Open Access Article
Open Access ArticleUnderstanding the antifouling mechanisms related to copper oxide and zinc oxide nanoparticles in anaerobic membrane bioreactors†
Hong
Cheng
a,
Qingtian
Guan
b,
Luis Francisco
Villalobos
 c,
Klaus-Viktor
Peinemann
c,
Klaus-Viktor
Peinemann
 c,
Arnab
Pain
b and
Pei-Ying
Hong
c,
Arnab
Pain
b and
Pei-Ying
Hong
 *a
*a
aKing Abdullah University of Science and Technology (KAUST), Biological and Environmental Sciences & Engineering Division (BESE), Water Desalination and Reuse Center (WDRC), Thuwal, 23955-6900, Saudi Arabia. E-mail: peiying.hong@kaust.edu.sa; Tel: +966 12 8082218
bKing Abdullah University of Science and Technology (KAUST), Biological and Environmental Sciences & Engineering Division (BESE), Pathogen Genomics Laboratory, Thuwal, 23955-6900, Saudi Arabia
cKing Abdullah University of Science and Technology (KAUST), Physical Sciences & Engineering Division (PSE), Advanced Membranes and Porous Materials Center (AMPMC), King Abdullah University of Science and Technology (KAUST), Thuwal, 23955-6900, Saudi Arabia
First published on 30th September 2019
Abstract
Biofouling impedes the performance of anaerobic membrane bioreactors. In this study, we aim to determine if copper oxide (CuO) and zinc oxide (ZnO) nanoparticles can effectively delay the biofouling of polyethersulfone (PES) membranes without disseminating emerging contaminants like antibiotic resistance genes (ARGs) and metal resistance genes (MRGs). A consequential decrease in biofilm composition related to total cells, polysaccharides, proteins, and bioactivity (i.e., adenosine triphosphate (ATP) and quorum sensing (QS) signal molecules) was observed in the presence of heavy metal nanoparticles. Metagenomic and metatranscriptomic analyses further attributed the delay of biofilm formation to the lower expression of QS-associated genes and biofilm formation genes. It was also determined that the expression of ARGs and MRGs was not stimulated in the presence of CuO and ZnO nanoparticles. These findings collectively suggest that CuO and ZnO nanoparticles embedded in membranes can delay biofouling with minimal potential for disseminating ARGs and MRGs post-treatment.
Environmental significanceMembrane bioreactors (MBRs) provide high quality treated wastewater for various reuse purposes. However, membrane biofouling remains a major bottleneck. Coating heavy metal nanoparticles (NPs) on membrane surfaces demonstrates potential to mitigate biofouling. It is however important to ensure that the presence of these heavy metal NPs does not select for other unintended contaminants like antibiotic resistance genes (ARGs) or heavy metal resistance genes (MRGs). Here, we demonstrate that CuO and ZnO NPs, which are cheaper than Ag NPs, are able to inhibit biofouling by suppressing expressions of genes related to quorum sensing and biofilm formation. There was no apparent increase in the abundance of expressed ARGs and MRGs, hence offering a safe, effective and low-cost approach to mitigate biofouling. |
1. Introduction
Anaerobic membrane bioreactors (AnMBRs) have become an emerging technology to use for sustainable wastewater treatment.1–3 The use of AnMBRs, which combines a membrane-based filtration process with anaerobic biodegradation, has several advantages over conventional aerobic processes typically used by most municipal wastewater treatment plants. For instance, using AnMBRs eliminates the need for aeration and lowers the energy consumption rate from ca. 2 kW h m−3 in aerobic MBRs to ca. 0.8 kW h m−3.4 Anaerobic biodegradation exhibits lower sludge production rates than activated sludge processes, and this would vastly alleviate the cost of sludge treatment. Moreover, the anaerobic biodegradation could convert organic carbon in municipal wastewater to methane while retaining total nitrogen and phosphorus in effluents which can be beneficial for agricultural irrigation.5However, biofouling, defined as the unwanted deposition of materials, particles, and bacteria on membrane surfaces during the filtration process,6 is particularly detrimental to the performance of membrane bioreactors as it can result in decreased permeate production, increased transmembrane pressure and a shorter lifetime of membrane modules.7 To overcome these problems caused by biofouling, various antifouling strategies have been devised. Examples of antifouling strategies include physical disruption of biofilm matrices using backwash, chemical wash or sonication,8 chemical modification of membranes (e.g. coating with nanoparticles and modifying membranes with organic functional groups)9 and biocidal control (e.g. quorum quenching and phage-based decomposition).10–12
In particular, coating heavy metal nanoparticles on the surface of membranes demonstrated promising results. An earlier study demonstrated that polysulfone membranes impregnated with silver (Ag) nanoparticles showed not only high antibacterial properties but also improved biofouling resistance.13 Zhang and his co-authors also reported that polyethersulfone (PES) membranes immobilized with Ag nanoparticles can inhibit biofilm formation on membrane surfaces.14 Despite these studies indicating the prospect of applying Ag nanoparticles to inhibit biofilm formation, another independent study observed that Ag nanoparticles would increase the amount of antibiotic resistance genes (ARGs) and heavy metal resistance genes (MRGs) in sequencing batch reactors.15 This would imply that if Ag nanoparticles were to be widely applied to AnMBRs to inhibit biofilm formation, there may be potential environmental risks associated with the dissemination of ARGs and MRGs in treated effluents since MBRs typically do not achieve good removal of extracellular DNA.16
Alternatives to Ag would include zinc oxide (ZnO) and copper oxide (CuO) nanoparticles. Compared to Ag nanoparticles, ZnO and CuO nanoparticles have relatively weaker antibacterial effects17 but the costs of both metals are at a lower price than that of Ag (Table S1†). Despite the weaker antibacterial effects, ZnO nanoparticles were found to effectively decrease the adsorption of pollutants on polyvinylidene fluoride (PVDF) membranes.18 Similarly, Booshehri et al. deposited CuO nanoparticles on PES membranes to achieve antibacterial activities against both Gram-positive and Gram-negative bacteria.19 However, the use of such membranes in an operational AnMBR and their efficacy to delay biofouling were not evaluated. No assessment was also made to determine if ARG or MGE abundance would be increased in the presence of both heavy metal nanoparticles.
In this study, we propose the use of CuO and ZnO nanoparticles functionalized on PES membranes to minimize anaerobic membrane biofouling. It is hypothesized that the use of these nanoparticles would achieve a delay in anaerobic membrane biofouling with no consequential increase in other unintended biological contaminants like ARGs and MRGs. To address this hypothesis, the impacts of these modified membranes on total bacterial cell numbers, adenosine triphosphate (ATP), proteins, polysaccharides and QS signal molecules in biofilm matrices were determined and compared against the respective membrane control. Furthermore, to elucidate the antifouling mechanism of nanoparticles at the molecular level and also to evaluate if genes related to ARGs and MGEs are increased, metatranscriptomic analysis was conducted. This study aims to demonstrate that a PES membrane modified with CuO and ZnO nanoparticles would achieve antifouling properties without disseminating ARGs and MRGs.
2. Materials and methods
2.1 Synthesis of modified PES membranes
To synthesize the PES membrane, 14 wt% Ultrason® PES powder (BASF, Germany) was dissolved in N-methyl-2-pyrrolidone (NMP) (Sigma-Aldrich, St. Louis, MO, US) at 60 °C under constant stirring at 500 rpm for 3 h. For nanoparticle-embedded membranes, CuO-NPs and ZnO-NPs (Sigma-Aldrich, St. Louis, MO, US) were dispersed in NMP using a probe sonicator to prepare 7.4% w/v and 3.3% w/v solutions, respectively. The concentration of each oxide was adjusted in order to obtain a similar surface loading of ZnO and CuO. The freshly prepared dispersion of CuO-NPs in NMP was mixed with PES powder to prepare 14% wt/v PES in NMP solution. For the case of ZnO-NPs, the freshly prepared solution was used to dilute 30 wt% PES in NMP solution to 14% wt/v. In both cases, the resulting nanoparticle-containing solutions were stirred at 60 °C for 3 h until all the PES powder was dissolved. All the solutions were cooled to room temperature and cast over a polyester non-woven support using a casting knife with a gap of 250 μm. Finally, they were immersed in a water bath to precipitate the membranes via non-solvent induced phase separation. The corresponding concentrations of the nanoparticles on the surface of the membranes were 1.3 mol% CuO-NPs (i.e., PES-CuO-NPs) and 1.8 mol% ZnO-NPs (i.e., PES-ZnO-NPs). All the membranes were kept in sterile deionized water before use.2.2 Membrane characterization
An FEI Nova Nano scanning electron microscope (SEM) equipped with an energy dispersive X-ray (EDX) analysis system was used to observe the surface of membranes and cross-section of biofilms at 5 kV. The concentration of the nanoparticles on the surface was also measured using EDX at 15 kV. To prepare sample for SEM, pristine and fouled membranes were respectively dried in air. Three nm thick iridium was sputtered onto the surface of the pristine membrane surface and the cross-section of the fouled membranes using a K575X Emitech sputter coater (Quorum Technologies, UK). To calculate the concentration of the nanoparticle on the surface of the membranes, EDX analysis was performed on three membranes and the average was reported.2.3 Reactor configuration and operating conditions
The AnMBR operated in this study was of similar configuration (Fig. S1†) to that operated in an earlier study.20 Briefly, the reactor had 2 L working volumes. Sludge was originally seeded with a mixture of camel manure and anaerobic sludge from a wastewater treatment plant in Riyadh, Saudi Arabia. The reactor was fed with synthetic wastewater of 800 mg L−1 chemical oxygen demand (COD) and operated at 35 °C (mesophilic conditions) and pH 7. The synthetic wastewater was made up of a mix of organic and inorganic compounds as well as trace metals, as shown in Table S2.†21 This equates to an organic loading rate (OLR) of 1.04 g COD L−1 day−1. Two separate runs were conducted (i.e., run 1 and run 2) in this study, approximately 3 months apart. For each run, three membranes (i.e., PES, PES-CuO-NPs and PES-ZnO-NPs), each individually housed in cassette holders, were connected in parallel to the anaerobic reactor. The AnMBR was operated at a 400 mL min−1 recirculation rate. Biogas was used to scour the membrane surface at a gas sparging rate of 250 mL min−1. Flux was maintained at ca. 4 L m−2 h−1 (LMH) while changes in the TMP were recorded using a pressure gauge connected to each membrane module. For each biological independent run (1 and 2), all the membranes were harvested at the same time as soon as any of the membranes reached the TMP that was indicative of critical fouling. These membranes were analyzed based on the procedures described in sections 2.5 through 2.8. No chemical cleaning was applied in this study as the main intention was to study the effect of heavy metal nanoparticles on fouling. The membranes were harvested using similar protocols detailed in an earlier study.9 The sludge in the reactor was sampled weekly for analyses. The COD in the effluent was quantified weekly.2.4 Cu and Zn concentration evaluation
The metal content (63Cu and 66Zn) in the sludge samples from the reactor was measured weekly on an inductively coupled plasma mass spectrometer (ICP-MS) (Agilent 7500). The sludge samples were vortexed for 1 min and then centrifuged at 8000g for 5 min. The supernatant was then filtered through 0.22 μm Whatman™ Puradisc 23 mm syringe filters (GE Healthcare, Little Chalfont, Buckinghamshire, UK) prior to measurement. Measurements were made against commercially available standards at 0, 1, 5, 10, 50 and 100 parts per billion in 2% HNO3 (CMS-5) (Inorganic Ventures, Christiansburg, VA, USA). Heavy metal ion concentrations were shown as averaged values from two reads on the ICP-MS.2.5 Cell count, total protein (PN) and polysaccharide (PS) determination
The total cells in the membrane biofilm were determined by flow cytometry on an Accuri C6 (BD Bioscience, NJ, US). Briefly, one mL of harvested biofilm liquid suspension was diluted 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000-fold with 1× PBS. The diluted samples were incubated in the dark for 10 min at 37 °C. Ten μL 100× SYBR green (Thermo Fisher Scientific, MA, US) was added into 1 mL of diluted samples to stain the cells, and then incubated in the dark for 10 min at 37 °C before flow cytometry. Fifty μL aliquots of the stained samples were injected into the Accuri C6 with a 35 μL min−1 flow rate to enumerate the total cells. Measurements were conducted in triplicate.
000-fold with 1× PBS. The diluted samples were incubated in the dark for 10 min at 37 °C. Ten μL 100× SYBR green (Thermo Fisher Scientific, MA, US) was added into 1 mL of diluted samples to stain the cells, and then incubated in the dark for 10 min at 37 °C before flow cytometry. Fifty μL aliquots of the stained samples were injected into the Accuri C6 with a 35 μL min−1 flow rate to enumerate the total cells. Measurements were conducted in triplicate.
The PS and PN concentrations of the soluble EPS fraction from the fouled membranes were quantified. The biofilm suspension was centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 10 min, and the supernatant was first filtered through a 0.22 μm syringe filter (VWR US, Radnor, PA, US) prior to the determination of its PN and PS. PN was quantified using a Total Protein kit (Sigma-Aldrich, St. Louis, MO, US) based on Peterson's modified micro Lowry method22 with 0 μg mL−1, 10 μg mL−1, 20 μg mL−1, 40 μg mL−1 and 80 μg mL−1 bovine serum albumin (BSA) as standards and measured in triplicate. PS was determined using the phenol–sulfuric acid method. 0 μg mL−1, 5 μg mL−1, 10 μg mL−1, 20 μg mL−1, 40 μg mL−1 and 80 μg mL−1 glucose were used as standards.
000g for 10 min, and the supernatant was first filtered through a 0.22 μm syringe filter (VWR US, Radnor, PA, US) prior to the determination of its PN and PS. PN was quantified using a Total Protein kit (Sigma-Aldrich, St. Louis, MO, US) based on Peterson's modified micro Lowry method22 with 0 μg mL−1, 10 μg mL−1, 20 μg mL−1, 40 μg mL−1 and 80 μg mL−1 bovine serum albumin (BSA) as standards and measured in triplicate. PS was determined using the phenol–sulfuric acid method. 0 μg mL−1, 5 μg mL−1, 10 μg mL−1, 20 μg mL−1, 40 μg mL−1 and 80 μg mL−1 glucose were used as standards.
2.6 Adenosine triphosphate (ATP), autoinducer-2 (AI-2) and N-acyl homoserine lactone (AHL) quantification
The fouled membranes with dimensions of 3 by 2.5 cm were respectively placed in 6 mL deionized water, ultrasonicated for 4 min at 25% amplitude with 2 s pulsating intervals. The ATP content in the suspension was quantified using the Celsis Amplified ATP™ reagent kit on an Advance luminometer (Celsis, Westminster, London, UK) with deionized water as a negative control. The relative AI-2 concentration of the liquid suspension of the harvested membranes was determined based on a previous protocol with slight modifications.23 Briefly, the AI-2 indicator Vibrio harveyi ATCC® strain BB170 was grown overnight in a sterile autoinducer bioassay (AB) medium. After the overnight growth, the AI-2 culture was diluted (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5000) with a fresh AB medium. Twenty μL sample was placed in a 96-well solid white microplate prior to addition of 180 μL of diluted AI-2 reporter. Twenty μL of deionized water in 180 μL of diluted AI-2 reporter was used as a negative control. The 96-well plate containing the samples was incubated in the dark on a 150 rpm shaker incubator platform at 30 °C. The bioluminescent intensity was detected with an Infinite M200 PRO microplate reader over time (Tecan, Männedorf, Switzerland). The relative concentration was calculated to be the intensity of the samples divided by the intensity of the negative control. For AHL, 20 μL samples and deionized water were loaded into a 96-well-plate with 80 μL of an overnight culture of Agrobacterium tumefaciens A136. The whole plate was then incubated for 90 min at 30 °C prior to addition of 100 μL of Beta-Glo (Promega, WI, USA) into each well and further incubated for 30 min at 30 °C. The bioluminescence intensity of each sample was recorded, and the relative concentration was calculated based on the same procedure as that described for AI-2 quantification. All the samples were measured in triplicate.
5000) with a fresh AB medium. Twenty μL sample was placed in a 96-well solid white microplate prior to addition of 180 μL of diluted AI-2 reporter. Twenty μL of deionized water in 180 μL of diluted AI-2 reporter was used as a negative control. The 96-well plate containing the samples was incubated in the dark on a 150 rpm shaker incubator platform at 30 °C. The bioluminescent intensity was detected with an Infinite M200 PRO microplate reader over time (Tecan, Männedorf, Switzerland). The relative concentration was calculated to be the intensity of the samples divided by the intensity of the negative control. For AHL, 20 μL samples and deionized water were loaded into a 96-well-plate with 80 μL of an overnight culture of Agrobacterium tumefaciens A136. The whole plate was then incubated for 90 min at 30 °C prior to addition of 100 μL of Beta-Glo (Promega, WI, USA) into each well and further incubated for 30 min at 30 °C. The bioluminescence intensity of each sample was recorded, and the relative concentration was calculated based on the same procedure as that described for AI-2 quantification. All the samples were measured in triplicate.
2.7 Total DNA extraction and sequencing
The genomic DNA of the biofilm samples from two independent runs (i.e., run 1 and run 2) was extracted using an UltraClean® soil DNA isolation kit (MoBio Laboratories, Carlsbad, USA) with slight modifications.24 DNA quality and quantity were determined using a 2100 Bioanalyzer (Agilent, Santa Clara, CA, USA) and Invitrogen's Qubit dsDNA BR assay kit (Thermo Fisher Scientific, NY, USA), respectively. 200 ng of total DNA was used for library preparation. The samples were sheared on a Covaris S220 (Covaris, Woburn, MA, USA) to ∼350![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) bp, following the manufacturer's recommendation, and uniquely tagged with one of Illumina's TruSeq LT DNA barcodes. DNA libraries were then sequenced in 2 lanes of an Illumina Hiseq 4000 platform.
bp, following the manufacturer's recommendation, and uniquely tagged with one of Illumina's TruSeq LT DNA barcodes. DNA libraries were then sequenced in 2 lanes of an Illumina Hiseq 4000 platform.
2.8 Total RNA extraction and sequencing
20 mL biofilm suspension samples from two independent runs (i.e., run 1 and run 2) were centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 10 minutes at 4 °C. The cell pellets were resuspended with RNAlater and then kept at −80 °C for further extraction. The total RNA was extracted using an RNeasy Midi kit (Qiagen, Hilden, Germany) with the optional on-column DNase treatment following the manufacturer's protocol. The RNA was eluted with RNase-free water, and then stored at −80 °C until sequencing. The quality and quantity of RNA were determined using a 2100 Bioanalyzer (Agilent, Santa Clara, CA, USA) and Invitrogen's Qubit RNA BR assay kit (Thermo Fisher Scientific, NY, USA), respectively. Prior to sequencing, the bacterial ribosomal RNA was removed and then converted to RNA-seq libraries using a TruSeq Stranded Total RNA with Ribo-Zero (bacteria) kit (Illumina, San Diego, CA, USA). The constructed library was sequenced on an Illumina HiSeq 4000 platform in the KAUST Genomics Core lab. All high-throughput sequencing files were deposited in the Short Read Archive (SRA) of the European Nucleotide Archive (ENA) under study accession number PRJEB33442.
000g for 10 minutes at 4 °C. The cell pellets were resuspended with RNAlater and then kept at −80 °C for further extraction. The total RNA was extracted using an RNeasy Midi kit (Qiagen, Hilden, Germany) with the optional on-column DNase treatment following the manufacturer's protocol. The RNA was eluted with RNase-free water, and then stored at −80 °C until sequencing. The quality and quantity of RNA were determined using a 2100 Bioanalyzer (Agilent, Santa Clara, CA, USA) and Invitrogen's Qubit RNA BR assay kit (Thermo Fisher Scientific, NY, USA), respectively. Prior to sequencing, the bacterial ribosomal RNA was removed and then converted to RNA-seq libraries using a TruSeq Stranded Total RNA with Ribo-Zero (bacteria) kit (Illumina, San Diego, CA, USA). The constructed library was sequenced on an Illumina HiSeq 4000 platform in the KAUST Genomics Core lab. All high-throughput sequencing files were deposited in the Short Read Archive (SRA) of the European Nucleotide Archive (ENA) under study accession number PRJEB33442.
2.9 Biofilm-associated gene analysis and annotation
The indexes, adapters and low-quality reads with a phred cutoff < Q30 were removed/trimmed using BBMap25 after sequencing. To further elucidate the effects of the nanoparticles on biofilm formation, a new QS database was manually curated for this study. QS-associated genes based on the QS pathway on the Kyoto Encyclopedia of Genes and Genomes (KEGG) map02024 (https://www.genome.jp/dbget-bin/www_bget?pathway+map02024) were collated. The filtered metagenomic DNA and metatranscriptomic RNA reads were then respectively aligned to the QS database using a basic local alignment search tool (version 2.8.1).26 A read was classified as a QS-like fragment with an E-value (≤10−5), a 90% nucleotide similarity and at least a 50 bp alignment length to the database. Owing to the various sequencing depths among the different samples (Table S3†), the abundance of QS-associated genes is shown as ppm unit (i.e., mapped reads per million total reads).To annotate the genes, the metagenomic DNA and metatranscriptomic RNA reads that passed through the quality control (QC) were respectively first assembled into contigs with MEGAHIT27 and annotated with PROKKA.28 The obtained contigs were then analyzed against the Kyoto Encyclopedia of Genes and Genomes (KEGG) database using KEGG pathways and modules29 with an E-value cutoff of 10−5. To better understand the effects of the nanoparticles on biofilm formation, this study focuses on the pathways which are related to biofilm formation (i.e., pathway 03070: bacterial secretion system; 02024: quorum sensing; 05111, 02025, 02026: biofilm formation; 02030: bacterial chemotaxis; 02040: flagellar assembly; 04810: regulation of actin-like cytoskeleton). The genes involved in each pathway are listed in the ESI.†
2.10 Analysis of antibiotic resistance genes and metal resistance genes
Similar to QS abundance analysis in section 2.9, the filtered metagenomic DNA and metatranscriptomic RNA reads were also aligned to the other local databases (i.e., structured antibiotic resistance genes (SARG, version 2.0) database,30 antibacterial biocide and metal resistance genes database (BacMet, version 1.1)31) through a basic local alignment search tool (version 2.8.1) with an E-value (≤10−5), a 90% nucleotide similarity and at least a 50 bp alignment length to the database. The abundance of ARGs and MRGs is shown in terms of ppm unit.3. Results and discussion
3.1 Reactor performance
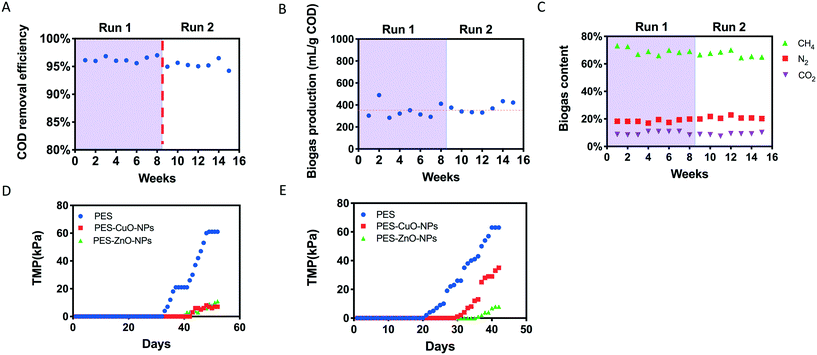
The reactor performed stably after connecting PES, PES-CuO-NP and PES-ZnO-NP membranes onto the system. It could achieve over 93% COD removal during the operation (Fig. 1A). This removal efficiency was similar to that achieved in earlier studies,20,23 implying that the nanoparticle embedded membranes did not affect anaerobic biodegradation. In addition, biogas production was stable with a ca. 70% methane content (v/v) within the biogas throughout the operation (Fig. 1B and C), suggesting that methanogenesis was not perturbed by the presence of heavy metal nanoparticles embedded on the membranes. These observations differ from that reported by an earlier study where CuO-NPs and ZnO-NPs were directly introduced to the anaerobic biodegradation stage, and detrimentally affected biogas and methane production.32 The lack of perturbation on anaerobic biodegradation and methanogenesis observed in this study may be due to fixation of the nanoparticles onto the polymeric membranes, which in turn minimizes leaching of nanoparticles into the reactor despite the high recirculation rate and biogas scouring. To verify this, we evaluated the concentration of Zn and Cu within our reactor. Our results indicated that both Zn and Cu concentrations were less than 300 μg L−1 (Fig. S4†), which were much lower than the nanoparticle dosage (i.e., 15 mg L−1 CuO and 120 mg L−1 ZnO) reported in the earlier study.32 In addition, biogenic sulfide could be generated under anaerobic conditions from the reduction of sulfate (present as trace elements in our synthetic wastewater, Table S2†), and sulfide could attenuate the toxicity of CuO and ZnO nanoparticles to methanogensis.33–353.2 Effect of CuO-NPs and ZnO-NPs on transmembrane pressure (TMP)
Although no significant effects were observed on anaerobic biodegradation and methanogenesis, the presence of CuO and ZnO nanoparticles resulted in distinct differences in the extent of membrane fouling compared to control membranes. In both runs, the PES control membrane achieved a faster TMP increment compared to the other two nanoparticle-embedded membranes (Fig. 1D). To illustrate, the PES membrane reached critical fouling on the 48th day in run 1, but not the PES-CuO-NP and PES-ZnO-NP membranes. Likewise, the PES-CuO-NP and PES-ZnO-NP membranes only showed 29 kPa and 10 kPa, respectively, for their TMPs while the PES membrane already reached critical fouling on the 40th day (>65 kPa) in run 2 (Fig. 1E). The TMP is perceived as one parameter to indicate the foulant layer.36 The slower increment in the TMPs of the PES-CuO-NP and PES-ZnO-NP membranes suggests that the CuO and ZnO nanoparticles can delay membrane biofouling, which could potentially be attributed to biocidal effects on microorganisms. Previous studies showed that the CuO and ZnO nanoparticles could inhibit the growth of both Gram-positive and Gram-negative microorganisms,37,38 leading to delayed biofilm formation.393.3 Effect on the biofilm matrix due to CuO and ZnO nanoparticles
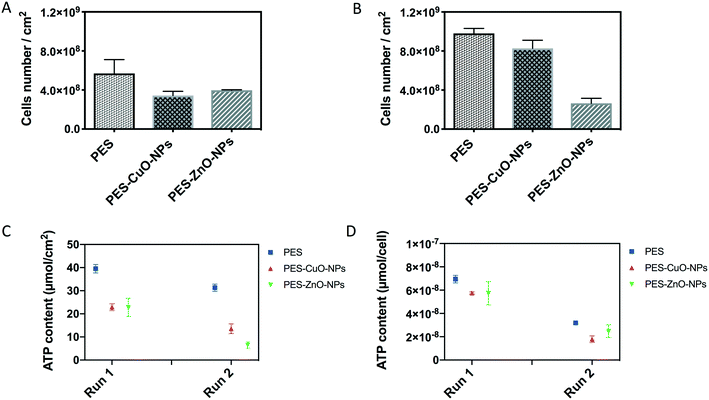
To further verify that the slower increment in the TMP was due to biocidal effects of CuO and ZnO nanoparticles, the biofilm attached on the membrane was harvested. Total cells, ATP, protein/polysaccharide contents and QS signal molecules were then quantified in the biofilm. The results showed that the PES membrane recovered from run 1 contained 5.69 × 108 cells per cm2. This cell number was significantly higher than those of the nanoparticle-embedded membranes (i.e., 3.43 × 108 and 2.61 × 108 cells per cm2 for the PES-CuO-NP and PES-ZnO-NP membranes, respectively (T-test, both P < 0.01)) (Fig. 2A). Although the cells in PES and PES-CuO-NPs in run 2 were higher than those observed in run 1 due to the biological variation within the AnMBR, a similar trend was still obtained in run 2, with significantly higher cell numbers on the control membrane than that on the nanoparticle-embedded membranes (t-test, both P < 0.01) (Fig. 2B). The total cells account for ca. 10% dry weight of a biofilm matrix.40 The lower microorganism counts on the PES-CuO-NP and PES-ZnO-NP membranes would imply that CuO and ZnO nanoparticles reduced the foulant on the membrane surfaces.However, the total cell count could not indicate the cell activity. Therefore, we quantified the ATP concentration in the biofilm matrix. The concentration of ATP is regarded as a proxy to estimate viable cells in biomass.41 The average ATP concentration in the biofilm matrix attached on the PES membrane was about 39.5 ± 1.9 nmol cm−2 in run 1 and 31.3 ± 1.6 nmol cm−2 in run 2. The presence of CuO and ZnO nanoparticles on the PES membrane significantly decreased the ATP concentration in both run 1 and run 2 (t-test, all P < 0.01) (Fig. 2C), indicating lower cell viability in the biofilm matrix of both nanoparticle modified membranes. The ATP concentration was further normalized with total cell numbers. The results indicated that there were 7.0 × 10−8 nmol per cell in the PES membrane, and this was higher than that in the PES-CuO-NP and PES-ZnO-NP membranes in run 1 (t-test, P = 0.00, 0.06, respectively). Similarly, CuO-NPs and ZnO-NPs significantly decreased the ATP concentration per cell by 43.5% and 22.4% compared to the control membrane (Fig. 2D). This decline in the ATP concentration per cell implied that both cell viability and microbial activity in the biofilm matrix were inhibited by the nanoparticles, reiterating the antibacterial effects of both CuO and ZnO nanoparticles.37,38
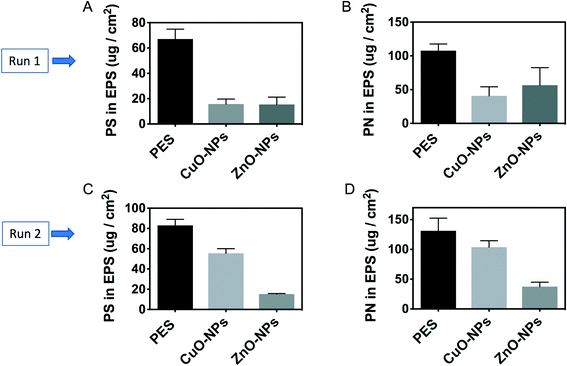
Besides microbial cells, polysaccharides and proteins account for approximately 90% dry weight of a biofilm matrix.40 The results in this study indicated that there were higher polysaccharide contents in the biofilm on the PES membrane than those on the nanoparticle-embedded membranes. To illustrate, the concentration of a polysaccharide in the soluble EPS of the biofilm attached onto PES was 67.1 ± 7.7 μg cm−2 in run 1. This concentration was significantly higher than those of the PES-CuO-NP (i.e., 16.2 ± 3.9 μg cm−2) and PES-ZnO-NP (i.e., 15.8 ± 5.7 μg cm−2) membranes (t-test, P < 0.01). In run 2, the concentrations of the polysaccharide on PES-CuO-NPs and PES-ZnO-NPs were 55.5 ± 4.6 μg cm−2 and 15.2 ± 0.7 μg cm−2, respectively. In contrast, 83.2 ± 6.1 μg cm−2 polysaccharide was detected on the PES control membrane (Fig. 3A and C). A similar decrease in the protein content in the biofilms on the nanoparticle-embedded membranes was observed in comparison to that in the control membranes (Fig. 3B and D). To further verify if this decrease in protein and polysaccharide contents on the overall membrane surface also translated to a lower EPS production per cell, we normalized the EPS content with the total cell number, and observed a significant decrease in the polysaccharide yield per cell (t-test, all P < 0.04) but no significant changes in the protein yield per cell across all the membranes (t-test, P > 0.05) (Fig. S3†). This suggested that the lower polysaccharide concentration on the nanoparticle-embedded membranes was due to both lower cell numbers and production per cell, while the lower protein yield was mainly due to lower cell numbers in the presence of CuO and ZnO nanoparticles.
Our observation was different from the previous studies which indicated that nanoparticles could stimulate EPS production.42,43 For instance, it was reported that 50 mg L−1 CuO nanoparticles could accelerate the EPS concentration in active sludge in wastewater treatment plants.42 ZnO nanoparticles at a 100 mg L−1 concentration were also observed to increase the EPS content in sludge from a sequencing batch reactor.43 However, these studies were conducted under aerobic conditions. In contrast, it was reported that 10–50 mg L−1 ZnO nanoparticles did not increase the EPS content in anaerobic granular sludge, while a higher concentration of 100 mg L−1 ZnO nanoparticles could significantly cause EPS to decrease.44 The reduction of total EPS is further attributed to the lower cell counts and microbial activity upon exposure to CuO and ZnO nanoparticles.44 The decline in protein and polysaccharide contents, along with total cell counts, indicated that CuO-NPs and ZnO-NPs could inhibit foulant layer formation onto the membrane surface. This phenomenon was further reiterated visually by the SEM images (Fig. S5†), which showed that the PES control membranes had a thicker cake layer than the modified membranes.
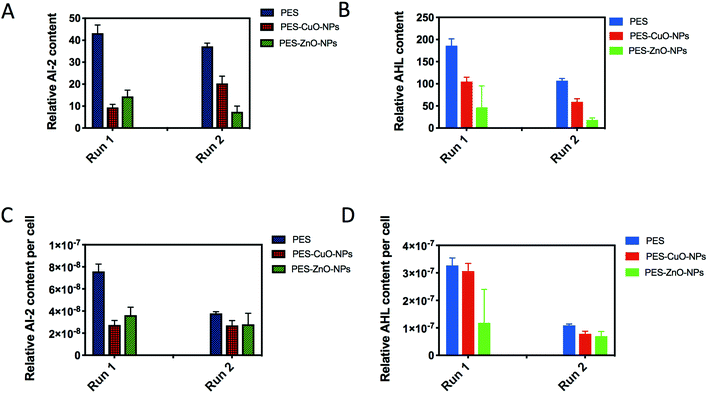
Lastly, we further examined AI-2 signals generated by both Gram-negative and Gram-positive bacteria for interspecies communication,45 as well as AHLs thought to be secreted mainly by Gram-negative bacteria for cell-to-cell communication.46 As a result of the lower cell counts, both PES-CuO-NPs and PES-ZnO-NPs showed significantly lower AI-2 and AHL contents in the biofilm (t-test, all P < 0.01) (Fig. 4A and B). After further normalization with total cells, a similar decline due to CuO and ZnO nanoparticles was observed (t-test, all P < 0.07) (Fig. 4C and D). The ZnO nanoparticle has been widely reported to decrease the QS concentration in the biofilm47,48 through disturbing QS-associated gene expression.49 However, whether CuO nanoparticles also impose a similar mechanism is not reported in any of the current literature.
3.4 Effects on biofilm-associated genes due to CuO and ZnO nanoparticles
Our results indicated that the CuO and ZnO nanoparticles could decrease the amount of QS molecules (i.e., AHL and AI-2) in the biofilm matrix. To further illustrate how these nanoparticles affect the production of signal molecules, the biofilm-associated genes were evaluated by omics-based approaches. As shown in Fig. 5A, there was no significant difference in the relative abundance of QS genes among all the membranes when analyzed through metagenomics (i.e., DNA-based). The metagenomics data were further annotated using the KEGG database, and it was found that the same subtypes of the biofilm-associated genes were detected for all the membranes (Fig. 5B). In contrast, when analyzed through metatranscriptomics (i.e., RNA-based), the relative abundance of QS-associated genes decreased from 22.8 ppm in the control membranes to 10.0 ppm (PES-CuO-NPs) and 7.5 ppm (PES-ZnO-NPs) (Fig. 5C). Furthermore, the number of annotated genes related to the QS pathway (pathway: 02024) decreased from 71 in the control membranes to 64 and 56 in the modified membranes (Fig. 5D). This result implied that although the diversity of QS-associated genes was not affected (as elucidated from metagenomics), the expression of these QS-associated genes was inhibited by the nanoparticles. Our metatranscriptomics analysis was further supported by the lower QS signal concentration in the biofilm exposed to the CuO and ZnO nanoparticles (Fig. 4).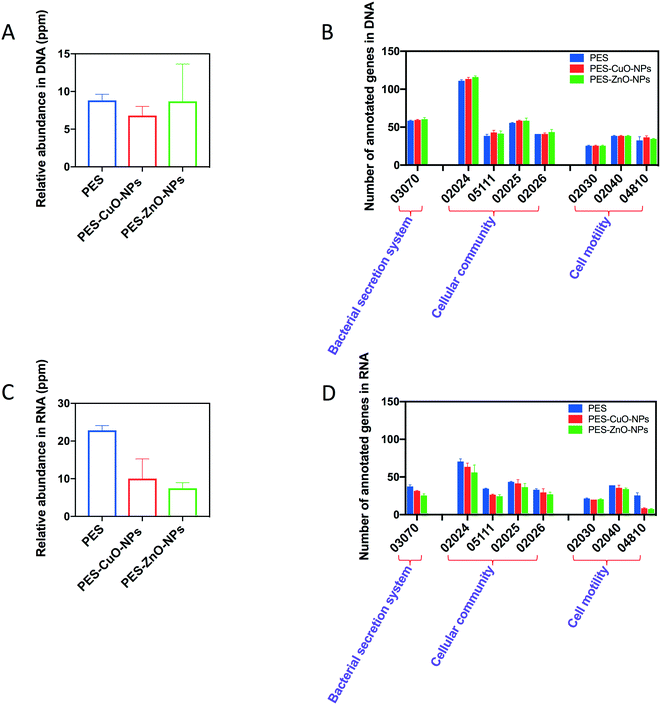 | ||
| Fig. 5 Relative abundance of quorum sensing (QS)-associated genes and the effects on KEGG biofilm formation associated pathways elucidated by metagenomics and metatranscriptomics. (A) Relative abundance of QS-associated genes by metagenomics, (B) relative abundance of KEGG annotated genes by metagenomics, (C) relative abundance of QS-associated gene transcripts by metatranscriptomics, and (D) relative abundance of KEGG annotated genes by metatranscriptomics. More information on the individual KEGG pathways can be found in the ESI.† | ||
Similarly, as shown in Fig. 5D, the nanoparticles also interrupted the expression of other biofilm-associated genes, and this could likely result in the decline of the EPS content in the biofilm matrix. To exemplify, gspI, gspJ, gspL and gspM, which encode genes related to protein secretion by the type II secretion system in Gram-negative bacteria,50 were not detected in PES-CuO-NPs and PES-ZnO-NPs but were present in the PES membrane. The lower expression of these genes would likely result in lower protein secretion and content in EPS of the biofilm matrix. In addition, the subtypes of cell motility (i.e., 02040 and 04810) also decreased and this would contribute to a delay in cell attachment and subsequent formation of the biofilm.51 A previous study observed that ZnO nanoparticles could inhibit EPS production and swarming motility.49 Our study reiterated this finding and further elucidated how nanoparticles affect EPS and motility in the biofilm.
3.5 Cu and Zn leached from membranes
Heavy metal nanoparticles exhibit antibacterial effects through four known mechanisms, namely, reactive oxygen species (ROS) production, release of metal ions, accumulation of nanoparticles on cell walls, and internalization of nanoparticles into cells which subsequently result in cell lysis.52 Given that our reactor was operated under anaerobic conditions, it is likely that the role of ROS in contributing to the antibacterial effect would be limited.53 Thus, it was hypothesized that other mechanisms (e.g. heavy metal ion release) would contribute to the antifouling effects. We quantified the amount of heavy metal ions in the reactor during the experiment. As shown in Fig. S4,† the Cu and Zn concentrations continuously increased after assembling the membranes (Fig. S4†), suggesting the gradual release of the heavy nanoparticles from the membranes into the reactor. When present in the ionic state, these metal ions induce toxicity because of their affinity for cellular components and biomolecules through the formation of metal–biomolecule complexes, eventually resulting in damage to cellular processes and enzymatic functions.54However, given that Cu and Zn were continuously released from the membranes, it means that the antifouling efficacy might decrease with time. This study only evaluated the membranes for 48 and 40 days (i.e., run 1 and run 2), and future studies should be conducted to assess the antifouling effect during the long-term operation. In addition, chemical cleaning is one of the most common strategies adopted by AnMBR operators to remove foulants on the membrane surface. Yet, this study did not apply chemical cleaning during the reactor operation, and therefore was unable to assess the stability of these nanoparticle-embedded membranes during chemical cleaning. Future studies should also include this study's limitation in their consideration.
3.6 Effects on the expression of ARGs and MRGs due to CuO and ZnO nanoparticles
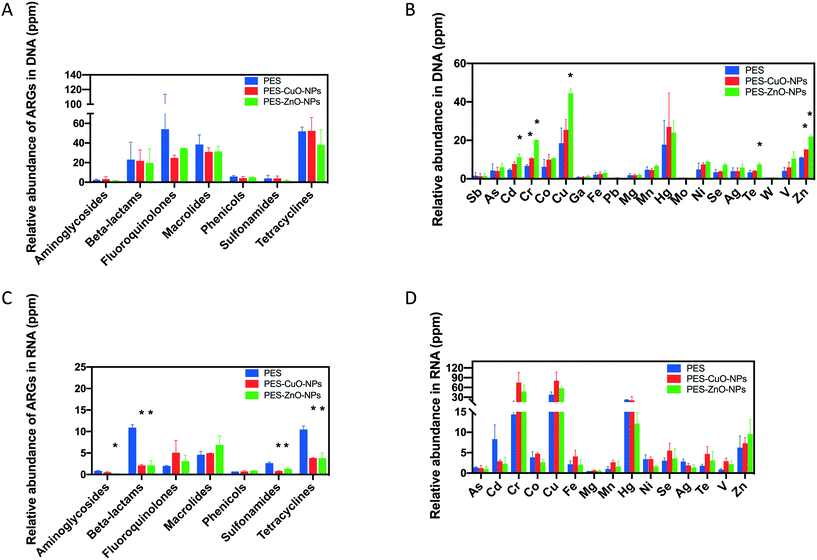
ARGs are increasingly recognized as emerging contaminants that can be disseminated into the environment through wastewater.55 Nanoparticles can produce ROS, which in turn promote the horizontal gene transfer of ARGs.56–58 Besides the contribution of ROS, the ubiquity of ARGs in wastewater can, in part, also be due to the presence of heavy metals that co-selects for both heavy metal and antibiotic resistance.59 In this study, although the presence of CuO and ZnO nanoparticles delayed membrane biofouling without adversely affecting anaerobic treatment of wastewater, it is equally important to assess whether the abundance of ARGs and MRGs changes due to the presence of these heavy metal nanoparticles.This study observed no difference in the detected ARG gene abundance among all the membranes by metagenomics (t-test, all P > 0.34) (Fig. 6A) and by metatranscriptomics (t-test, P > 0.07) (Fig. 6C). It is likely that ROS production in anaerobic systems might be negligible and did not contribute to dissemination of ARGs in this instance. Instead, the antibacterial effect of the nanoparticles under anaerobic conditions may be due to the release of heavy metal ions as observed in Fig. S4.† Our observation is different from a previous study which indicated that CuO and ZnO nanoparticles could increase ARGs60 at 200 mg L−1. However, this discrepancy in observations might be reasonable when considering the lower dosage of nanoparticles released from our membranes (Fig. S4†). Huang and his co-authors showed that low dosage of CuO and ZnO nanoparticles (i.e., 50 mg L−1) has no significant effects on the relative abundance of ARGs (i.e., sul I, sul II, tet Q and tet C).60 Shi et al. also concluded that less than 100 mg L−1 ZnO nanoparticles did not change the ARG abundance even after 21 days of exposure.61 Our study, along with the findings of earlier studies, suggests that exposure to heavy metal nanoparticles with low concentrations had no apparent stimulatory effects on the ARG abundance in the anaerobic system. Therefore, a key consideration when incorporating heavy metal nanoparticles as an antifouling strategy would be to consider their efficacy at the lowest possible concentration to prevent unintentional effects on ARG abundance.
For MRGs, it was found that both nanoparticles only significantly increased the relative abundance of MRG-Cu and MRG-Zn (t-test, P ≤ 0.05) (Fig. 6B). This observation coincided with the previous study which indicated that both CuO and ZnO nanoparticles could increase MRGs.60 However, the earlier study did not evaluate the MRG expression by metatranscriptomics. Interestingly, when metatranscriptomics was conducted, the results suggested that the nanoparticles applied in this study did not significantly stimulate the MRG expression (Fig. 6D). This is in contrast with an earlier study that noted an increase in copper resistance gene expression upon exposure to 1 mg L−1 CuO nanoparticles. However, the earlier study based their findings on a pure P. aeruginosa culture,62 which is not representative of the overall response exhibited by the mixed anaerobic microbial community in this study.
The variability in metagenomic and metatranscriptomic data could be explained by the metabolic costs associated with expressing MRGs gained through horizontal transfer or inheritance. Given that the microorganisms are only exposed to heavy metals with low concentrations, it may be metabolically expensive to express these genes. Gene expression costs bioenergy (e.g. ATP),63 and this energy currency was decreased by the presence of the nanoparticles (Fig. 2D) and hence may limit the ability of bacteria to switch on these genes.
4. Conclusions
Findings from this study suggest that the use of CuO and ZnO NP-embedded membranes can act as a potentially effective strategy to delay membrane biofouling in AnMBR. In the presence of CuO and ZnO NPs, a slower increment in the TMP was associated with a decrease in total attached cells, cell viability, and protein and polysaccharide contents, as well as microbial activity within the anaerobic biofilm matrix. The metatranscriptomic analysis revealed that both nanoparticles could inhibit the expression of not only QS-related genes, but also EPS and bacteria motility associated genes. Our results further indicated no apparent increment in the abundance of expressed ARGs and MRGs, suggesting that it can suffice as a safe and effective antifouling strategy that will not contribute to further stimulation of ARGs and MRGs within the reactor. This may also mean a low probability of disseminating ARGs and MRGs through the post-AnMBR effluent although more studies would have to be performed to verify this.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This study is supported by KAUST Center Competitive Funding FCC/1/1971-32-01 awarded to P.-Y. Hong.References
- B. Lew, S. Tarre, M. Beliavski, C. Dosoretz and M. Green, Anaerobic membrane bioreactor (AnMBR) for domestic wastewater treatment, Desalination, 2009, 243, 251–257 CrossRef CAS.
- H. Ozgun, R. K. Dereli, M. E. Ersahin, C. Kinaci, H. Spanjers and J. B. van Lier, A review of anaerobic membrane bioreactors for municipal wastewater treatment: integration options, limitations and expectations, Sep. Purif. Technol., 2013, 118, 89–104 CrossRef CAS.
- C.-H. Wei, M. Harb, G. Amy, P.-Y. Hong and T. Leiknes, Sustainable organic loading rate and energy recovery potential of mesophilic anaerobic membrane bioreactor for municipal wastewater treatment, Bioresour. Technol., 2014, 166, 326–334 CrossRef CAS PubMed.
- I. Martin, M. Pidou, A. Soares, S. Judd and B. Jefferson, Modelling the energy demands of aerobic and anaerobic membrane bioreactors for wastewater treatment, Environ. Technol., 2011, 32, 921–932 CrossRef CAS PubMed.
- A. L. Prieto, H. Futselaar, P. N. Lens, R. Bair and D. H. Yeh, Development and start up of a gas-lift anaerobic membrane bioreactor (Gl-AnMBR) for conversion of sewage to energy, water and nutrients, J. Membr. Sci., 2013, 441, 158–167 CrossRef CAS.
- P. Le-Clech, Membrane bioreactors and their uses in wastewater treatments, Appl. Microbiol. Biotechnol., 2010, 88, 1253–1260 CrossRef CAS PubMed.
- H.-C. Flemming, Reverse osmosis membrane biofouling, Exp. Therm. Fluid Sci., 1997, 14, 382–391 CrossRef CAS.
- A. Lim and R. Bai, Membrane fouling and cleaning in microfiltration of activated sludge wastewater, J. Membr. Sci., 2003, 216, 279–290 CrossRef CAS.
- H. Cheng, Y. Xie, L. F. Villalobos, L. Song, K.-V. Peinemann, S. Nunes and P.-Y. Hong, Antibiofilm effect enhanced by modification of 1, 2, 3-triazole and palladium nanoparticles on polysulfone membranes, Sci. Rep., 2016, 6, 24289 CrossRef CAS PubMed.
- J.-H. Kim, D.-C. Choi, K.-M. Yeon, S.-R. Kim and C.-H. Lee, Enzyme-immobilized nanofiltration membrane to mitigate biofouling based on quorum quenching, Environ. Sci. Technol., 2011, 45, 1601–1607 CrossRef CAS PubMed.
- M. Simoes, L. C. Simoes and M. J. Vieira, A review of current and emergent biofilm control strategies, LWT–Food Sci. Technol., 2010, 43, 573–583 CrossRef CAS.
- G. Scarascia, S. A. Yap, A. H. Kaksonen and P.-Y. Hong, Bacteriophage infectivity against Pseudomonas aeruginosa in saline conditions, Front. Microbiol., 2018, 9, 875 CrossRef PubMed.
- K. Zodrow, L. Brunet, S. Mahendra, D. Li, A. Zhang, Q. Li and P. J. Alvarez, Polysulfone ultrafiltration membranes impregnated with silver nanoparticles show improved biofouling resistance and virus removal, Water Res., 2009, 43, 715–723 CrossRef CAS PubMed.
- M. Zhang, R. W. Field and K. Zhang, Biogenic silver nanocomposite polyethersulfone UF membranes with antifouling properties, J. Membr. Sci., 2014, 471, 274–284 CrossRef CAS.
- Y. Ma, J. W. Metch, Y. Yang, A. Pruden and T. Zhang, Shift in antibiotic resistance gene profiles associated with nanosilver during wastewater treatment, FEMS Microbiol. Ecol., 2016, 92, fiw022 CrossRef PubMed.
- S. Zhou, Y. Zhu, Y. Yan, W. Wang and Y. Wang, Deciphering extracellular antibiotic resistance genes (eARGs) in activated sludge by metagenome, Water Res., 2019, 610–620 CrossRef CAS PubMed.
- O. Bondarenko, K. Juganson, A. Ivask, K. Kasemets, M. Mortimer and A. Kahru, Toxicity of Ag, CuO and ZnO nanoparticles to selected environmentally relevant test organisms and mammalian cells in vitro: a critical review, Arch. Toxicol., 2013, 87, 1181–1200 CrossRef CAS PubMed.
- S. Liang, K. Xiao, Y. Mo and X. Huang, A novel ZnO nanoparticle blended polyvinylidene fluoride membrane for anti-irreversible fouling, J. Membr. Sci., 2012, 394, 184–192 CrossRef.
- A. Y. Booshehri, R. Wang and R. Xu, Simple method of deposition of CuO nanoparticles on a cellulose paper and its antibacterial activity, Chem. Eng. J., 2015, 262, 999–1008 CrossRef.
- M. Harb, Y. Xiong, J. Guest, G. Amy and P.-Y. Hong, Differences in microbial communities and performance between suspended and attached growth anaerobic membrane bioreactors treating synthetic municipal wastewater, Environ. Sci.: Water Res. Technol., 2015, 1, 800–813 RSC.
- I. Nopens, C. Capalozza and P. A. Vanrolleghem, Stability analysis of a synthetic municipal wastewater, DEPARTMENT OF APPLIED MATHEMATICS, BIOMETRICS AND PROCESS CONTROL, 2001 Search PubMed.
- O. H. Lowry, N. J. Rosebrough, A. L. Farr and R. J. Randall, Protein measurement with the Folin phenol reagent, J. Biol. Chem., 1951, 193, 265–275 CAS.
- Y. Xiong, M. Harb and P.-Y. Hong, Characterization of biofoulants illustrates different membrane fouling mechanisms for aerobic and anaerobic membrane bioreactors, Sep. Purif. Technol., 2016, 157, 192–202 CrossRef CAS.
- P.-Y. Hong, E. Wheeler, I. K. Cann and R. I. Mackie, Phylogenetic analysis of the fecal microbial community in herbivorous land and marine iguanas of the Galapagos Islands using 16S rRNA-based pyrosequencing, ISME J., 2011, 5, 1461–1470 CrossRef PubMed.
- B. Bushnell, BBMap: a fast, accurate, splice-aware aligner, Lawrence Berkeley National Lab. (LBNL), Berkeley, CA (United States), 2014 Search PubMed.
- C. Camacho, G. Coulouris, V. Avagyan, N. Ma, J. Papadopoulos, K. Bealer and T. L. Madden, BLAST+: architecture and applications, BMC Bioinf., 2009, 10, 421 CrossRef PubMed.
- D. Li, C.-M. Liu, R. Luo, K. Sadakane and T.-W. Lam, MEGAHIT: an ultra-fast single-node solution for large and complex metagenomics assembly via succinct de Bruijn graph, Bioinformatics, 2015, 31, 1674–1676 CrossRef CAS PubMed.
- T. Seemann, Prokka: rapid prokaryotic genome annotation, Bioinformatics, 2014, 30, 2068–2069 CrossRef CAS PubMed.
- M. Kanehisa, M. Araki, S. Goto, M. Hattori, M. Hirakawa, M. Itoh, T. Katayama, S. Kawashima, S. Okuda and T. Tokimatsu, KEGG for linking genomes to life and the environment, Nucleic Acids Res., 2007, 36, D480–D484 CrossRef PubMed.
- X. Yin, X.-T. Jiang, B. Chai, L. Li, Y. Yang, J. R. Cole, J. M. Tiedje and T. Zhang, ARGs-OAP v2. 0 with an expanded SARG database and Hidden Markov Models for enhancement characterization and quantification of antibiotic resistance genes in environmental metagenomes, Bioinformatics, 2018, 34, 2263–2270 CrossRef CAS PubMed.
- C. Pal, J. Bengtsson-Palme, C. Rensing, E. Kristiansson and D. J. Larsson, BacMet: antibacterial biocide and metal resistance genes database, Nucleic Acids Res., 2013, 42, D737–D743 CrossRef PubMed.
- M. Luna-delRisco, K. Orupõld and H.-C. Dubourguier, Particle-size effect of CuO and ZnO on biogas and methane production during anaerobic digestion, J. Hazard. Mater., 2011, 189, 603–608 CrossRef CAS PubMed.
- E. Lombi, E. Donner, E. Tavakkoli, T. W. Turney, R. Naidu, B. W. Miller and K. G. Scheckel, Fate of zinc oxide nanoparticles during anaerobic digestion of wastewater and post-treatment processing of sewage sludge, Environ. Sci. Technol., 2012, 46, 9089–9096 CrossRef CAS PubMed.
- J. Gonzalez-Estrella, D. Puyol, R. Sierra-Alvarez and J. A. Field, Role of biogenic sulfide in attenuating zinc oxide and copper nanoparticle toxicity to acetoclastic methanogenesis, J. Hazard. Mater., 2015, 283, 755–763 CrossRef CAS PubMed.
- L. Otero-González, J. A. Field and R. Sierra-Alvarez, Inhibition of anaerobic wastewater treatment after long-term exposure to low levels of CuO nanoparticles, Water Res., 2014, 58, 160–168 CrossRef PubMed.
- B. Cho and A. Fane, Fouling transients in nominally sub-critical flux operation of a membrane bioreactor, J. Membr. Sci., 2002, 209, 391–403 CrossRef CAS.
- N. Jones, B. Ray, K. T. Ranjit and A. C. Manna, Antibacterial activity of ZnO nanoparticle suspensions on a broad spectrum of microorganisms, FEMS Microbiol. Lett., 2008, 279, 71–76 CrossRef CAS PubMed.
- A. Azam, A. S. Ahmed, M. Oves, M. Khan and A. Memic, Size-dependent antimicrobial properties of CuO nanoparticles against Gram-positive and-negative bacterial strains, Int. J. Nanomed., 2012, 7, 3527 CrossRef CAS PubMed.
- M. Eshed, J. Lellouche, S. Matalon, A. Gedanken and E. Banin, Sonochemical coatings of ZnO and CuO nanoparticles inhibit Streptococcus mutans biofilm formation on teeth model, Langmuir, 2012, 28, 12288–12295 CrossRef CAS PubMed.
- H.-C. Flemming and J. Wingender, The biofilm matrix, Nat. Rev. Microbiol., 2010, 8, 623 CrossRef CAS PubMed.
- J. W. Patterson, P. L. Brezonik and H. D. Putnam, Measurement and significance of adenosine triphosphate in activated sludge, Environ. Sci. Technol., 1970, 4, 569–575 CrossRef CAS.
- J. Hou, L. Miao, C. Wang, P. Wang, Y. Ao and B. Lv, Effect of CuO nanoparticles on the production and composition of extracellular polymeric substances and physicochemical stability of activated sludge flocs, Bioresour. Technol., 2015, 176, 65–70 CrossRef CAS PubMed.
- N.-Q. Puay, G. Qiu and Y.-P. Ting, Effect of Zinc oxide nanoparticles on biological wastewater treatment in a sequencing batch reactor, J. Cleaner Prod., 2015, 88, 139–145 CrossRef CAS.
- H. Mu, X. Zheng, Y. Chen, H. Chen and K. Liu, Response of anaerobic granular sludge to a shock load of zinc oxide nanoparticles during biological wastewater treatment, Environ. Sci. Technol., 2012, 46, 5997–6003 CrossRef CAS PubMed.
- M. B. Miller and B. L. Bassler, Quorum sensing in bacteria, Annu. Rev. Microbiol., 2001, 55, 165–199 CrossRef CAS PubMed.
- H. Withers, S. Swift and P. Williams, Quorum sensing as an integral component of gene regulatory networks in Gram-negative bacteria, Curr. Opin. Microbiol., 2001, 4, 186–193 CrossRef CAS PubMed.
- B. García-Lara, M. Saucedo-Mora, J. Roldán-Sánchez, B. Pérez-Eretza, M. Ramasamy, J. Lee, R. Coria-Jimenez, M. Tapia, V. Varela-Guerrero and R. García-Contreras, Inhibition of quorum-sensing-dependent virulence factors and biofilm formation of clinical and environmental P seudomonas aeruginosa strains by ZnO nanoparticles, Lett. Appl. Microbiol., 2015, 61, 299–305 CrossRef PubMed.
- J.-H. Lee, Y.-G. Kim, M. H. Cho and J. Lee, ZnO nanoparticles inhibit Pseudomonas aeruginosa biofilm formation and virulence factor production, Microbiol. Res., 2014, 169, 888–896 CrossRef CAS PubMed.
- N. A. Al-Shabib, F. M. Husain, F. Ahmed, R. A. Khan, I. Ahmad, E. Alsharaeh, M. S. Khan, A. Hussain, M. T. Rehman and M. Yusuf, Biogenic synthesis of Zinc oxide nanostructures from Nigella sativa seed: prospective role as food packaging material inhibiting broad-spectrum quorum sensing and biofilm, Sci. Rep., 2016, 6, 36761 CrossRef CAS PubMed.
- A. P. Pugsley, The complete general secretory pathway in gram-negative bacteria, Microbiol. Mol. Biol. Rev., 1993, 57, 50–108 CAS.
- J. D. Shrout, D. L. Chopp and C. L, Just, M. Hentzer, M. Givskov and M. R. Parsek, The impact of quorum sensing and swarming motility on Pseudomonas aeruginosa biofilm formation is nutritionally conditional, Mol. Microbiol., 2006, 62, 1264–1277 CrossRef CAS PubMed.
- A. B. Djurisic, Y. H. Leung, A. M. Ng, X. Y. Xu, P. K. Lee, N. Degger and R. Wu, Toxicity of metal oxide nanoparticles: mechanisms, characterization, and avoiding experimental artefacts, Small, 2015, 11, 26–44 CrossRef CAS PubMed.
- Y. Yang, C. Zhang and Z. Hu, Impact of metallic and metal oxide nanoparticles on wastewater treatment and anaerobic digestion, Environ. Sci.: Processes Impacts, 2012, 15, 39–48 Search PubMed.
- J. L. Hobman and L. C. Crossman, Bacterial antimicrobial metal ion resistance, J. Med. Microbiol., 2015, 64, 471–497 CrossRef CAS PubMed.
- P.-Y. Hong, T. Julian, M.-L. Pype, S. Jiang, K. Nelson, D. Graham, A. Pruden and C. Manaia, Reusing treated wastewater: consideration of the safety aspects associated with antibiotic-resistant bacteria and antibiotic resistance genes, Water, 2018, 10, 244 CrossRef.
- Y. Wang, J. Lu, L. Mao, J. Li, Z. Yuan, P. L. Bond and J. Guo, Antiepileptic drug carbamazepine promotes horizontal transfer of plasmid-borne multi-antibiotic resistance genes within and across bacterial genera, ISME J., 2019, 13, 509 CrossRef CAS PubMed.
- D. Mantilla-Calderon, M. J. Plewa, G. Michoud, S. Fodelianakis, D. Daffonchio and P. Hong, Water disinfection byproducts increase natural transformation rates of environmental DNA in Acinetobacter baylyi ADP1, Environ. Sci. Technol., 2019, 6520–6528 CrossRef CAS PubMed.
- N. Augsburger, D. Mantilla-Calderon, D. Daffonchio and P. Hong, Acquisition of Extracellular DNA by Acinetobacter baylyi ADP1 in response to Solar and UV-C254nm Disinfection, Environ. Sci. Technol., 2019, 10312–10319 CrossRef CAS PubMed.
- C. Baker-Austin, M. S. Wright, R. Stepanauskas and J. McArthur, Co-selection of antibiotic and metal resistance, Trends Microbiol., 2006, 14, 176–182 CrossRef CAS PubMed.
- H. Huang, Y. Chen, S. Yang and X. Zheng, CuO and ZnO nanoparticles drive the propagation of antibiotic resistance genes during sludge anaerobic digestion: possible role of stimulated signal transduction, Environ. Sci.: Nano, 2019, 6, 528–539 RSC.
- J. Shi, Y. Su, Z. Zhang, H. Wei and B. Xie, How do zinc oxide and zero valent iron nanoparticles impact the occurrence of antibiotic resistance genes in landfill leachate?, Environ. Sci.: Nano, 2019, 2141–2151 RSC.
- J. Guo, S.-H. Gao, J. Lu, P. L. Bond, W. Verstraete and Z. Yuan, Copper oxide nanoparticles induce lysogenic bacteriophage and metal-resistance genes in Pseudomonas aeruginosa PAO1, ACS Appl. Mater. Interfaces, 2017, 9, 22298–22307 CrossRef CAS PubMed.
- M. Lynch and G. K. Marinov, The bioenergetic costs of a gene, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 15690–15695 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Table: commercial price of nanoparticles; composition of synthetic wastewater; sequencing depth of each sample. Figures: schematic diagram of anaerobic membrane reactor configuration; surface images of membranes obtained by scanning electron microscopy; EPS content normalized by cells; metal concentration in the AnMBR; cross-sectional images of membranes obtained by scanning electron microscopy. Information: genes included in the various pathways examined in this study. See DOI: 10.1039/c9en00872a |
| This journal is © The Royal Society of Chemistry 2019 |