 15° C to 3° C, Environ. Sci.: Water Res. Technol., 2015, 1, 56–64 RSC.
- H. Ozgun, J. B. Gimenez, M. E. Ersahin, Y. Tao, H. Spanjers and J. B. Van Lier, Impact of membrane addition for effluent extraction on the performance and sludge characteristics of upflow anaerobic sludge blanket reactors treating municipal wastewater, J. Membr. Sci., 2015, 479, 95–104 CrossRef CAS.
- A. L. Smith, S. J. Skerlos and L. Raskin, Psychrophilic anaerobic membrane bioreactor treatment of domestic wastewater, Water Res., 2013, 47, 1655–1665 CrossRef CAS PubMed.
- D. Martinez-Sosa, B. Helmreich, T. Netter, S. Paris, F. Bischof and H. Horn, Anaerobic submerged membrane bioreactor (AnSMBR) for municipal wastewater treatment under mesophilic and psychrophilic temperature conditions, Bioresour. Technol., 2011, 102, 10377–10385 CrossRef CAS PubMed.
- B. Lew, S. Tarre, M. Beliavski, C. Dosoretz and M. Green, Anaerobic membrane bioreactor (AnMBR) for domestic wastewater treatment, Desalination, 2009, 243, 251–257 CrossRef CAS.
- Y. Gu, Y. Li, X. Li, P. Luo, H. Wang, X. Wang, J. Wu and F. Li, Energy self-sufficient wastewater treatment plants: feasibilities and challenges, Energy Procedia, 2017, 105, 3741–3751 CrossRef.
- A. L. Smith, L. B. Stadler, N. G. Love, S. J. Skerlos and L. Raskin, Perspectives on anaerobic membrane bioreactor treatment of domestic wastewater: A critical review, Bioresour. Technol., 2012, 122, 149–159 CrossRef CAS PubMed.
- B. D. Shoener, C. Zhong, A. D. Grainer, W. O. Khunjar, P.-Y. Hong and J. S. Guest, Design of anaerobic membrane bioreactors for the valorization of dilute organic carbon waste streams, Energy Environ. Sci., 2016, 9, 1102–1112 RSC.
- M. V. Simankova, O. R. Kotsyurbenko, T. Lueders, A. N. Nozhevnikova, B. Wagner, R. Conrad and M. W. Friedrich, Isolation and characterization of new strains of methanogens from cold terrestrial habitats, Syst. Appl. Microbiol., 2003, 26, 312–318 CrossRef PubMed.
- A. N. Nozhevnikova, K. Zepp, F. Vazquez, A. J. B. Zehnder and C. Holliger, Evidence for the existence of psychrophilic methanogenic communities in anoxic sediments of deep lakes, Appl. Environ. Microbiol., 2003, 69, 1832–1835 CrossRef CAS PubMed.
- E. Petropoulos, J. Dolfing, R. J. Davenport, E. J. Bowen and T. P. Curtis, Developing cold-adapted biomass for the anaerobic treatment of domestic wastewater at low temperatures (4, 8 and 15 °C) with inocula from cold environments, Water Res., 2017, 112, 100–109 CrossRef CAS PubMed.
- L. T. Angenent, G. C. Banik and S. Sung, Anaerobic migrating blanket reactor treatment of low-strength wastewater at low temperatures, Water Environ. Res., 2001, 73, 567–574 CrossRef CAS PubMed.
- M. J. Hahn and L. A. Figueroa, Pilot scale application of anaerobic baffled reactor for biologically enhanced primary treatment of raw municipal wastewater, Water Res., 2015, 87, 494–502 CrossRef CAS PubMed.
- E. Petropoulos, Investigating the true limits of anaerobic treatment of wastewater at low temperature using a cold-adapted inoculum, Ph.D. Thesis, Newcastle University (UK), 2015 Search PubMed.
- APHA, Standard methods for the examination of water and wastewater, APHA, 1985 Search PubMed.
- B. E. Rittman and P. L. McCarty, Environmental biotechnology: principles and applications, McCraw-Hill, Inc., USA, 2001, pp. 6–11 Search PubMed.
- D. A. C. Manning and A. Bewsher, Determination of anions in landfill leachates by ion chromatography, J. Chromatogr. A, 1997, 770, 203–210 CrossRef CAS.
- S. Sanders, D. Bergen, S. Buijs, R. Corstanje, M. Gerrits, T. Hoogerwerf, S. Kanwar, G. Zeeman, J. Groenestijn and G. Lettinga, Treatment of waste activated sludge in an anaerobic hydrolysis upflow sludge bed reactor, 1996 Search PubMed.
- E. G. Bligh and W. J. Dyer, A rapid method of total lipid extraction and purification, Can. J. Biochem. Physiol., 1959, 37, 911–917 CrossRef CAS PubMed.
- J. E. Hedge, B. T. Hofreiter and R. L. Whistler, Carbohydrate chemistry, Academic Press, New York, 1962, p. 17 Search PubMed.
- R. I. Griffiths, A. S. Whiteley, A. G. O'Donnell and M. J. Bailey, Rapid method for coextraction of DNA and RNA from natural environments for analysis of ribosomal DNA-and rRNA-based microbial community composition, Appl. Environ. Microbiol., 2000, 66, 5488–5491 CrossRef CAS PubMed.
- J. J. Kozich, S. L. Westcott, N. T. Baxter, S. K. Highlander and P. D. Schloss, Development of a dual-index sequencing strategy and curation pipeline for analyzing amplicon sequence data on the MiSeq Illumina sequencing platform, Appl. Environ. Microbiol., 2013, 79, 5112–5120 CrossRef CAS PubMed.
- J. G. Caporaso, J. Kuczynski, J. Stombaugh, K. Bittinger, F. D. Bushman, E. K. Costello, N. Fierer, A. G. Pena, J. K. Goodrich and J. I. Gordon, QIIME allows analysis of high-throughput community sequencing data, Nat. Methods, 2010, 7, 335 CrossRef CAS PubMed.
- C. Quince, T. P. Curtis and W. T. Sloan, The rational exploration of microbial diversity, ISME J., 2008, 2, 997 CrossRef CAS PubMed.
- A. K. Shaw, A. L. Halpern, K. Beeson, B. Tran, J. C. Venter and J. B. H. Martiny, It's all relative: ranking the diversity of aquatic bacterial communities, Environ. Microbiol., 2008, 10, 2200–2210 CrossRef PubMed.
- L. M. Steinberg and J. M. Regan, Phylogenetic comparison of the methanogenic communities from an acidic, oligotrophic fen and an anaerobic digester treating municipal wastewater sludge, Appl. Environ. Microbiol., 2008, 74, 6663–6671 CrossRef CAS PubMed.
- I. Bodik and M. Kubaska, Energy and sustainability of operation of a wastewater treatment plant, Environ. Prot. Eng., 2013, 39, 15–24 CAS.
- Y. Li, Q. Qi., X. He and J. Li, Energy use project and conversion efficiency analysis on biogas produced in breweries, Proceedings in World Renewable Energy Congress, Linköping, Sweden, 2011 Search PubMed.
- J. Dolfing and J.-W. Mulder, Comparison of methane production rate and coenzyme F420 content of methanogenic consortia in anaerobic granular sludge, Appl. Environ. Microbiol., 1985, 49, 1142–1145 CAS.
- T. P. H. Van Den Brand, K. Roest, G. H. Chen, D. Brdjanovic and M. C. M. van Loosdrecht, Potential for beneficial application of sulfate reducing bacteria in sulfate containing domestic wastewater treatment, World J. Microbiol. Biotechnol., 2015, 31, 1675–1681 CrossRef CAS PubMed.
- G. Tchobanoglous, F. Burton and H. D. Stensel, Wastewater Engineering: Treatment and Reuse, ed. Metcalf & Eddy Inc.; Mc Graw Hill, 2002 Search PubMed.
- E. Petropoulos, J. Dolfing, Y. Yu, M. J. Wade, E. Bowen, R. J. Davenport and T. Curtis, Lipolysis of Domestic Wastewater in Anaerobic Reactors Operating at Low Temperatures, Environ. Sci.: Water Res. Technol., 2018, 7, 1002–1013 RSC.
- R. R. Dague, G. C. Banik and T. G. Ellis, Anaerobic sequencing batch reactor treatment of dilute wastewater at psychrophilic temperatures, Water Environ. Res., 1998, 70, 155–160 CrossRef CAS.
- Y. Miron, G. Zeeman, J. B. Van Lier and G. Lettinga, The role of sludge retention time in the hydrolysis and acidification of lipids, carbohydrates and proteins during digestion of primary sludge in CSTR systems, Water Res., 2000, 34, 1705–1713 CrossRef CAS.
- L. Zhang, J. D. Vrieze, T. L. G. Hendrickx, W. Wei, H. Temmink, H. Rijnaarts and G. Zeeman, Anaerobic treatment of raw domestic wastewater in a UASB-digester at 10 °C and microbial community dynamics, Chem. Eng. J., 2018, 334, 2088–2097 CrossRef CAS.
- A. R. M. van der Last and G. Lettinga, Anaerobic Treatment of Domestic Sewage under Moderate Climatic (Dutch) Conditions Using Upflow Reactors at Increased Superficial Velocities, Water Sci. Technol., 1992, 25, 167–178 CrossRef CAS.
- H. Lin, J. Chen, F. Wang, L. Ding and H. Hong, Feasibility evaluation of submerged anaerobic membrane bioreactor for municipal secondary wastewater treatment, Desalination, 2011, 280, 120–126 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8ew00410b |
| This journal is © The Royal Society of Chemistry 2019 |