Open Access Article
Open Access ArticleHow reversible are the effects of silver nanoparticles on macrophages? A proteomic-instructed view†
Bastien
Dalzon‡
a,
Anaelle
Torres‡
a,
Hélène
Diemer
b,
Stéphane
Ravanel
c,
Véronique
Collin-Faure
a,
Karin
Pernet-Gallay
d,
Pierre-Henri
Jouneau
e,
Jacques
Bourguignon
 c,
Sarah
Cianférani
b,
Marie
Carrière
c,
Sarah
Cianférani
b,
Marie
Carrière
 f,
Catherine
Aude-Garcia§
a and
Thierry
Rabilloud
f,
Catherine
Aude-Garcia§
a and
Thierry
Rabilloud
 *a
*a
aChemistry and Biology of Metals, Univ. Grenoble Alpes, CNRS UMR5249, CEA, IRIG, CBM-ProMD, 17 Rue des Martyrs, F-38054 Grenoble Cedex 9, France. E-mail: thierry.rabilloud@cnrs.fr; Tel: +33 438 783 212
bLaboratoire de Spectrométrie de Masse BioOrganique (LSMBO), Université de Strasbourg, CNRS, IPHC UMR 7178, 67000 Strasbourg, France
cUniv. Grenoble Alpes, INRA, CNRS UMR5168, CEA, IRIG, Laboratory of Plant Cellular Physiology, Grenoble, F-38000 France
dUniv. Grenoble Alpes, Inserm U1216 Grenoble Institut des Neurosciences, 38000 Grenoble, France
eUniv. Grenoble Alpes, Modelization and Exploration of Materials, CEA-DRF-IRIG-DEPHY-MEM-LEMMA, F-38000 Grenoble, France
fUniv. Grenoble-Alpes, CEA, CNRS UMR 5819, IRIG, SyMMES, Chimie Interface Biologie pour l'Environnement, la Santé et la Toxicologie (CIBEST), F-38054 Grenoble, France
First published on 20th September 2019
Abstract
Silver nanoparticles are known to strongly affect biological systems, and numerous toxicological studies have investigated their effects. Most of these studies examine the effects immediately following acute exposure. In this work, we have conducted further investigation by studying not only the acute, post-exposure response, but also the cellular response after a 72 hour-recovery-phase post exposure. As a biological model we have used macrophages, which are very important cells with respect to their role in the immune response to particulate materials. To investigate the response of macrophages to nanoparticles and their recovery post exposure, we have used a combination of proteomics and targeted experiments. These experiments provided evidence that the cellular reaction to nanoparticles, including the reaction during the recovery phase, is a very active process involving massive energy consumption. Pathways such as the oxidative stress response, central and lipid metabolism, protein production and quality control are strongly modulated during the cellular response to nanoparticles, and restoration of basic cellular homeostasis occurs during the recovery period. However, some specialized macrophage functions, such as lipopolysaccharide-induced cytokine and nitric oxide production, did not return to their basal levels even 72 hours post exposure, showing that some effects of silver nanoparticles persist even after exposure has ceased.
Environmental significanceSilver nanoparticles are known to have profound effects on living cells. Because of their widespread use, contamination is almost unavoidable. In this context, it is important not only to assess the immediate effects of silver nanoparticles on cells, but also how they recover after exposure. Using macrophages as the target cell type and a combination of proteomic and targeted experiments, we show here that the recovery phase is not just a “return to normal” condition. For example, some of the specialized macrophage functions are still not restored after this time, showing that subtle but sustained effects can occur after a single exposure. Although cell survival is not affected, such effects may impact the health status of living beings. |
1. Introduction
Silver nanoparticles (AgNPs) are used as a biocide in a variety of consumer and medical products, because of the toxic effects that they have on microorganisms. However, as an unwanted side effect, AgNPs also show toxicity to mammalian cells. Thus, numerous toxicological studies have been conducted both in vitro in bacterial and eukaryotic model cell systems and in vivo in animal models, including at the microbiome level.1–3 Although some studies, especially those using animal models, have examined the effects of NPs after repeated, low-dose exposures,4–8 most of the cellular, toxicological studies have been conducted in an acute exposure mode, meaning that high doses have been used and exposure times have been short, spanning less than or equal to 24 hours. In these types of studies, toxicological endpoints are investigated at the end of the exposure period.9–16 In terms of exposure, the acute schemes relate most closely to accidental, acute exposures, while chronic schemes correspond highly to occupational exposures. It is of interest to study how biological systems recover after a single, high, non-lethal exposure. This is not a trivial question, as some mineral toxicants such as beryllium or crystalline silica show very prolonged effects, even after the exposure has ceased.17,18 This poor recovery has also been observed for silver nanomaterials such as silver nanowires19 and silver nanoparticles.20Many toxicological studies have reported pro-inflammatory responses5,11,12,21 and/or immunological effects15,22 pointing to macrophages as a cell type of major interest in toxicological studies of AgNPs. This is in accordance with the important scavenging function of macrophages in many different tissues.
This work investigates the recovery of macrophages after a unique, acute, but subtoxic dose of AgNPs. Targeted experiments carried out primarily on macrophages showed that several functional effects (e.g. LPS-induced cytokine and NO production, mitochondrial transmembrane potential and phagocytosis) were altered immediately after exposure to AgNPs. However, these functions tended to return to normal after a 72 h recovery period.23 We therefore decided to get a broader view of cellular recovery using a proteomic approach.
Proteomic approaches have been used to study cellular responses to AgNPs in several models such as intestinal cells9,24,25 or liver cells.10 This allowed several important pathways modulated in response to acute exposures to AgNPs to be highlighted. These included mitochondrial proteins, pointing at potential mitochondrial dysfunction,9,10,24 and proteins involved in intermediate metabolism9,10,24 or in inflammatory responses.10 The latter findings reinforced the interest in studying macrophages, which are major players in inflammation. We therefore decided to use proteomics to study the cellular responses using a macrophage J774 cell line, directly after acute exposure and after a recovery period had lapsed. This model cell line was selected because, unlike human monocyte cell lines,26,27 mouse macrophage cell lines such as RAW264.7 and J774 do not need to be chemically differentiated into macrophages, a process that deeply alters cell physiology and causes some cell death. Furthermore, as opposed to primary macrophages derived from human blood, these mouse cell lines do not show extensive variability from one experiment to another. This explains why these cell lines are extensively used as models for testing of a wide variety of nanomaterials,28–31 including large scale projects.32–34
2. Experimental
Most experiments have been performed essentially as described in previous publications.35–37 Details are given here for the sake of the consistency of the paper. All biological experiments were carried out at least on three independent biological replicates.2.1 Nanoparticles
PVP-coated silver nanoparticles were purchased from Sigma, directly as a concentrated suspension (catalog number #758329). Their characteristics have been published previously.23,38 These experiments confirmed the manufacturer's data and showed that the nanoparticles are spherical, with a diameter in the 50–110 nm range, and did not aggregate upon dilution in water or culture medium.2.2 Cell culture
The mouse macrophage cell line J774 was obtained from the European Cell Culture Collection (Salisbury, UK). The cells were cultured in DMEM + 10% fetal bovine serum (FBS). For routine culture, cells were seeded in non-adherent flasks (e.g. suspension culture flasks from Greiner) at 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per ml and harvested 48 hours later, at 1
000 cells per ml and harvested 48 hours later, at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000
000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per ml. Cell viability was measured by a dye exclusion assay, either with eosin (1 mg ml−1)39 under a microscope or with propidium iodide (1 μg ml−1)40 in a flow cytometry mode.
000 cells per ml. Cell viability was measured by a dye exclusion assay, either with eosin (1 mg ml−1)39 under a microscope or with propidium iodide (1 μg ml−1)40 in a flow cytometry mode.
For determination of the useful dose, cells were seeded at 500![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per ml. They were treated with nanoparticles on the following day and harvested after a further 24 hours in culture.
000 cells per ml. They were treated with nanoparticles on the following day and harvested after a further 24 hours in culture.
For treatment with nanoparticles, cells were seeded in classical cell culture flasks and left for 24 hours at 37 °C for cell adhesion and confluence. For the recovery condition, cells were treated with nanoparticles on the following day. After 24 hours of exposure, the cell culture medium was removed and replaced by fresh medium. For consistency reasons, this operation was also carried out on cells used for control and for acute exposure. Another medium culture change was carried out 36 hours after the initial medium change, i.e. mid-term of the 72 hour recovery period. Finally, acute exposure was carried out for the final 24 hours and the cells were used immediately afterwards.
2.3 Phagocytosis and particle internalization assay
The phagocytic activity was measured using fluorescent latex beads (1 μm diameter, green labelled, catalog number L4655 from Sigma). The beads were pre-incubated at a final concentration of 55 μg mL−1 for 30 minutes at 37 °C in PBS/FBS (v/v). Then, they were incubated with cells (5 μg mL−1) for 2 h 30 at 37 °C. Cells were harvested and washed with PBS. Cells were resuspended by vortexing with addition of 3/4 water volume and then 1/4 NaCl (35 mg mL−1) volume was added under vortexing in order to clean the cell surface of adsorbed particles. Cells were harvested in PBS with propidium iodide (1 μg mL−1). Viability and phagocytic activity were measured simultaneously by flow cytometry on a FacsCalibur instrument (Beckton Dickinson). The dead cells (propidium positive) were excluded from the analysis.2.4 Mitochondrial transmembrane potential measurement
The mitochondrial transmembrane potential was assessed by rhodamine 123 uptake. Cells were incubated with rhodamine 123 (80 nM) for 30 minutes at 37 °C, 5% CO2 then rinsed twice in cold glucose (1 mg mL−1)–PBS (PBSG) and harvested in cold PBSG supplemented with propidium iodide (1 μg mL−1). The mitochondrial potential of the cells was analysed by flow cytometry on a FacsCalibur instrument (Beckton Dickinson). The dead cells (propidium positive) were excluded from the analysis. The low rhodamine concentration was used to avoid intramitochondrial fluorescence quenching that would result in a poor estimation of the mitochondrial potential.412.5 Enzyme assays
The enzymes were assayed according to published procedures (see below).The cell extracts for enzyme assays were prepared by lysing the cells for 20 minutes at 0 °C in Hepes (20 mM, pH 7.5), MgCl2 (2 mM), KCl (50 mM), EGTA (1 mM), and tetradecyldimethylammonio propane sulfonate (SB 3-14) (0.15% (w/v)), followed by centrifugation at 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 15 minutes to clear the extract. The protein concentration was determined by a dye-binding assay.42 The dehydrogenase or dehydrogenase-coupled activities were assayed at 500 nm using the phenazine methosulfate/iodonitrotetrazolium coupled assay.43 The enzyme assay buffer contained 25 mM Hepes, NaOH (pH 7.5), 5 mM magnesium acetate, 100 mM potassium nitrate and 1% Triton X-100. It also contained 30 μM phenazine methosulfate, 200 μM iodonitrotetrazolium chloride, 250 μM of the adequate cofactor (NAD or NADP) and 1–5 mM of the organic substrate, which was used to start the reaction. For phosphate-dependent enzymes such as glyceraldehyde dehydrogenase (GAPDH) and purine phosphorylase (PNPH), 50 mM potassium phosphate (pH 7.5) was added to the enzyme assay buffer. Triose phosphate isomerase was assayed with dihydroxyacetone phosphate and a glyceraldehyde dehydrogenase-coupled assay.44 Purine phosphorylase (PNPH) was assayed by a xanthine oxidase-coupled assay.45 Hexokinase was assayed by a glucose phosphate dehydrogenase (G6PDH)-coupled assay.46 Biliverdine reductase was assayed at 450 nm as described.47 Pyridoxal kinase was assayed directly at 388 nm.48 Enolase was assayed at 340 nm by a pyruvate kinase–lactate dehydrogenase-coupled assay.49
000g for 15 minutes to clear the extract. The protein concentration was determined by a dye-binding assay.42 The dehydrogenase or dehydrogenase-coupled activities were assayed at 500 nm using the phenazine methosulfate/iodonitrotetrazolium coupled assay.43 The enzyme assay buffer contained 25 mM Hepes, NaOH (pH 7.5), 5 mM magnesium acetate, 100 mM potassium nitrate and 1% Triton X-100. It also contained 30 μM phenazine methosulfate, 200 μM iodonitrotetrazolium chloride, 250 μM of the adequate cofactor (NAD or NADP) and 1–5 mM of the organic substrate, which was used to start the reaction. For phosphate-dependent enzymes such as glyceraldehyde dehydrogenase (GAPDH) and purine phosphorylase (PNPH), 50 mM potassium phosphate (pH 7.5) was added to the enzyme assay buffer. Triose phosphate isomerase was assayed with dihydroxyacetone phosphate and a glyceraldehyde dehydrogenase-coupled assay.44 Purine phosphorylase (PNPH) was assayed by a xanthine oxidase-coupled assay.45 Hexokinase was assayed by a glucose phosphate dehydrogenase (G6PDH)-coupled assay.46 Biliverdine reductase was assayed at 450 nm as described.47 Pyridoxal kinase was assayed directly at 388 nm.48 Enolase was assayed at 340 nm by a pyruvate kinase–lactate dehydrogenase-coupled assay.49
2.6 NADP/NADPH and glucose assays
The glucose concentration in conditioned media was determined by a hexokinase–G6PDH assay.50 Briefly, culture media collected at the end of the exposure period were centrifuged for 15 min at 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g to pelletize the particulate material. The supernatant was collected, and diluted 100-fold in the dehydrogenase assay buffer described above, containing also 1 mM EGTA, 30 μM phenazine methosulfate, 200 μM iodonitrotetrazolium chloride, 10 IU ml−1 G6PDH and 7.5 IU ml−1 hexokinase. The reaction was started by the addition of ATP (1 mM final concentration) and the increase in absorbance at 500 nm read for 1 minute. The linear part of the absorbance curve was used to determine the reaction speed, which was proportional to the glucose concentration. Fresh culture media supplemented with 10% fetal calf serum, i.e. high glucose DMEM (4.5 g L−1 glucose), RPMI1640 (2 g L−1 glucose) and 199 medium (1 g L−1 glucose) were used as standards.
000g to pelletize the particulate material. The supernatant was collected, and diluted 100-fold in the dehydrogenase assay buffer described above, containing also 1 mM EGTA, 30 μM phenazine methosulfate, 200 μM iodonitrotetrazolium chloride, 10 IU ml−1 G6PDH and 7.5 IU ml−1 hexokinase. The reaction was started by the addition of ATP (1 mM final concentration) and the increase in absorbance at 500 nm read for 1 minute. The linear part of the absorbance curve was used to determine the reaction speed, which was proportional to the glucose concentration. Fresh culture media supplemented with 10% fetal calf serum, i.e. high glucose DMEM (4.5 g L−1 glucose), RPMI1640 (2 g L−1 glucose) and 199 medium (1 g L−1 glucose) were used as standards.
The NADP–NADPH concentration was determined using an adapted alkaline extraction buffer.51 Briefly, at the end of the exposure period, cells were collected by scraping, rinsed twice in PBS and pelleted. The packed cell pellet (PCP) volume was estimated, and the cells were lysed in 10 PCP volumes of 10 mM CAPS, 1 mM EGTA and 2 mM MgCl2 for 10 minutes on ice with occasional vortexing. The suspension was centrifuged (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g, 5 minutes, 4 °C), the viscous cell pellet discarded and the supernatant collected and split into two aliquots. The first aliquot was neutralized on ice by adding 0.1 volume of 1 M tricine. This aliquot contained both the oxidized and reduced forms of the pyridine nucleotides, and was also used to determine the protein concentration by a dye-binding assay.42
000g, 5 minutes, 4 °C), the viscous cell pellet discarded and the supernatant collected and split into two aliquots. The first aliquot was neutralized on ice by adding 0.1 volume of 1 M tricine. This aliquot contained both the oxidized and reduced forms of the pyridine nucleotides, and was also used to determine the protein concentration by a dye-binding assay.42
The other aliquot was heated at 60 °C for 30 minutes in a thermostated water bath to destroy the oxidized forms of the pyridine nucleotides.51 It was then cooled on ice, neutralized by adding 0.1 volume of 1 M tricine, and centrifuged for 10 minutes at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g at 4 °C to eliminate any particulate material.
000g at 4 °C to eliminate any particulate material.
The NADP–NADPH concentration was then determined by using an enzyme cycling assay and standard NADP solutions.52
2.7 NO production and cytokine production
The cells were grown to confluence in a 6-well plate and pre-treated with nanoparticles as described above. For the final 18 hours of culture, half of the wells were treated with 100 ng ml−1 LPS (from salmonella, purchased from Sigma), and arginine monohydrochloride was added to all the wells (5 mM final concentration) to give a high concentration of the substrate for nitric oxide synthase. After 18 hours of incubation, the cell culture medium was recovered and centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 10 minutes to remove cells and debris, and the nitrite concentration in the supernatants was read at 540 nm after addition of an equal volume of Griess reagent and incubation at room temperature for 30 minutes.
000g for 10 minutes to remove cells and debris, and the nitrite concentration in the supernatants was read at 540 nm after addition of an equal volume of Griess reagent and incubation at room temperature for 30 minutes.
For cytokine production, a commercial kit (BD Cytometric Bead Array, catalog number 552364 from BD Biosciences) was used.
2.8 F-actin staining
The experiments were performed essentially as previously described.53 The cells were recovered at the mid-term medium change (see above) and cultured on coverslips placed in 6-well plates. For the acute exposure condition, the cells were exposed to silver nanoparticles for 24 h at 37 °C. At the end of the exposure time, the cells were washed twice for 5 min at 4 °C in PBS and fixed in 4% paraformaldehyde for 30 min at room temperature. After two washes (5 min/4 °C in PBS), they were permeabilized in 0.1% Triton X100 for 5 min at room temperature. After two more washes in PBS, 500 nM Phalloidin-Atto 550 (Sigma) was added to the cells and left for 20 min at room temperature in the dark. Coverslip-attached cells were washed, placed on microscope slides (Thermo Scientific) using a Vectashield mounting medium containing DAPI (Eurobio) and imaged using a Zeiss LSM 800 confocal microscope. The images were processed using ImageJ software.2.9 Silver assay by ICP-MS
For measuring the Ag-NP uptake and release in cells, 2 mL cell cultures in 6-well plates were used, with the exposure scheme described above in section 2.2. At the end of the exposure period, the culture medium was removed and saved and the cell layer was gently rinsed twice with culture medium without serum.The cells were then lysed by scraping in 2 ml of lysis buffer (50 mM Hepes (pH 7.5), 4 mM magnesium acetate, 200 mM sorbitol, and 0.1% (w/v) tetradecyldimethylammonio propane sulfonate (SB 3-14)). The lysate was pipetted into a microtube and incubated for 20 minutes on ice to complete cell lysis. For determination of soluble silver, aliquots of the culture media and of the cell lysates were centrifuged for 45 minutes at 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g, and the upper half of the supernatant was collected.
000g, and the upper half of the supernatant was collected.
The samples were mineralized by the addition of one volume of suprapure 65% HNO3 and incubation on a rotating wheel at room temperature for 18 h.
Mineralized samples were diluted in 0.5% (v/v) HNO3 and analysed using an iCAP RQ quadrupole mass instrument (Thermo Fisher Scientific GmbH, Germany) equipped with an ASX-560 auto-sampler (Teledyne CETAC Technologies, Omaha, USA). The instrument was used with a MicroMist U-Series glass concentric nebulizer, a quartz spray chamber cooled at 3 °C, a Qnova quartz torch, a nickel sample cone, and a nickel skimmer cone equipped with a high-sensitivity insert. 24Mg, 25Mg, 107Ag and 109Ag concentrations were determined using standard curves and corrected using an internal standard solution of 103Rh added online. Data integration was done using the Qtegra software (version 2.8.2944.115). The results were normalized using the Mg concentration (4 mM in the cellular extracts and 0.82 mM in culture medium). To take into account the cellular concentration effects, the protein concentration of the extracts was determined by a dye-binding assay.42
2.10 Proteomics
The 2D gel-based proteomic experiments were essentially carried out as previously described,35 at least on independent biological triplicates. However, detailed materials and methods are provided for the sake of paper consistency.![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g at room temperature for 1 h), and the protein concentration in the supernatant was determined by a dye-binding assay.42 Carrier ampholytes (Pharmalytes, pH 3–10) were added to a final concentration of 0.4% (w/v), and the samples were kept frozen at −20 °C until use.
000g at room temperature for 1 h), and the protein concentration in the supernatant was determined by a dye-binding assay.42 Carrier ampholytes (Pharmalytes, pH 3–10) were added to a final concentration of 0.4% (w/v), and the samples were kept frozen at −20 °C until use.
The strips were then placed in a Multiphor plate (GE Healthcare), and IEF was carried out with the following electrical parameters: 100 V for 1 hour, then 300 V for 3 hours, then 1000 V for 1 hour, then 3400 V up to 60–70 kVh. After IEF, the gels were equilibrated for 20 minutes in 125 mM Tris, 100 mM HCl, 2.5% SDS, 30% glycerol and 6 M urea.57 They were then transferred on top of the SDS gels and sealed in place with 1% agarose dissolved in 125 mM Tris, 100 mM HCl, 0.4% SDS and 0.005% (w/v) bromophenol blue.
For the global analysis of the spot abundance data, we used directly the spot abundance data as provided by the gel analysis software. The software directly normalizes each spot abundance by the sum of all spot abundances detected on the gel. These relative abundance data were used directly for global analysis using the PAST software suite64 without any transformation. No limitation in the number of principal components was implemented in the principal component analysis.
NanoLC-MS/MS analysis was performed using a nanoACQUITY Ultra-Performance-LC (Waters Corporation, Milford, USA) coupled to a Synapt™ High Definition Mass Spectrometer™ (Waters Corporation, Milford, USA), or to a TripleTOF 5600 (Sciex, Ontario, Canada).
The nanoLC system was composed of an ACQUITY UPLC® CSH130 C18 column (250 mm × 75 μm with a 1.7 μm particle size, Waters Corporation, Milford, USA) and a symmetry C18 precolumn (20 mm × 180 μm with a 5 μm particle size, Waters Corporation, Milford, USA). The solvent system consisted of 0.1% formic acid in water (solvent A) and 0.1% formic acid in acetonitrile (solvent B). 4 μL of the sample were loaded into the enrichment column over 3 min at 5 μL min−1 with 99% of solvent A and 1% of solvent B. Elution of the peptides was performed at a flow rate of 300 nL min−1 with a 8–35% linear gradient of solvent B in 9 minutes.
The Synapt™ High Definition Mass Spectrometer™ (Waters Corporation, Milford, USA) was equipped with a Z-spray ion source and a lock mass system. The system was fully controlled by MassLynx 4.1 SCN639 (Waters Corporation, Milford, USA). The capillary voltage was set at 2.8 kV and the cone voltage at 35 V. Mass calibration of the TOF was achieved using fragment ions from Glu-fibrino-peptide B in the [50;2000] m/z range. Online correction of this calibration was performed with Glu-fibrino-peptide B as the lock-mass. The ion (M + 2H)2+ at m/z 785.8426 was used to calibrate MS data and the fragment ion (M + H)+ at m/z 684.3469 was used to calibrate MS/MS data during the analysis.
For tandem MS experiments, the system was operated with automatic switching between the MS (0.5 s per scan in the m/z range [150;1700]) and MS/MS mode (0.5 s per scan in the m/z range [50;2000]). The two most abundant peptides (intensity threshold 20 counts per s), preferably doubly and triply charged ions, were selected in each MS spectrum for further isolation and CID fragmentation using a collision energy profile. Fragmentation was performed using argon as the collision gas.
Mass data collected during analysis were processed and converted into .pkl files using ProteinLynx Global Server 2.3 (Waters Corporation, Milford, USA). Normal background subtraction was used for both MS and MS/MS with a 5% threshold and polynomial correction of order 5. Smoothing was performed on MS/MS spectra (Savitzky–Golay, 2 iterations, window of 3 channels). Deisotoping was applied for MS (medium deisotoping) and for MS/MS (fast deisotoping).
The TripleTOF 5600 (Sciex, Ontario, Canada) was operated in positive mode, with the following settings: ion spray voltage floating (ISVF) 2300 V, curtain gas (CUR) 10, interface heater temperature (IHT) 150, ion source gas 1 (GS1) 2, declustering potential (DP) 80 V. The information-dependent acquisition (IDA) mode was used with Top 10 MS/MS scans. The MS scan had an accumulation time of 250 ms in the m/z [400;1250] range and the MS/MS scans 100 ms in the m/z [150;1800] range in high sensitivity mode. Switching criteria were set to ions with a charge state of 2–4 and an abundance threshold of more than 500 counts; the exclusion time was set at 4 s. The IDA rolling collision energy script was used for automatically adapting the CE. Mass calibration of the analyser was achieved using peptides from digested BSA. The complete system was fully controlled by AnalystTF 1.7 (Sciex). Raw data collected were processed and converted with MSDataConverter in the .mgf peak list format.
For protein identification, the MS/MS data were interpreted using a local Mascot server with the MASCOT 2.4.1 algorithm (Matrix Science, London, UK) against UniProtKB/SwissProt (version 2016_01, 550![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 299 sequences). The research was carried out in all species. Spectra were searched with a mass tolerance of 15 ppm for MS and 0.05 Da for MS/MS data, allowing a maximum of one trypsin missed cleavage. Carbamidomethylation of cysteine residues and oxidation of methionine residues were specified as variable modifications. Protein identifications were validated with at least two peptides with a Mascot ion score above 30.
299 sequences). The research was carried out in all species. Spectra were searched with a mass tolerance of 15 ppm for MS and 0.05 Da for MS/MS data, allowing a maximum of one trypsin missed cleavage. Carbamidomethylation of cysteine residues and oxidation of methionine residues were specified as variable modifications. Protein identifications were validated with at least two peptides with a Mascot ion score above 30.
2.11 Electron microscopy
J774 macrophage cells were grown on coverslips and treated with AgNPs as described above. Cells were allowed to recover for 72 hours before being fixed with 2.5% glutaraldehyde in 0.1 M cacodylate buffer (pH 7.4) for 2 hours at room temperature. Cells were then washed with buffer, post fixed with 1% osmium tetroxide in the same buffer for 1 hour at 4°, and washed with water. Cells were then dehydrated through a series of graded alcohol (30–60–90–100–100–100%) and infiltrated with a mix of 1/1 epon/alcohol (100%) for 1 hour before several baths of fresh epon (Fluka) for 3 hours. Finally, a capsule full of epon was placed on the surface of the cells and the resin was allowed to polymerise for 72 h at 60 °C. The polymerised bloc was then detached from the coverslip with HF (48%) over 1 hour. Ultrathin sections of the cell monolayer were cut with an ultramicrotome (Leica). For transmission electron microscopy (TEM) observation, the sections were post-stained with 5% uranyl acetate and 0.4% lead citrate and observed with a transmission electron microscope at 80 kV (JEOL 1200EX). Images were acquired with a digital camera at magnification ranging from 15k to 50k (Veleta, Olympus). For scanning transmission electron microscopy observation (STEM) and energy dispersive spectroscopic analysis (EDS), the sections were not post-stained. They were coated with a thin layer of carbon and observed and analyzed with an OSIRIS microscope (TECNAI) operating at 200 kV.3. Results
3.1 Silver accumulation upon treatment with silver nanoparticles and after recovery
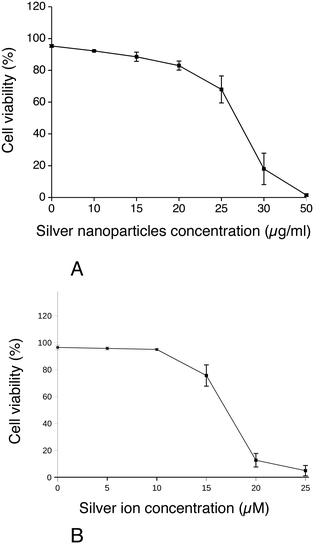
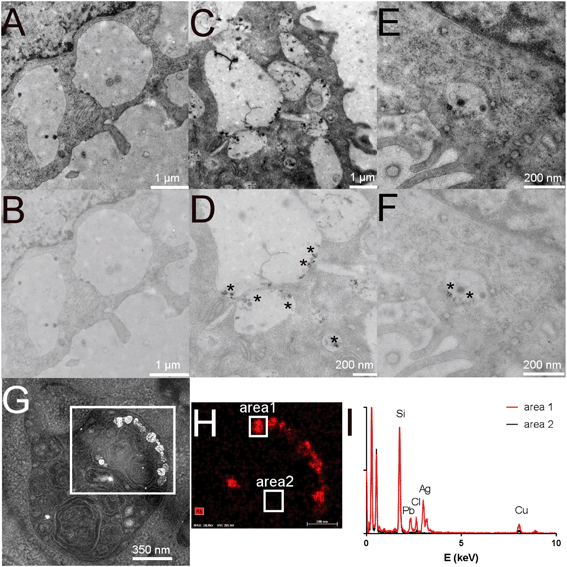
First, the effect of silver nanoparticles and soluble silver on cell viability was assayed, and the results are shown in Fig. 1. For all the subsequent experiments, a dose of 20 μg ml−1 was selected, corresponding to the lethal dose 20% (LD20). This dose ensures strong effects on cells while keeping cellular mortality at an acceptable level for subsequent experiments. We also checked the presence of nanoparticles in cells by electron microscopy. Control cells did not show any electron-dense materials (Fig. 2A and B), while cells exposed to Ag nanoparticles acutely showed some electron-dense particles located in large membrane compartments called macropinosomes (Fig. 2C and D). After the recovery period, these electron dense particles were still observed in the macropinosomes (Fig. 2E and F). Energy dispersive spectroscopic analysis of these electron-dense features confirmed that they contained Ag (Fig. 2G–I).Silver accumulation in cells was measured by ICP-MS. The results are summarized in Table 1. They showed a strong accumulation of silver in cells at the end of the 24 hour exposure followed by a moderate loss of total silver. The cells were able to cycle once during the 72 h recovery period, as shown by the increase in the protein amount in the extracts, which resulted in a further reduction of the cellular silver content. A moderate but detectable silver excretion was also evidenced. It should be also noted that the protein concentration is used in toxicology as an indicator for cell number,67,68
| Unexposed | Exposed to 20 μg ml−1 AgNPs | 72 h recovery after exposure to AgNPs | |
|---|---|---|---|
| a This value contains both the soluble silver that is actively excreted by cells during the exposure and the soluble silver that arises from nanoparticle dissolution directly in the culture medium. b Defined as soluble silver in medium/total silver (i.e. silver in medium + silver in cells). | |||
| Protein concentration in the cell extract (mg ml−1) | 0.352 ± 0.02 | 0.322 ± 0.02 | 0.657 ± 0.04 |
| Total silver concentration in the cell extract (μg l−1) | 0 | 9794 ± 1159 | 8258 ± 1536 |
| Soluble silver concentration in the cell extract (μg l−1) | 0 | 39.86 ± 11.83 | 47.57 ± 3.11 |
| Soluble silver concentration in culture medium (μg l−1) | 457 ± 64a | 142 ± 4 | |
| Normalized silver content (ng Ag mg−1 protein) | 0 | 30![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 608 ± 5055 608 ± 5055 |
12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 530 ± 1984 530 ± 1984 |
| Soluble silver fraction in cells | 0 | 0.0041 ± 0.0014 | 0.0059 ± 0.001 |
| Fraction of silver excretedb | 0 | ND | 0.017 ± 0.003 |
3.2 Global analysis of the proteomic results
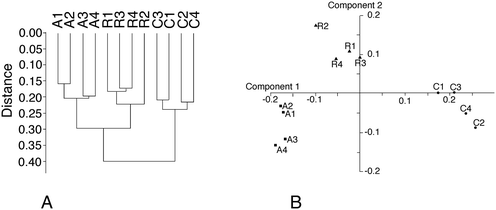
From the total protein expression data obtained from the gel analysis software, a subset of variable proteins (p < 0.25 in either the acute vs. control or the recovery vs. control comparisons) was extracted. This allowed the noise brought by proteins that do not show a consistent variation in the biological phenomena under investigation to be decreased. This subset was then tested using the PAST statistical suite.64 As a first test, a hierarchical clustering was performed, and the results are shown in Fig. 3A. Clustering clearly separates the control group from the two silver-treated ones, but gives no information regarding which silver treatment condition is closer to the control. Principal component analysis gave a more valuable indication, as shown in Fig. 3B. The recovery group appeared to be in an intermediate position between the control and acute groups along component one, which carries most of the variance. This is an expected position in a “return to normal” model during the recovery phase. However, the recovery group separated from both the control and the acute groups in component 2. This suggested that the three conditions were fairly different from each other. This was further confirmed by analysis of similarities (ANOSIM), which gave the following results for the p-values of the pairwise comparisons:| p = 0.031 for the acute vs. control comparison |
| p = 0.032 for the recovery vs. control comparison |
| p = 0.027 for the acute vs. recovery comparison |
We then divided this dataset of 239 spots into lists (Table S2†). The first list, labelled as “returning spots”, contained the spots for which the amplitude of the change in abundance between the recovery and control stages is lower than the amplitude of the change in abundance between the acute exposure and control stages. 135 spots belonged to this list. It was then divided into subsets. The first subset contained “fast returning spots”. It was defined as spots for which the quantitative change in the acute vs. control comparison shows an amplitude at least twice that in the recover vs. control comparison (i.e. more than 50% convergence). 44 spots belonged to this subset. The other subset of this list contained the 91 “slow returning spots”, for which the return-to-normal trend was lower than 50% in the 72 hour recovery period.
The other list, labelled as “diverging spots”, contained the 104 spots for which the amplitude of the change in abundance between the recovery and control stages is higher than the amplitude of the change in abundance between the acute exposure and control stages. This list was also divided into subsets. The “fast diverging spots” subset was defined as the spots for which the quantitative change in the recover vs. control comparison was at least 1.5 higher than the quantitative change in the acute vs. control comparison (i.e. more than 50% divergence). 53 spots belonged to this subset. The other subset of this list contained the 51 “slow diverging spots”, for which the diverging trend was lower than 50% in the 72 hour recovery period.
3.3 Detailed analysis of the proteomic results
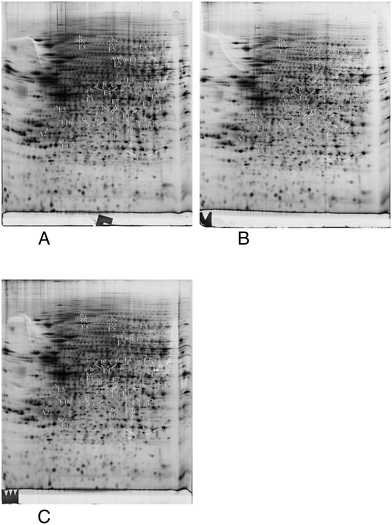
In order to gain more precise insights, we investigated which proteins were significantly altered (p < 0.05) between the control group and at least one of the silver-treated groups. The list of modulated proteins is given in Table 2 and the corresponding 2D gel images are shown in Fig. 4. As 2D gels separate protein forms, a variation in a spot does not necessarily mean that the whole amount of the protein is modulated. In fact, a single protein can be represented by various 2D gel spots, which do not necessarily all change upon the treatment with nanoparticles. To alleviate this problem, we systematically identified by mass spectrometry not only the modulated spots, but also the neighboring spots to check for such isoform issues. The quantitative results have been integrated in Table 2 and the mass spectrometry identification parameters are detailed in Table S3.† The results for some classes (apoptosis, cell fate and energy metabolism) are also shown in gel images in Fig. 4, and the results for the other classes are shown in Fig. S1–S5.† For global analysis of the modulated proteins, we used the DAVID annotation tool,69,70 available at https://david.ncifcrf.gov/, and the results are shown in Tables S4 and S5.† The results showed that some functional classes such as energy metabolism, cytoskeleton especially actin cytoskeleton, protein folding and quality control (especially the ubiquitin/proteasome pathway), nucleotide and nucleic acid metabolism, cell signaling, oxidative stress response and detoxification proteins are strongly represented among the modulated proteins. This prompted us to carry out validation experiments on some of these proteins or pathways.| Spot number | Protein name | Accession number | Ratio acute/ctrl | T test acute vs. ctrl | Ratio recov/ctrl | T test recov vs. ctrl |
|---|---|---|---|---|---|---|
| ∑: sum of the different spots identified for the same protein. | ||||||
| A1 | Casp3 | P70677 | 0.59 | 0.04 | 1.00 | 0.99 |
| A2 | Efhd2 | Q9D8Y | 1.24 | 0.02 | 1.18 | 0.23 |
| C1 | Actin | P60710 | 1.51 | <0.01 | 1.67 | 0.01 |
| C2a | Actn4/1 | P57780 | 1.45 | 0.04 | 1.18 | 0.16 |
| C2b | Actn4/2 | P57780 | 1.43 | 0.04 | 1.38 | 0.01 |
| C2c | Actn4/3 | P57780 | 1.25 | 0.33 | 1.17 | 0.03 |
| C2d | Actn4/4 | P57780 | 1.44 | 0.20 | 1.28 | 0.05 |
| C2S | ∑ Actn4 | P57780 | 1.36 | 0.16 | 1.23 | 0.02 |
| C3a | Arp2/1 | P61161 | 0.90 | 0.06 | 0.76 | <0.01 |
| C3b | Arp2/2 | P61161 | 1.10 | 0.41 | 0.94 | 0.63 |
| C3c | Arp2/3 | P61161 | 1.19 | 0.04 | 1.32 | <0.01 |
| C3S | ∑ Arp2 | P61161 | 1.15 | 0.07 | 1.14 | 0.06 |
| C4a | Arpc2/1 | Q9CVB6 | 0.57 | 0.03 | 0.52 | 0.02 |
| C4b | Arpc2/2 | Q9CVB6 | 0.76 | 0.04 | 0.78 | 0.06 |
| C4c | Arpc2/3 | Q9CVB6 | 0.91 | 0.51 | 1.12 | 0.41 |
| C4S | ∑Arpc2 | Q9CVB6 | 0.80 | 0.11 | 0.90 | 0.39 |
| C5a | Arpc5/1 | Q9CPW4 | 0.59 | 0.03 | 0.25 | 0.01 |
| C5b | Arpc5/2 | Q9CPW4 | 1.06 | 0.51 | 0.99 | 0.90 |
| C5S | ∑ Arpc5 | Q9CPW4 | 0.88 | 0.30 | 0.71 | 0.04 |
| C6a | Capg/1 | P24452 | 0.81 | 0.01 | 1.00 | 0.97 |
| C6b | Capg/2 | P24452 | 1.16 | 0.15 | 1.04 | 0.58 |
| C6S | ∑ capg | P24452 | 0.35 | 0.39 | 0.35 | 0.39 |
| C7 | Capza2 | P47754 | 1.30 | 0.01 | 1.28 | 0.01 |
| C8a | Capzb/1 | P47757 | 1.07 | 0.37 | 1.20 | 0.06 |
| C8b | Capzb/2 | P47757 | 0.80 | 0.02 | 1.01 | 0.95 |
| C8S | ∑ capzb | P47757 | 0.98 | 0.60 | 1.13 | 0.14 |
| C9a | Cof/1 | P18760 | 0.67 | 0.05 | 0.60 | 0.03 |
| C9b | Cof/2 | P18760 | 1.49 | 0.01 | 1.26 | 0.10 |
| C9c | Cof/3 | P18760 | 0.74 | <0.01 | 0.73 | <0.01 |
| C9S | ∑ Cof | P18760 | 0.86 | 0.02 | 0.79 | <0.01 |
| C10a | Dyncli2/1 | O88487 | 1.21 | 0.49 | 2.20 | <0.01 |
| C10b | Dyncli2/2 | O88487 | 1.43 | 0.26 | 1.69 | 0.04 |
| C10S | ∑ dyncli2 | O88487 | 1.36 | 0.30 | 1.84 | 0.01 |
| C11a | Gelsolin/1 | P13020 | 1.16 | 0.44 | 0.94 | 0.71 |
| C11b | Gelsolin/2 | P13020 | 1.16 | 0.36 | 1.00 | 0.97 |
| C11c | Gelsolin/3 | P13020 | 1.39 | 0.16 | 1.27 | 0.26 |
| C11d | Gelsolin/4 | P13020 | 1.57 | 0.05 | 1.32 | 0.13 |
| C11S | ∑ gelsolin | P13020 | 1.33 | 0.15 | 1.15 | 0.39 |
| C12 | Gmfg | Q9ERL7 | 0.54 | <0.01 | 0.66 | <0.01 |
| C13 | Lasp1 | Q61792 | 0.41 | <0.01 | 0.38 | <0.01 |
| C14 | Ml12b | Q3THE2 | 0.92 | 0.51 | 0.81 | 0.01 |
| C15a | Moesin/1 | P26041 | 0.94 | 0.75 | 0.86 | 0.41 |
| C15b | Moesin/2 | P26041 | 0.90 | 0.53 | 0.78 | 0.08 |
| C15c | Moesin/3 | P26041 | 1.22 | 0.17 | 0.89 | 0.13 |
| C15d | Moesin/4 | P26041 | 1.18 | 0.07 | 1.09 | 0.10 |
| C15e | Moesin/5 | P26041 | 1.52 | 0.03 | 1.24 | 0.04 |
| C15S | ∑ Moesin | P26041 | 1.20 | 0.08 | 1.01 | 0.91 |
| C16 | Mtap | Q9CQ65 | 1.04 | 0.83 | 1.30 | 0.02 |
| C17 | RhoA | Q9QUI0 | 0.59 | <0.01 | 0.76 | 0.02 |
| C18a | Rhogdi1/1 | Q99PT1 | 0.71 | <0.01 | 0.67 | <0.01 |
| C18b | Rhogdi1/2 | Q99PT1 | 1.16 | 0.13 | 1.11 | 0.08 |
| C18S | ∑ RhoGdi1 | Q99PT1 | 0.99 | 0.85 | 0.95 | 0.28 |
| C19a | Rhogdi2/1 | Q61599 | 0.64 | <0.01 | 0.54 | <0.01 |
| C19b | Rhogdi2/2 | Q61599 | 1.00 | 0.99 | 0.95 | 0.17 |
| C19S | ∑ rhogdi2 | Q61599 | 0.87 | 0.02 | 0.80 | <0.01 |
| C20a | Stmn/1 | P54227 | 0.84 | 0.19 | 0.70 | 0.01 |
| C20b | Stmn/2 | P54227 | 0.60 | 0.02 | 0.53 | 0.01 |
| C20c | Stmn/3 | P54227 | 0.88 | 0.14 | 0.76 | 0.01 |
| C20S | ∑ Stmn | P54227 | 0.83 | 0.05 | 0.71 | 0.01 |
| C21a | Sw70/1 | Q6A028 | 1.63 | 0.02 | 1.50 | 0.20 |
| C21b | Sw70/2 | Q6A028 | 2.02 | 0.02 | 1.76 | 0.08 |
| C21S | ∑ Sw70 | Q6A028 | 1.77 | 0.01 | 1.60 | 0.14 |
| C22 | Tbcb | Q9D1E6 | 1.31 | 0.03 | 1.11 | 0.45 |
| C23a | Tctp/1 | P63028 | 0.60 | <0.01 | 0.54 | <0.01 |
| C23b | Tctp/2 | P63028 | 0.84 | 0.03 | 0.85 | 0.05 |
| C23S | ∑ Tctp | P63028 | 0.84 | 0.03 | 0.72 | <0.01 |
| C24a | Twf1/1 | Q91YR1 | 0.58 | <0.01 | 0.93 | 0.39 |
| C24b | Twf1/2 | Q91YR1 | 0.25 | 0.01 | 0.86 | 0.52 |
| C24S | ∑ Twf1 | Q91YR1 | 0.40 | <0.01 | 0.89 | 0.47 |
| C25 | Twf2 | Q9Z0P5 | 0.70 | 0.01 | 0.60 | 0.01 |
| C26 | Vime | P20152 | 1.54 | 0.05 | 1.35 | 0.04 |
| C27a | Vinculin/1 | Q64727 | 1.28 | 0.25 | 1.24 | 0.11 |
| C27b | Vinculin/2 | Q64727 | 1.32 | 0.07 | 1.16 | 0.24 |
| C27c | Vinculin/3 | Q64727 | 1.63 | 0.01 | 1.12 | 0.28 |
| C27S | ∑ Vinculin | Q64727 | 1.39 | 0.07 | 1.18 | 0.16 |
| D1 | Cbx1 | P83917 | 0.91 | 0.27 | 0.72 | <0.01 |
| D2a | Ddb1/1 | Q3U1J4 | 1.74 | 0.04 | 1.40 | 0.16 |
| D2b | Ddb1/2 | Q3U1J4 | 0.93 | 0.65 | 0.78 | 0.21 |
| D2S | ∑ Ddb1 | Q3U1J4 | 1.31 | 0.14 | 1.07 | 0.73 |
| D3 | Nt5c | Q9JM14 | 1.18 | 0.08 | 1.39 | 0.03 |
| D4a | Pcna/1 | P17918 | 0.57 | <0.01 | 0.57 | <0.01 |
| D4b | Pcna/2 | P17918 | 0.82 | 0.02 | 0.80 | 0.03 |
| D4c | Pcna/3 | P17918 | 1.00 | 0.92 | 0.98 | 0.73 |
| D4S | ∑ Pcna | P17918 | 0.84 | <0.01 | 0.82 | 0.02 |
| D5 | Ruvb2 | Q9WTM5 | 1.46 | 0.06 | 1.42 | 0.04 |
| E1a | 6pgd/1 | Q9DCD0 | 0.88 | 0.05 | 0.95 | 0.42 |
| E1b | 6pgd/2 | Q9DCD0 | 1.05 | 0.32 | 1.01 | 0.90 |
| E1S | ∑ 6pgd | Q9DCD0 | 0.98 | 0.45 | 0.99 | 0.82 |
| E2a | Aacs/1 | Q9D2R0 | 1.79 | 0.11 | 1.39 | 0.52 |
| E2b | Aacs/2 | Q9D2R0 | 3.12 | 0.01 | 2.56 | <0.01 |
| E2S | ∑ Aacs | Q9D2R0 | 2.17 | 0.01 | 1.73 | 0.16 |
| E3 | Acadl | P51174 | 1.13 | 0.14 | 1.27 | 0.01 |
| E4a | Eno1a/1 | P17182 | 0.99 | 0.96 | 0.89 | 0.28 |
| E4b | Eno1a/2 | P17182 | 1.10 | 0.50 | 0.89 | 0.42 |
| E4c | Eno1a/3 | P17182 | 1.18 | 0.10 | 1.21 | 0.10 |
| E4d | Eno1a/4 | P17182 | 1.50 | <0.01 | 1.42 | <0.01 |
| E4S | ∑ Eno1A | P17182 | 1.26 | 0.02 | 1.19 | 0.06 |
| E5 | GalK | Q9R0N0 | 1.45 | <0.01 | 1.50 | 0.03 |
| E6 | Gapdh | P16858 | 0.56 | <0.01 | 0.60 | <0.01 |
| E7 | Gpd1L | Q3ULJ0 | 2.27 | <0.01 | 1.42 | 0.14 |
| E8a | Hxk3/1 | Q3TRM8 | 1.91 | 0.02 | 1.80 | 0.03 |
| E8b | Hxk3/2 | Q3TRM8 | 1.23 | 0.04 | 1.22 | 0.09 |
| E8c | Hxk3/3 | Q3TRM8 | 1.52 | 0.02 | 1.52 | 0.02 |
| E8S | ∑ Hxk3 | Q3TRM8 | 1.51 | 0.01 | 1.49 | 0.01 |
| E9 | Idhc | O88844 | 1.24 | 0.02 | 1.19 | <0.01 |
| E10a | Ldha/1 | P06151 | 0.79 | 0.02 | 0.86 | 0.06 |
| E10b | Ldha/2 | P06151 | 0.97 | 0.81 | 0.89 | 0.40 |
| E10S | ∑ Ldha | P06151 | 0.89 | 0.26 | 0.88 | 0.21 |
| E11a | Mdhc/1 | P14152 | 0.47 | 0.02 | 0.72 | 0.11 |
| E11b | Mdhc/2 | P14152 | 0.83 | 0.01 | 0.84 | 0.01 |
| E11c | Mdhc/3 | P14152 | 1.38 | 0.02 | 1.40 | <0.01 |
| E11S | ∑ Mdhc | P14152 | 1.01 | 0.91 | 1.05 | 0.20 |
| E12 | Pckgm | Q8BH04 | 1.30 | 0.20 | 0.61 | 0.01 |
| E13 | Pfkap | Q9WUA3 | 0.84 | 0.01 | 0.86 | 0.01 |
| E14 | Pfkl | P12382 | 1.83 | 0.01 | 1.62 | 0.03 |
| E15a | Pgls/1 | Q9CQ60 | 0.78 | 0.02 | 0.97 | 0.75 |
| E15b | Pgls/2 | Q9CQ60 | 1.11 | 0.16 | 1.22 | 0.01 |
| E16 | Pgp | Q8CHP8 | 1.37 | 0.02 | 1.60 | 0.05 |
| E17a | Taldo1 | Q93092 | 0.80 | 0.02 | 0.66 | <0.01 |
| E17b | Taldo/2 | Q93092 | 1.17 | 0.12 | 1.18 | 0.08 |
| E17c | Taldo/3 | Q93092 | 1.24 | 0.02 | 1.23 | <0.01 |
| E17d | Taldo/4 | Q93092 | 0.81 | 0.06 | 0.79 | 0.02 |
| E17e | Taldo/5 | Q93092 | 1.22 | 0.06 | 1.36 | 0.02 |
| E17S | ∑ Taldo | Q93092 | 1.08 | 0.16 | 1.09 | 0.06 |
| E18a | Tpis/1 | P17751 | 0.74 | 0.01 | 0.82 | 0.04 |
| E18b | Tpis/2 | P17751 | 1.04 | 0.61 | 1.20 | 0.09 |
| E18c | Tpis/3 | P17751 | 1.45 | 0.04 | 1.43 | 0.05 |
| E18S | ∑ Tpis | P17751 | 1.09 | 0.29 | 1.16 | 0.17 |
| F1 | Cdk4 | P30285 | 0.69 | <0.01 | 0.71 | <0.01 |
| F2 | Cdk6 | Q64261 | 0.71 | 0.04 | 0.77 | 0.02 |
| F3a | Ndrg1/1 | Q62433 | 1.32 | 0.09 | 1.52 | 0.04 |
| F3b | Ndrg1/2 | Q62433 | 1.38 | 0.01 | 1.33 | 0.02 |
| F3S | ∑ ndrg1 | Q62433 | 1.34 | 0.02 | 1.44 | 0.03 |
| F4 | Pa2 g4 | P50580 | 1.50 | <0.01 | 1.31 | 0.01 |
| F5a | Vma5a/1 | Q99KC8 | 1.60 | 0.06 | 0.99 | 0.97 |
| F5b | Vma5a/2 | Q99KC8 | 1.31 | 0.11 | 1.19 | 0.10 |
| F5c | Vma5a/3 | Q99KC8 | 1.84 | <0.01 | 1.59 | 0.02 |
| F5S | ∑ Vma5a | Q99KC8 | 1.54 | 0.02 | 1.25 | 0.12 |
| G1 | GalE | Q8R059 | 0.50 | 0.03 | 1.09 | 0.50 |
| G2 | Gnpda1 | O88958 | 0.47 | <0.01 | 0.63 | 0.01 |
| G3 | Gt25c | Q8K297 | 1.53 | 0.01 | 1.48 | 0.01 |
| G4 | Mlec | Q6ZQI3 | 1.49 | 0.02 | 1.79 | <0.01 |
| G5a | Naga/1 | Q8JZV7 | 1.19 | 0.07 | 1.19 | 0.06 |
| G5b | Naga/2 | Q8JZV7 | 1.05 | 0.56 | 1.18 | 0.08 |
| G5S | ∑ Naga | Q8JZV7 | 1.12 | 0.16 | 1.19 | 0.06 |
| H1 | Adh5 | P28474 | 0.72 | 0.09 | 0.74 | 0.11 |
| H2a | Aldr/1 | P45376 | 0.57 | 0.01 | 0.66 | 0.01 |
| H2b | Aldr/2 | P45376 | 0.84 | 0.09 | 0.73 | 0.04 |
| H2c | Aldr/3 | P45376 | 1.18 | 0.04 | 1.25 | 0.01 |
| H2S | ∑ Aldr | P45376 | 0.85 | 0.02 | 0.89 | 0.10 |
| H3 | Aldr2 | P47738 | 1.16 | 0.08 | 1.23 | <0.01 |
| H4a | Bvra/1 | Q9CY64 | 0.54 | 0.02 | 0.62 | 0.03 |
| H4b | Bvra/2 | Q9CY64 | 1.23 | 0.08 | 1.30 | 0.06 |
| H4S | ∑ Bvra | Q9CY64 | 0.98 | 0.81 | 1.06 | 0.54 |
| H5 | Bvrb | Q923D2 | 1.45 | 0.01 | 0.96 | 0.77 |
| H6 | Ca13 | Q9D6N1 | 0.62 | 0.01 | 0.83 | 0.11 |
| H7a | Esd/1 | Q9R0P3 | 0.56 | <0.01 | 0.64 | 0.01 |
| H7b | Esd/2 | Q9R0P3 | 0.78 | <0.01 | 0.92 | 0.15 |
| H7S | ∑ Esd | Q9R0P3 | 0.73 | <0.01 | 0.86 | 0.02 |
| H8 | Frih | P09528 | 0.61 | 0.01 | 0.67 | 0.02 |
| H9 | Gclm | O09172 | 1.32 | 0.01 | 0.98 | 0.86 |
| H10 | Hmox2 | O70252 | 1.53 | <0.01 | 1.03 | 0.69 |
| H11a | Lgul/1 | Q9CPU0 | 0.80 | 0.04 | 0.77 | 0.05 |
| H11b | Lgul/2 | Q9CPU0 | 1.10 | 0.24 | 1.10 | 0.16 |
| H11S | ∑ Lgul | Q9CPU0 | 1.00 | 0.99 | 1.00 | 0.96 |
| L1 | Anxa1 | P10107 | 0.78 | 0.02 | 1.11 | 0.06 |
| L2a | Anxa2/1 | P07356 | 0.79 | 0.01 | 0.60 | <0.01 |
| L2b | Anxa2/2 | P07356 | 0.44 | <0.01 | 0.56 | <0.01 |
| L2S | ∑ Anxa2 | P07356 | 0.67 | <0.01 | 0.59 | <0.01 |
| L3 | Anxa3 | O35639 | 1.34 | 0.01 | 1.30 | 0.02 |
| L4 | Anxa4 | P97429 | 1.30 | 0.01 | 1.33 | <0.01 |
| L5a | Anxa5/1 | P48036 | 0.88 | 0.02 | 0.81 | <0.01 |
| L5b | Anxa5/2 | P48036 | 1.09 | 0.34 | 1.07 | 0.37 |
| L5S | ∑ Anxa5 | P48036 | 0.95 | 0.35 | 0.90 | 0.08 |
| L6 | Anxa6 | P14824 | 1.54 | 0.02 | 1.80 | <0.01 |
| L7 | Anxa7 | Q07076 | 1.14 | 0.44 | 1.57 | 0.02 |
| L8 | Idi1 | P58044 | 1.40 | 0.02 | 1.34 | 0.01 |
| L9 | Lypla2 | Q9WTL7 | 1.07 | 0.28 | 1.22 | 0.02 |
| L10 | Mbd3 | Q9Z2D8 | 1.52 | 0.02 | 1.77 | <0.01 |
| L11 | Pipna | P53810 | 1.22 | 0.11 | 1.32 | 0.01 |
| L12 | Ppt1 | O88531 | 1.24 | 0.14 | 1.34 | 0.03 |
| M1a | Clic4/1 | Q9QYB1 | 0.90 | 0.12 | 0.95 | 0.58 |
| M1b | Clic4/2 | Q9QYB1 | 1.27 | 0.02 | 1.27 | 0.04 |
| M1S | ∑ Clic4 | Q9QYB1 | 1.12 | 0.04 | 1.14 | 0.01 |
| M2 | Clybl | Q8R4N0 | 1.80 | 0.03 | 1.41 | 0.19 |
| M3 | Gatm | Q9D964 | 0.68 | 0.01 | 0.54 | 0.01 |
| M4 | Hmgcl | P38060 | 0.61 | <0.01 | 0.76 | 0.07 |
| M5 | Nduv2 | Q9D6J6 | 1.24 | 0.01 | 1.49 | 0.05 |
| M6 | Oat | P29758 | 1.30 | 0.05 | 1.50 | 0.01 |
| M7 | Odba | P50136 | 1.54 | 0.02 | 1.28 | 0.12 |
| M8 | Phb | P67778 | 1.12 | 0.16 | 1.18 | 0.05 |
| M9 | Tmem11 | Q8BK08 | 1.78 | 0.04 | 1.46 | 0.41 |
| M10 | Vdac2 | Q60930 | 0.63 | 0.02 | 0.72 | 0.05 |
| M11a | Atpb ac | P56480 | 1.23 | 0.16 | 1.41 | 0.03 |
| M11b | Atpb bas | P56480 | 1.60 | 0.04 | 1.58 | <0.01 |
| M11S | ∑Atpb | P56480 | 1.41 | 0.06 | 1.49 | <0.01 |
| M12 | Clpp | O88696 | 1.28 | 0.33 | 1.84 | 0.02 |
| M13a | Trap1/1 | Q9CQN1 | 1.74 | 0.02 | 1.32 | 0.13 |
| M13b | Trap1/2 | Q9CQN1 | 1.21 | 0.25 | 0.92 | 0.64 |
| M13S | ∑Trap1 | Q9CQN1 | 1.47 | 0.03 | 1.11 | 0.51 |
| N1a | Aprt/1 | P08030 | 0.52 | <0.01 | 0.72 | <0.01 |
| N1b | Aprt/2 | P08030 | 1.06 | 0.43 | 1.08 | 0.39 |
| N1S | ∑ Aprt | P08030 | 0.92 | 0.19 | 0.99 | 0.85 |
| N2 | Bpnt1 | Q9Z0S1 | 1.77 | 0.02 | 1.87 | 0.01 |
| N3 | Guaa | Q3THK7 | 1.42 | 0.02 | 1.12 | 0.25 |
| N4 | Hint1 | P70349 | 1.46 | 0.30 | 0.64 | 0.02 |
| N5 | Ndka | P15532 | 0.42 | 0.03 | 0.75 | 0.22 |
| N6 | Paps1 | Q60967 | 5.54 | 0.04 | 2.44 | 0.01 |
| N7a | Pnph/1 | P23492 | 0.83 | 0.12 | 0.72 | 0.05 |
| N7b | Pnph/2 | P23492 | 1.25 | 0.02 | 1.22 | 0.08 |
| N7S | ∑ Pnph | P23492 | 1.08 | 0.17 | 1.02 | 0.76 |
| N8a | Prps1/1 | Q9D7G0 | 0.69 | 0.01 | 0.92 | 0.48 |
| N8b | Prps1/2 | Q9D7G0 | 0.57 | <0.01 | 0.77 | 0.16 |
| N8c | Prps1/3 | Q9D7G0 | 0.52 | 0.01 | 0.83 | 0.21 |
| N8S | ∑ Prps1 | Q9D7G0 | 0.60 | <0.01 | 0.83 | 0.14 |
| N9a | Pur4/1 | Q5SUR0 | 1.71 | 0.14 | 1.27 | 0.07 |
| N9b | Pur4/2 | Q5SUR0 | 2.53 | 0.01 | 2.01 | 0.01 |
| N9c | Pur4/3 | Q5SUR0 | 1.15 | 0.53 | 1.25 | 0.22 |
| N9S | ∑ Pur4 | Q5SUR0 | 1.65 | 0.07 | 1.43 | 0.01 |
| N10 | Pur9 | Q9CWJ9 | 1.16 | 0.27 | 1.23 | 0.01 |
| N11 | Nt5c3b | Q3UFY7 | 1.55 | <0.01 | 1.64 | 0.01 |
| O1 | Prx1ox | P35700 | 0.58 | 0.02 | 0.70 | 0.04 |
| O2a | Prx3/1 | P20108 | 0.93 | 0.69 | 1.13 | 0.08 |
| O2b | Prx3/2 | P20108 | 1.23 | 0.06 | 1.32 | 0.01 |
| O2S | ∑ Prx3 | P20108 | 1.16 | 0.22 | 1.27 | 0.01 |
| O3 | Prx4 | O08807 | 0.75 | 0.06 | 0.56 | 0.01 |
| O4a | Prx6/1 | O08709 | 0.56 | 0.02 | 0.45 | 0.01 |
| O4b | Prx6/2 | O08709 | 1.05 | 0.64 | 0.97 | 0.76 |
| O4S | ∑ Prx6 | O08709 | 0.96 | 0.63 | 0.87 | 0.23 |
| P1 | Aars | Q8BGQ7 | 1.65 | 0.01 | 1.18 | 0.20 |
| P2a | Eef2/1 | P58252 | 0.77 | 0.02 | 0.63 | <0.01 |
| P2b | Eef2/2 | P58252 | 1.06 | 0.41 | 0.96 | 0.39 |
| P2c | Eef2/3 | P58252 | 0.84 | 0.07 | 0.90 | 0.20 |
| P2d | Eef2/4 | P58252 | 0.99 | 0.88 | 0.98 | 0.61 |
| P2e | Eef2/5 | P58252 | 1.39 | 0.01 | 1.29 | 0.01 |
| P2S | ∑ Eef2 | P58252 | 1.02 | 0.72 | 0.97 | 0.43 |
| P3 | If3g | Q9Z1D1 | 0.77 | 0.31 | 0.56 | 0.03 |
| P4a | If5a/1 | P63242 | 0.54 | <0.01 | 0.62 | <0.01 |
| P4d | If5a/2 | P63242 | 1.07 | 0.26 | 1.03 | 0.58 |
| P4S | ∑ If5a | P63242 | 0.86 | 0.04 | 0.87 | 0.04 |
| P5 | If4a1 | P60843 | 1.55 | 0.01 | 1.51 | 0.01 |
| P6 | If4a3 | Q91VC3 | 1.27 | 0.05 | 1.09 | 0.28 |
| P7 | Rla0 | P14869 | 1.28 | 0.07 | 1.33 | 0.04 |
| P8 | Rs4y1 | P62702 | 0.50 | 0.04 | 0.62 | 0.07 |
| P9a | Sars/1 | P26638 | 1.42 | 0.03 | 1.05 | 0.59 |
| P9b | Sars/2 | P26638 | 1.52 | 0.04 | 1.10 | 0.42 |
| P9S | ∑ Sars | P26638 | 1.48 | 0.02 | 1.08 | 0.46 |
| Q1a | Dpp3/1 | Q99KK7 | 1.28 | 0.22 | 1.75 | <0.01 |
| Q1b | Dpp3/2 | Q99KK7 | 1.55 | 0.04 | 1.84 | <0.01 |
| Q1S | ∑ Dpp3 | Q99KK7 | 1.40 | 0.09 | 1.79 | <0.01 |
| Q2a | Hsp74/1 | Q61316 | 0.77 | 0.22 | 0.77 | 0.23 |
| Q2b | Hsp74/2 | Q61316 | 1.33 | 0.08 | 1.18 | 0.23 |
| Q2c | Hsp74/3 | Q61316 | 1.31 | 0.09 | 1.14 | 0.29 |
| Q2d | Hsp74/4 | Q61316 | 1.26 | 0.15 | 1.31 | <0.01 |
| Q2S | ∑ Hsp74 | Q61316 | 1.07 | 0.58 | 1.03 | 0.80 |
| Q3a | Hyou1/1 | Q9JKR6 | 1.18 | 0.25 | 0.83 | 0.20 |
| Q3b | Hyou1/2 | Q9JKR6 | 1.38 | 0.01 | 1.18 | 0.03 |
| Q3c | Hyou1/3 | Q9JKR6 | 1.35 | 0.01 | 1.21 | 0.05 |
| Q3d | Hyou1/4 | Q9JKR6 | 0.80 | 0.23 | 0.79 | 0.11 |
| Q3S | ∑ Hyou1 | Q9JKR6 | 1.04 | 0.67 | 0.93 | 0.27 |
| Q4 | Lxn | P70202 | 0.98 | 0.80 | 1.26 | <0.01 |
| Q5 | Pfd2 | O70591 | 0.72 | 0.01 | 0.70 | 0.01 |
| Q6a | Ppce/1 | Q9QUR6 | 1.40 | 0.15 | 1.78 | 0.01 |
| Q6b | Ppce/2 | Q9QUR6 | 1.58 | 0.04 | 1.66 | 0.01 |
| Q6S | ∑ Ppce | Q9QUR6 | 1.50 | 0.07 | 1.71 | 0.01 |
| Q7a | Ppia/1 | P17742 | 0.68 | 0.11 | 0.57 | 0.05 |
| Q7b | Ppia/2 | P17742 | 0.80 | 0.21 | 0.79 | 0.21 |
| Q7c | Ppia/3 | P17742 | 0.81 | 0.09 | 0.99 | 0.97 |
| Q7S | ∑ Ppia | P17742 | 0.78 | 0.09 | 0.83 | 0.21 |
| Q8a | Spb6/1 | Q60854 | 1.66 | 0.02 | 1.49 | 0.01 |
| Q8b | Spb6/2 | Q60854 | 1.43 | 0.01 | 1.42 | 0.01 |
| Q8S | ∑ Spb6 | Q60854 | 1.57 | 0.01 | 1.46 | <0.01 |
| Q9 | Stip1 | Q60864 | 1.72 | <0.01 | 0.94 | 0.67 |
| Q10 | Tcpa | P11983 | 1.55 | 0.02 | 1.09 | 0.55 |
| R1a | Btf3/1 | Q64152 | 0.56 | 0.01 | 0.50 | 0.01 |
| R1b | Btf3/2 | Q64152 | 0.76 | 0.05 | 0.76 | 0.05 |
| R1S | ∑ Btf3 | Q64152 | 0.66 | 0.02 | 0.64 | 0.01 |
| R2 | Dcps | Q9DAR7 | 1.30 | <0.01 | 1.20 | 0.02 |
| R3 | Ddx39a | Q8VDW0 | 1.64 | 0.03 | 1.31 | 0.02 |
| R4 | Ddx39b | Q9Z1N5 | 1.59 | 0.03 | 1.35 | 0.11 |
| R5 | Mgn | P61327 | 0.96 | 0.60 | 0.56 | 0.01 |
| R6a | Sf3b2/1 | Q3UJB0 | 1.03 | 0.91 | 0.87 | 0.49 |
| R6b | Sf3b2/2 | Q3UJB0 | 1.16 | 0.44 | 1.32 | <0.01 |
| R6c | Sf3b2/3 | Q3UJB0 | 1.26 | 0.38 | 1.80 | <0.01 |
| R6S | ∑ Sf3b2 | Q3UJB0 | 1.13 | 0.54 | 1.27 | 0.03 |
| R7 | ExoS4 | Q921I9 | 0.88 | 0.42 | 0.51 | 0.02 |
| R8 | ExoS6 | Q8BTW3 | 1.50 | 0.02 | 1.53 | 0.01 |
| S1a | 14-3-3 gam/1 | P61982 | 0.85 | 0.28 | 0.67 | 0.02 |
| S1b | 14-3-3 gam/2 | P61982 | 0.83 | 0.06 | 0.91 | 0.24 |
| S1S | ∑ 14-3-3 gam | P61982 | 0.84 | 0.10 | 0.82 | 0.07 |
| S2 | 14-3-3 th | P68254 | 1.31 | 0.01 | 1.19 | 0.04 |
| S3 | Cab39 | Q06138 | 0.70 | 0.03 | 0.62 | <0.01 |
| S4 | Fam49b | Q921M7 | 1.47 | 0.02 | 1.59 | 0.05 |
| S5 | Gbb1 | P62874 | 1.17 | 0.03 | 1.18 | 0.03 |
| S6a | Gnai2/1 | P08752 | 1.33 | 0.06 | 1.42 | 0.05 |
| S6b | Gnai2/2 | P08752 | 1.51 | <0.01 | 1.67 | 0.01 |
| S6S | ∑ Gnai2 | P08752 | 1.45 | 0.01 | 1.59 | 0.02 |
| S7 | Gnb2L1 | P68040 | 0.89 | 0.02 | 0.81 | <0.01 |
| S8 | Grb2 | Q60631 | 1.06 | 0.83 | 1.46 | 0.02 |
| S9 | Igbp1 | Q61249 | 1.36 | 0.06 | 1.36 | 0.04 |
| S10 | In35 | Q9D8C4 | 1.25 | 0.04 | 1.21 | 0.14 |
| S11 | Inpp | P49442 | 2.46 | 0.01 | 1.76 | 0.17 |
| S12 | Ppp1ca | P62137 | 1.17 | 0.07 | 1.27 | 0.02 |
| S13a | Ppp1r7/1 | Q3UM45 | 0.30 | 0.02 | 0.38 | 0.04 |
| S13b | Ppp1r7/2 | Q3UM45 | 0.67 | 0.02 | 0.73 | 0.08 |
| S13c | Ppp1r7/3 | Q3UM45 | 0.90 | 0.51 | 0.94 | 0.72 |
| S13S | ∑ Ppp1r7 | Q3UM45 | 0.61 | 0.03 | 0.68 | 0.06 |
| S14 | Ppp6 | O00743 | 1.48 | 0.03 | 1.04 | 0.82 |
| S15a | Snd1/1 | Q78PY7 | 0.65 | 0.03 | 0.66 | 0.04 |
| S15b | Snd1/2 | Q78PY7 | 0.59 | 0.02 | 0.50 | 0.01 |
| S15c | Snd1/3 | Q78PY7 | 0.89 | 0.34 | 0.78 | 0.18 |
| S15d | Snd1/4 | Q78PY7 | 1.11 | 0.34 | 0.91 | 0.47 |
| S15S | ∑ Snd1 | Q78PY7 | 0.79 | <0.01 | 0.70 | 0.01 |
| U1 | Chip | Q9WUD1 | 0.59 | <0.01 | 0.73 | 0.11 |
| U2 | Csn4 | O88544 | 1.34 | 0.03 | 1.37 | 0.03 |
| U3a | Prs8/1 | P62196 | 0.60 | <0.01 | 0.65 | <0.01 |
| U3b | Prs8/2 | P62196 | 0.85 | 0.04 | 0.85 | 0.22 |
| U3S | ∑ Prs8 | P62196 | 0.31 | 0.33 | 0.32 | 0.34 |
| U4a | Psa2/1 | P49722 | 0.58 | <0.01 | 0.58 | <0.01 |
| U4b | Psa2/2 | P49722 | 1.15 | 0.12 | 1.28 | 0.07 |
| U4S | ∑ Psa2 | P49722 | 0.86 | 0.04 | 0.92 | 0.36 |
| U5a | Psa5/1 | Q9Z2U1 | 0.80 | 0.01 | 0.84 | 0.11 |
| U5b | Psa5/2 | Q9Z2U1 | 0.92 | 0.33 | 0.99 | 0.83 |
| U5S | ∑ Psa5 | Q9Z2U1 | 0.86 | 0.07 | 0.92 | 0.22 |
| U6 | Psb10 | O35955 | 0.73 | 0.12 | 0.43 | 0.02 |
| U7a | Psb2/1 | Q9R1P3 | 0.67 | 0.02 | 0.76 | 0.14 |
| U7b | Psb2/2 | Q9R1P3 | 0.90 | 0.36 | 1.01 | 0.91 |
| U7S | ∑ Psb2 | Q9R1P3 | 0.82 | 0.09 | 0.92 | 0.50 |
| U8 | Psb3 mod | Q9R1P1 | 0.47 | 0.01 | 0.65 | <0.01 |
| U9a | Psb4/1 | P99026 | 0.74 | 0.05 | 0.69 | 0.03 |
| U9b | Psb4/2 | P99026 | 0.98 | 0.71 | 0.94 | 0.25 |
| U9S | ∑ Psb4 | P99026 | 0.90 | 0.05 | 0.86 | 0.01 |
| U10 | Psmd14 | O35593 | 1.03 | 0.61 | 1.32 | <0.01 |
| U11a | Psmd2/1 | Q8VDM4 | 0.82 | 0.22 | 1.12 | 0.24 |
| U11b | Psmd2/2 | Q8VDM4 | 1.10 | 0.11 | 1.09 | 0.02 |
| U11c | Psmd2/3 | Q8VDM4 | 1.39 | 0.02 | 1.13 | 0.17 |
| U11S | ∑ Psmd2 | Q8VDM4 | 1.03 | 0.70 | 1.12 | 0.12 |
| U12 | Psmd7 | P26516 | 0.68 | 0.03 | 0.68 | 0.02 |
| U13 | Psme1 | P97371 | 1.40 | <0.01 | 1.40 | <0.01 |
| U14a | Psme2/1 | P97372 | 1.00 | 0.98 | 0.71 | <0.01 |
| U14b | Psme2/2 | P97372 | 1.22 | <0.01 | 1.07 | 0.34 |
| U14S | ∑ Psme2 | P97372 | 1.15 | <0.01 | 0.96 | 0.50 |
| U15 | Ubl7 | Q91W67 | 0.41 | 0.02 | 0.88 | 0.55 |
| V1 | Asna1 | O54984 | 1.29 | 0.09 | 1.46 | <0.01 |
| V2 | Chm2a | Q9DB34 | 0.83 | <0.01 | 0.92 | 0.09 |
| V3 | CopE | O89079 | 1.35 | 0.01 | 1.41 | 0.07 |
| V4 | Emc2 | Q9CRD2 | 0.56 | 0.01 | 0.84 | 0.07 |
| V5 | Erp29 | P57759 | 0.78 | 0.12 | 0.68 | 0.05 |
| V6 | Mss4 | Q91X96 | 0.52 | 0.04 | 0.64 | 0.10 |
| V7 | Nsf1c | Q9CZ44 | 1.55 | 0.01 | 1.51 | 0.01 |
| V8 | Pef1 | Q8BFY6 | 0.64 | 0.05 | 0.50 | 0.02 |
| V9 | Snaa | Q9DB05 | 1.53 | <0.01 | 1.99 | 0.05 |
| V10 | Snap23 | O09044 | 0.53 | 0.02 | 0.34 | 0.01 |
| V11 | Stxb2 | Q64324 | 2.07 | <0.01 | 1.30 | 0.09 |
| V12a | Tera/1 | Q01853 | 1.31 | 0.13 | 1.18 | 0.16 |
| V12b | Tera/2 | Q01853 | 1.49 | 0.02 | 1.06 | 0.38 |
| V12c | Tera/3 | Q01853 | 1.39 | 0.02 | 1.29 | 0.04 |
| V12d | Tera/4 | Q01853 | 1.48 | 0.10 | 1.35 | <0.01 |
| V12S | ∑ Tera | Q01853 | 1.43 | 0.02 | 1.25 | 0.01 |
| V13 | Tpd52 | Q62393 | 1.42 | 0.04 | 1.31 | 0.10 |
| V14 | Vat1 | Q62465 | 1.48 | 0.01 | 1.22 | 0.15 |
| V15 | Vps29 | Q9QZ88 | 1.30 | 0.05 | 1.29 | 0.08 |
| V16a | Vps35/1 | Q9EQH3 | 2.44 | 0.16 | 2.33 | <0.01 |
| V16b | Vps35/2 | Q9EQH3 | 1.80 | 0.17 | 2.03 | 0.01 |
| V16c | Vps35/3 | Q9EQH3 | 1.53 | 0.33 | 2.25 | <0.01 |
| V16S | ∑ Vps35 | Q9EQH3 | 1.79 | 0.18 | 2.20 | <0.01 |
| V17 | Vta1 | Q9CR26 | 2.84 | 0.01 | 2.97 | 0.02 |
| X1 | Pdxk | Q8K183 | 1.29 | <0.01 | 1.45 | 0.02 |
| X2 | Spee | Q64674 | 1.55 | 0.02 | 1.43 | 0.07 |
| X3 | Sps1 | Q8BH69 | 1.49 | 0.02 | 1.95 | 0.01 |
| X4a | Ran/1 | P62827| | 0.83 | 0.04 | 0.87 | 0.12 |
| X4b | Ran/2 | P62827| | 1.18 | 0.08 | 1.11 | 0.17 |
| X4S | ∑ Ran | P62827| | 1.03 | 0.62 | 1.00 | 0.98 |
| Y1 | CatD | P18242 | 1.38 | 0.01 | 1.48 | <0.01 |
| Y2 | CatZ | Q9WUU7 | 1.79 | 0.04 | 1.08 | 0.85 |
| Y3 | Ncf4 | P97369 | 1.15 | 0.02 | 1.19 | 0.01 |
3.4 Enzymatic activities
For some of the modulated proteins, we first checked whether their enzyme activities (Table 3), i.e. the relevant parameter in terms of cell physiology, followed or not the trends observed via proteomics. There is no obvious inference between the two parameters, as the activity may be modulated by PTM and thus correlate with one or several spots, or with the sum of the spots representing the same protein. One good example is represented by malate dehydrogenase, which is known to be up-regulated by acetylation.71 In our case, the activity correlated with the amount of the acidic, modified spots and not with the total amount of the protein. For other activities, e.g. lactate dehydrogenase, triose phosphate isomerase and purine phosphorylase, the activity correlated with the total protein amount and not with a single spot. For several other enzymes such as enolase, hexokinase, pyridoxal kinase and isocitrate dehydrogenase, we detected an increase of the protein amount by proteomics and a stable or decreased enzyme activity.| Enzyme | Control | Acute | Recov | Ctrl + silver ion |
|---|---|---|---|---|
| All the activities are expressed in nmol substrate converted per min per mg total protein. Statistical significance of the results in the Student T test: *p < 0.05. ND: not determined. Abbreviations: LDH: lactate dehydrogenase; MDH: malate dehydrogenase; TPIS: triose phosphate isomerase; BVR: biliverdine reductase; PDXK: pyridoxal kinase; GAPDH: glyceraldehyde phosphate dehydrogenase; 6PGDH: 6-phosphogluconate dehydrogenase; PNPH: purine nucleoside phosphorylase; HXK: hexokinase; ENO: enolase; IDHC: NADPH-dependent isocitrate dehydrogenase. | ||||
| LDH | 74.4 ± 2.75 | 68.9 ± 1.14* | 73.8 ± 4.13 | ND |
| MDH | 37.4 ± 0.95 | 31.3 ± 2.85* | 31.7 ± 2.36* | ND |
| TPIS | 70.7 ± 8.75 | 76.6 ± 5.58 | 76.3 ± 7.80 | ND |
| BVR | 0.67 ± 0.04 | 0.53 ± 0.1* | 0.79 ± 0.06* | ND |
| PDXK | 7.4 ± 0.95 | 5.9 ± 0.6* | 7.5 ± 0.68* | ND |
| GAPDH | 157.12 ± 14.5 | 144.25 ± 9.77 | 157.44 ± 5.91 | ND |
| 6PGDH | 30.69 ± 3.17 | 39.37 ± 4.92* | 38.75 ± 1.24* | ND |
| PNPH | 8.97 ± 1.17 | 9.12 ± 1.6 | 9.06 ± 0.77 | ND |
| HXK | 23.4 ± 3.04 | 16.3 ± 4* | 23.3 ± 2.45 | (1 μM) 14.2 ± 2.93 |
| ENO | 214.5 ± 13.6 | 166.4 ± 28.1* | 184 ± 41 | (10 μM) 167 ± 12.4 |
| IDHC | 16.37 ± 1.48 | 17.62 ± 2.79 | 23.75 ± 2.97** | (5 μM) 8.06 ± 1.09 |
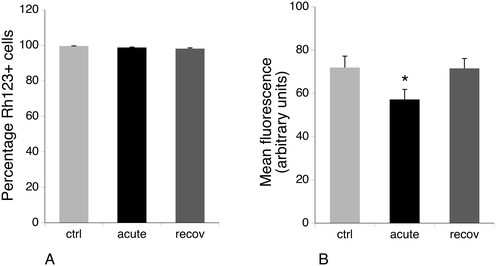
3.5 Mitochondrial potential and glucose consumption
Several mitochondrial proteins appeared modulated upon exposure of macrophages to silver nanoparticles, among which are a few subunits of the respiratory complexes (NDUV2 and ATPB), some components of the mitochondrial protein quality control (TRAP1 and CLPP) and an antioxidant protein (PRX3). As this may suggest perturbations in the mitochondrial functions, we investigated the mitochondrial transmembrane potential. The results, shown in Fig. 5, indicated that all viable cells accumulated rhodamine 123 and thus had a strong mitochondrial transmembrane potential. However, the amount of accumulated rhodamine 123 and thus the level of the mitochondrial transmembrane potential were lower for the acute condition and went back to normal after 72 hours of recovery. As a lower transmembrane potential is indicative of a less efficient oxidative phosphorylation chain, we investigated the global metabolism under the three experimental conditions.We thus measured the glucose consumption during the last 36 hours of the experiments by measuring the remaining glucose in the culture medium at the end of the experiment. The initial medium contained 4.1 g of glucose per liter due to the dilution brought by the addition of fetal serum to obtain the complete medium. In the untreated, control cells, 1.8 ± 0.2 g of glucose per liter remained in the culture medium after 36 hours of culture. In contrast, 1 ± 0.05 g of glucose and 0.9 ± 0.04 g of glucose remained in the medium for the acute and recovery conditions, respectively. These differences were statistically significant (Mann Whitney U test, p < 0.05).
The increased glucose consumption in the acute exposure condition could be correlated with the less efficient mitochondria via a Warburg effect. However, this correlation did not hold for the recovery condition, in which the mitochondria appear as efficient as those of the control cells. This may suggest that the silver expulsion process that takes place during the recovery period consumes a lot of energy.
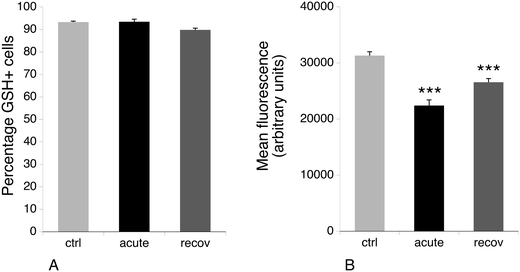
3.6 Glutathione levels
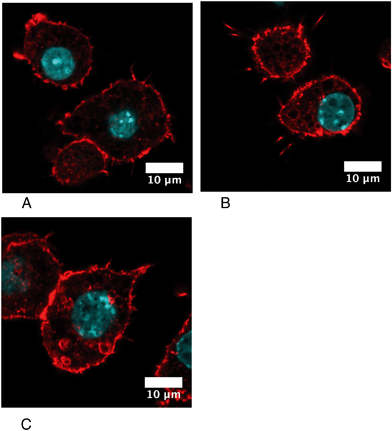
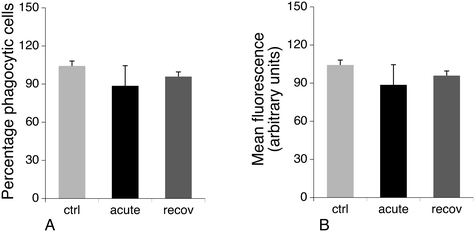
One of the proteins that is induced after acute exposure to silver nanoparticles is GCLM, i.e. the regulatory subunit of the enzyme involved in the first step of glutathione biosynthesis, which is the limiting step of the pathway.72 We thus investigated the levels of free glutathione, as studies on primary macrophages had shown that treatment with silver nanoparticles decreased the levels of free glutathione.23 This result may be due to the formation of silver–glutathione complexes.73 We thus measured by flow cytometry the proportion of cells with high levels of reduced glutathione, and the level of glutathione in this population. The results, shown in Fig. 6, indicated that the glutathione level is only 70% of the normal one just after the exposure to silver nanoparticles, and is 85% of the normal after 3 days of recovery. Thus, here again, the increase in GCLM amount can be interpreted as a cellular mechanism to compensate for the decrease of free glutathione caused by silver.3.7 Actin cytoskeleton and phagocytosis
Cytoskeletal proteins, especially proteins implicated in the actin cytoskeleton dynamics, were among the most important classes emerging from the proteomic screen. Of note, many of these proteins have activities that are modulated by phosphorylation, such as cofilin74,75 or Rho-GDIs.76,77 As the regulation patterns that emerged from the proteomic screen for these proteins were very complex, with some spots modulated while others were constant, we directly examined the consequences on the actin cytoskeleton. The results, shown in Fig. 7, indicated a strong vesicularization of the cells during the recovery period.Phagocytosis is one of the specialized functions of the macrophages that is highly dependent on the actin cytoskeleton. We thus tested the phagocytic activity of the cells. The results, shown in Fig. 8, indicated that the proportion of phagocytic cells does not change upon treatment with silver nanoparticles. However, the number of internalized particles per cell, as described by the mean fluorescence, slightly decreased immediately after exposure to nanoparticles and went back to quasi-normal values after 72 hours of recovery.
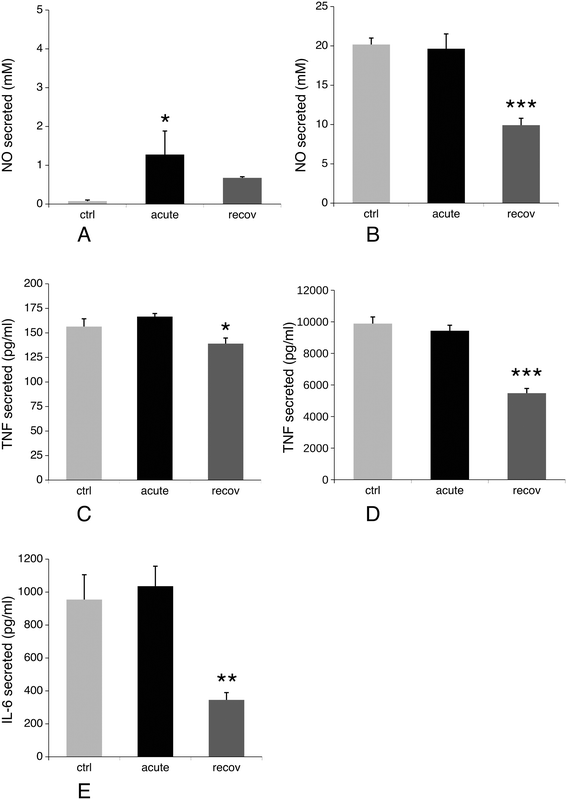
3.8 NO and cytokine production, redox balance
Production of nitric oxide and pro-inflammatory cytokines such as IL-6 and TNF upon stimulation is another specialized function of macrophages. We thus investigated their production after treatment of macrophages with silver nanoparticles under two schemes: either after treatment with nanoparticles only, or after treatment with nanoparticles and lipopolysaccharide (LPS) for the last 24 hours of culture. The first scheme investigated the intrinsic pro-inflammatory action of the nanoparticles, while the second one investigated the interference of nanoparticles with a standard pro-inflammatory response induced by a bacterial stimulus. The results, shown in Fig. 9, indicated that silver nanoparticles have a weak but significant intrinsic pro-inflammatory effect, as detected from NO and TNF production, which returned to normal values after the recovery period. The situation was however very different for the combined nanoparticle–LPS treatment. In this case, the acute exposure condition did not differ significantly from the control cells (exposed to LPS only), while the cytokine and NO production were significantly lower after the recovery period. As the NO production is NADPH-dependent, we investigated the NADP–NADPH levels after treatment with nanoparticles. The results, shown in Table 4, indicated that both the total NADP + NADPH and NADPH levels were similar for the three conditions tested.| Ctrl | Acute | Recov | |
|---|---|---|---|
| All the concentrations are expressed in nmol nucleotide per mg total protein. | |||
| NADPH | 1.87 ± 0.12 | 1.88 ± 0.03 | 1.85 ± 0.01 |
| NADP + NADPH | 2.5 ± 0.09 | 2.33 ± 0.04 | 2.62 ± 0.06 |
| NADPH/(NADP + NADPH) | 0.75 ± 0.07 | 0.81 ± 0.02 | 0.71 ± 0.02 |
4. Discussion
Silver nanoparticles are usually produced through a wet route and need to be stabilized as soon as prepared to avoid irreversible aggregation. Stabilization of AGNPs is usually achieved through interaction with complex ions such as citrate, or by polymers such as polyethylene glycol or polyvinylpyrrolidone (PVP). As such, PVP-coated silver nanoparticles are industrial products used in several applications including as antibacterial compounds,78 conductive inks,79–81 and sensors,82 and in electromechanics.83 In the course of their use, nanoparticles can be aerosolized. In this case they can contaminate living organisms without being modified. In the specific case of inhalation contamination, the nanoparticles reach the alveolae where there will be internalized by the lung macrophages, which is the reason why we made the choice to use this cell type in this study. Regarding other environmental dispersion routes, PVP-coated silver nanoparticles have been shown to be the most resistant in freshwater environments,84 and thus recommended for ecotoxicological testing.84 The PVP-coated silver nanoparticles, therefore, appear to be relevant for assessing the toxicity of silver nanoparticles.Most of the toxicological studies on nanoparticles use an acute exposure scheme; they expose cells to a high, but non-lethal dose. Such a scheme does not represent chronic daily exposure under, for example, occupational conditions, but may represent the dose following an accidental exposure. To fully evaluate the toxicity of AgNPs, it is therefore important to know not only the acute effects post exposure, but also how cells recover from such exposures to high dosages of AgNPs. To accomplish this, a ternary comparison must be made between cells before and immediately after treatment and also between cells at the end of a recovery period. In this experimental frame, the acute exposure point is not the primary focus of attention. We therefore decided not to investigate in detail the role of silver ions in the acute response, based on the fact that the rather large silver nanoparticles used in this study dissolve to a very low extent,23 producing free silver ion concentrations that are far below the LD20 observed for silver ions. Furthermore, this aspect has been investigated in numerous studies (e.g. in ref. 8, 15, 59 and 85) and the general outcome of these studies is that the mechanisms observed for silver nanoparticles cannot be explained by silver ions, at least for large nanoparticles.15
In this frame, the proteomic approach is of interest because it can be used to explore not only a few parameters, as targeted approaches do, but also can be used to investigate a few hundred parameters at the same time, i.e. the abundances of the various proteins analyzed in a proteomic screen. In this respect, the use of a 2D gel-based proteomic approach may appear as a medium scale approach, compared to deep shotgun proteomic approaches. However, compared to shotgun proteomics, 2D gels have the unique ability to separate protein forms, without the requirement that they appear as a single product. With growing recognition of the importance of post-translational modifications, the ability to do this is biologically relevant. Indeed, we have shown both in this study and in previous ones53,86 that some enzyme activities correlate with a single protein form and not with the sum of all the protein forms, showing the relevance of this parameter.
First, we performed a global analysis of the proteomic results. Both the principal component analysis and the analysis of similarity (ANOSIM) indicated that the recovery state was not just an intermediate state between the unexposed cells and the acutely-exposed cells. If this were the case, the recovery state should not have been separated from the other two states as it is in the principal component analysis. This is further confirmed by ANOSIM. The fact that all the p-values in the binary comparison are low and rather similar shows that the three states are significantly different from each other. If the recovery stage was just between the other two stages, at least one of the p-values between the recovery stage and one of the other two stages should be much higher than the p-value between the control and the acute stage. As this is not the case, we can reject the hypothesis that the recovery phase is only an intermediate between the control and acutely-treated stages.
We then performed a more detailed analysis of the proteomic results. The first important challenge was to determine to what extent the changes observed through proteomics are specific to silver nanoparticles and which are due to the simple presence of a PVP-coated nanoparticle. We have addressed this question in previously published studies.35,53 In these studies, we showed that PVP-coated zirconium oxide nanoparticles induced minimal changes at the proteomic level. This ruled out the possibility that the changes that we observe in the present study could be due either to the internalization process per se or are due to the effect of the addition of PVP.
As a further step, we compared our results obtained immediately after exposure to those published in the literature.9,10,24 It should be noted that this comparison is challenging because these other studies did not use the same cell type as was used here (although two of them use human intestinal cell lines, however different ones9,24) or the same silver nanoparticles. Citrate-coated silver nanoparticles were used in one study,24 while the other two used surfactant-coated nanoparticles,9,10 and in this study PVP-coated nanoparticles were used. These differences are very likely to explain the wide differences observed between the studies in which proteins were differentially modulated by treatment, with a minimal overlap between the results. Despite this general trend, several convergences could be observed between our study and those previously published. For example, increases in the abundances of Ran, Moesin, RuvB2, EfhD2, Pfkl, Anxa3, Tcpa and Tbcb were observed both in our study and the one published by Verano-Braga et al.24
However, proteomics is prone to multiple testing issues, so that the results obtained by proteomics must be verified by independent, targeted experiments. To accomplish this, we analyzed enzyme activities. Our enzyme activity results were consistent with observations made about changes at the proteomic level. However, a few discrepancies occurred, which were always of the same type: increases in the protein level by proteomics, corresponding to stable or decreased activities. We could, however, show that these enzyme activities are very sensitive to the silver ion, which was released within cells during their exposure to silver nanoparticles. Thus, the increase in protein levels can be seen in such cases as a cellular mechanism to compensate for the decrease of activity brought by silver.
We then carried out indirect validation studies, for example on the free glutathione levels, which were decreased immediately after exposure, a phenomenon that was previously observed for copper nanopartlcles.35 We also tested phagocytosis, which is an important function of macrophages and is important for clearing bacteria. For this assay we used micron-sized fluorescent beads, which functioned as bacterial mimics. This allowed us to investigate whether nanoparticle-treated cells were still able to clear bacteria. The proteomic screen suggested that phagocytosis might be altered after treatment with silver nanoparticles, which was confirmed in our assay testing the ability of the cell to clear bacteria. These results were previously observed for primary macrophages.23
The key question asked in this study, however, does not revolve around the acute response to silver nanoparticles, but instead asks to what extent and by which means do cells recover after such an exposure. In a simple model for recovery, protein changes should exhibit trends that return to normal, i.e. the amplitude of the change in abundance between the recovery and control stages should be lower than the amplitude of the change in abundance between the acute exposure and control stages. Out of the 239 spots that are highlighted in the proteomic screen, 135 (56%) show such a trend. This means that 104 (44%) show a stronger response at the recovery phase than immediately after exposure. With such a split trend, it is interesting to evaluate the results of targeted experiments during the recovery phase.
Interestingly, this split trend was also apparent in the results of targeted experiments. General metabolic processes seem to return to normal (mitochondrial potential, glutathione levels, and most enzymatic activities), as well as phagocytosis and basal NO production (without LPS stimulation). Oppositely, the LPS-induced activities (e.g. NO, IL-6 and TNF production) worsen during the recovery phase, as well as the isocitrate dehydrogenase activity, and the glucose consumption, which stays much higher at both the recovery and acute exposure phases than in unexposed cells.
It was the necessary to check in detail how proteomic changes correlate with observed functional changes. For example, for mitochondrial proteins, the proteomic screen detects an increase in the beta subunit of ATP synthase (ATPB) and an increase in one subunit of respiratory complex I (NDUV2) upon cell exposure to silver nanoparticles. At the same time the mitochondrial transmembrane potential decreases slightly, showing that this increase in mitochondrial proteins could be an attempt to restore normal energy production in cells. Upon recovery, one of the ATPB spots slightly decreases compared to the value immediately after exposure, while the other spot further increases. The NDUV2 spot is also enhanced. However, even the ATPB spot that decreased is still significantly more abundant at this stage of recovery than in control cells. This shows that the cellular situation is still not back to normal, as can be expected from the still high intracellular silver content, and that restoring the metabolic activity of a cell is an active process requiring an upregulation of several proteins.
Opposite to the delayed return to control values, GCLM, for example, shows an increase just after exposure. When the glutathione demand is high and the free glutathione levels are low, there is a return to control values at the end of the recovery period, at which point free glutathione levels are close to normal again.
In contrast to this simple case, the regulation of the actin cytoskeleton is much more complex. Many of the proteins interacting with actin and regulating its dynamics are regulated by phosphorylation. This holds true for cofilin,74 actinin 4,87 vinculin,88 swap70,89 arpc5,90 gelsolin91 and Arp2.92 While this further demonstrates the interest of taking into account modified protein forms, there are no obvious rules to predict which modified spot(s) is (are) effector(s) on the actin cytoskeleton. For example, the effector spot for cofilin is the median spot.93 Consistent with what has been described in the literature, we observe mostly modulations on acidic, modified forms of the above-cited proteins.
In line with the sustained changes in the actin cytoskeleton observed even at the recovery stage, we observe sustained changes in these modified spots, as exemplified by spots 2 of actinin 4 and cofilin, and spots 1 for Arp2 and Arpc5. It must be recalled, however, that such changes are not induced by the particle internalization process per se, as they were not observed for cells treated with the non-toxic zirconium oxide nanoparticles.53 They are also not induced by every toxic nanoparticle, as they were observed neither with zinc oxide53 nor with copper oxide, except for arpc5 which also decreased in response to treatment with copper oxide.37
Overall, the proteomic results suggest that the recovery process is a slow process. To evaluate this aspect, we determined the number of spots that showed a fast recovery. This category was defined as spots for which the quantitative change in the acute vs. control comparison showed an amplitude at least twice that in the recovery vs. control comparison. Only 44 spots were identified that fell in this category (including GCLM).
A striking result of our study was the observation that some changes in protein levels that were not immediately significant after exposure became significant at the end of the 72 h recovery phase. Such changes were observed both in targeted experiments (e.g. for the LPS-dependent responses) and in the proteomic screen. 51 spots showed such an expression profile, i.e. 21% of the total variable spots.
Functionally speaking, the results obtained when evaluating LPS-induced activities contrast with those previously described on primary macrophages,23 in which the cytokine and TNF productions also tended to return to normal values during the recovery phase. This discrepancy may have several origins. One such cause for the discrepancy could be differences in the persistence of silver between differing experimental conditions. In the experiments on primary macrophages, measurements of silver by PIXE showed a strong decrease (50%) in cellular silver content upon recovery, without any cellular multiplication (primary macrophages are post-mitotic in vitro). In the current experiments using the J774 cell line, the total silver content decreased by only 15%, and the decrease in cellular silver content is due mostly to cell division. The simple fact that the cells are able to divide shows that the silver concentration present in the cells is not toxic and explains also why, generally, the cellular parameter tends to return to normal levels. The remaining silver concentration may, however, may be high enough to inhibit specialized macrophage functions that depend on cell signaling, such as LPS-dependent activities.
In this context, it is tempting to dismiss the results obtained on the cell lines and favor those obtained using primary cells. The situation may, however, not be so simple. First, primary macrophages in vitro survive for only a few days once differentiated, while resident macrophages, such as those used in experiments using cell lines, have a very long lifespan,94,95 Furthermore, the condition in which the results between cell lines and primary macrophages diverge corresponds to an acute exposure to silver nanoparticles followed by a massive exposure to bacteria (mimicked by exposure to LPS), i.e. conditions that have not been tested in vivo. Therefore, we are currently unable to determine which system best represents the in vivo situation.
5. Conclusions
Overall, our results obtained both by targeted measurements and by proteomic experiments involving hundreds of proteins show that recovery after an acute exposure to silver nanoparticles is a very active process that involves a massive energy consumption, 50% higher than normal. This process leads to the quasi restoration of cellular homeostasis within 72 hours post exposure, as confirmed by targeted experiments testing the activity of enzymes, on mitochondrial function and on phagocytosis. Proteomic experiments also show, however, that this restoration of cellular homeostasis involves changes in the levels of many proteins, of which many are sustained, or even amplified, at the end of the 72 hour recovery period studied. The persistence of proteomic changes might be linked with the persistence of intracellular silver during the recovery period, which may require a long-lasting cellular adaptation process. Some other specialized macrophage functions, such as LPS-induced cytokine or nitric oxide production, did not return to normal within the 72 hour recovery period.Author contributions
AT performed the phagocytosis, rhodamine uptake and glutathione assay experiments. BD performed the cytokine assays and confocal microscopy experiments. HD, SC, VSF and TR performed the proteomic experiments. TR performed in addition the enzymatic, NADP(H) and glucose assays. SR and JB performed the ICP-MS experiments. MC, KPG and PHJ performed the electron microscopy experiments.Funding
This work was funded by ANSES (Agence Nationale de Sécurité Alimentaire, Environementale et du Travail) (PNREST 2011/25, Innimmunotox project) and by the CEA toxicology program (Nanostress grant). It is a contribution to the Labex Serenade (no. ANR-11-LABX-0064) funded by the “Investissements d'Avenir” French Government program of the French National Research Agency (ANR) through the A*MIDEX project (no. ANR-11-IDEX-0001-02). STEM-EDS analyses were conducted with the TEM OSIRIS, Nano-Safety Platform, CEA-Grenoble, operated by P. H. Jouneau. This project was managed by Agence National de la Recherche (ANR), program ‘Investissements d'Avenir’, reference ANR-10-EQPX-39.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
We thank Anne Bertrand and the electron microscopy facility of the Grenoble Institute of Neurosciences for the preparation of the samples and her kind help during the acquisition of the electron microscopy images.References
- S. Chernousova and M. Epple, Silver as antibacterial agent: ion, nanoparticle, and metal, Angew. Chem., Int. Ed., 2013, 52, 1636–1653 CrossRef CAS PubMed.
- C. A. Dos Santos, M. M. Seckler, A. P. Ingle, I. Gupta, S. Galdiero, M. Galdiero, A. Gade and M. Rai, Silver nanoparticles: therapeutical uses, toxicity, and safety issues, J. Pharm. Sci., 2014, 103, 1931–1944 CrossRef CAS PubMed.
- S. van den Brule, J. Ambroise, H. Lecloux, C. Levard, R. Soulas, P. J. De Temmerman, M. Palmai-Pallag, E. Marbaix and D. Lison, Dietary silver nanoparticles can disturb the gut microbiota in mice, Part. Fibre Toxicol., 2016, 13, 38 CrossRef PubMed.
- J. H. Sung, J. H. Ji, J. D. Park, J. U. Yoon, D. S. Kim, K. S. Jeon, M. Y. Song, J. Jeong, B. S. Han, J. H. Han, Y. H. Chung, H. K. Chang, J. H. Lee, M. H. Cho, B. J. Kelman and I. J. Yu, Subchronic inhalation toxicity of silver nanoparticles, Toxicol. Sci., 2009, 108, 452–461 CrossRef CAS PubMed.
- R. Ebabe Elle, S. Gaillet, J. Vide, C. Romain, C. Lauret, N. Rugani, J. P. Cristol and J. M. Rouanet, Dietary exposure to silver nanoparticles in Sprague-Dawley rats: effects on oxidative stress and inflammation, Food Chem. Toxicol., 2013, 60, 297–301 CrossRef CAS PubMed.
- R. Pecoraro, F. Marino, A. Salvaggio, F. Capparucci, G. Di Caro, C. Iaria, A. Salvo, A. Rotondo, D. Tibullo, G. Guerriero, E. M. Scalisi, M. Zimbone, G. Impellizzeri and M. V. Brundo, Evaluation of Chronic Nanosilver Toxicity to Adult Zebrafish, Front. Physiol., 2017, 8, 1011 CrossRef PubMed.
- S. Juling, L. Bohmert, D. Lichtenstein, A. Oberemm, O. Creutzenberg, A. F. Thunemann, A. Braeuning and A. Lampen, Comparative proteomic analysis of hepatic effects induced by nanosilver, silver ions and nanoparticle coating in rats, Food Chem. Toxicol., 2018, 113, 255–266 CrossRef CAS PubMed.
- W. H. De Jong, L. T. Van Der Ven, A. Sleijffers, M. V. Park, E. H. Jansen, H. Van Loveren and R. J. Vandebriel, Systemic and immunotoxicity of silver nanoparticles in an intravenous 28 days repeated dose toxicity study in rats, Biomaterials, 2013, 34, 8333–8343 CrossRef CAS PubMed.
- A. Oberemm, U. Hansen, L. Bohmert, C. Meckert, A. Braeuning, A. F. Thunemann and A. Lampen, Proteomic responses of human intestinal Caco-2 cells exposed to silver nanoparticles and ionic silver, J. Appl. Toxicol., 2016, 36, 404–413 CrossRef CAS PubMed.
- A. Braeuning, A. Oberemm, J. Gorte, L. Bohmert, S. Juling and A. Lampen, Comparative proteomic analysis of silver nanoparticle effects in human liver and intestinal cells, J. Appl. Toxicol., 2018, 38(5), 638–648 CrossRef CAS PubMed.
- F. Martinez-Gutierrez, E. P. Thi, J. M. Silverman, C. C. de Oliveira, S. L. Svensson, A. Vanden Hoek, E. M. Sanchez, N. E. Reiner, E. C. Gaynor, E. L. Pryzdial, E. M. Conway, E. Orrantia, F. Ruiz, Y. Av-Gay and H. Bach, Antibacterial activity, inflammatory response, coagulation and cytotoxicity effects of silver nanoparticles, Nanomedicine, 2012, 8, 328–336 CrossRef CAS PubMed.
- R. P. Nishanth, R. G. Jyotsna, J. J. Schlager, S. M. Hussain and P. Reddanna, Inflammatory responses of RAW 264.7 macrophages upon exposure to nanoparticles: role of ROS-NFkappaB signaling pathway, Nanotoxicology, 2011, 5, 502–516 CrossRef CAS PubMed.
- E.-J. Park, J. Yi, Y. Kim, K. Choi and K. Park, Silver nanoparticles induce cytotoxicity by a Trojan-horse type mechanism, Toxicol. In Vitro, 2010, 24, 872–878 CrossRef CAS PubMed.
- M. J. Piao, K. A. Kang, I. K. Lee, H. S. Kim, S. Kim, J. Y. Choi, J. Choi and J. W. Hyun, Silver nanoparticles induce oxidative cell damage in human liver cells through inhibition of reduced glutathione and induction of mitochondria-involved apoptosis, Toxicol. Lett., 2011, 201, 92–100 CrossRef CAS PubMed.
- A. Pratsinis, P. Hervella, J. C. Leroux, S. E. Pratsinis and G. A. Sotiriou, Toxicity of silver nanoparticles in macrophages, Small, 2013, 9, 2576–2584 CrossRef CAS PubMed.
- S. Arora, J. Jain, J. M. Rajwade and K. M. Paknikar, Cellular responses induced by silver nanoparticles: In vitro studies, Toxicol. Lett., 2008, 179, 93–100 CrossRef CAS PubMed.
- D. B. Warheit, T. A. McHugh and M. A. Hartsky, Differential pulmonary responses in rats inhaling crystalline, colloidal or amorphous silica dusts, Scand. J. Work, Environ. Health, 1995, 21(Suppl 2), 19–21 CAS.
- B. M. Levy and G. M. Higgins, Experimental berylliosis. I. Effects of a single intradermal injection of beryllium nitrate into guinea pigs, J. Invest. Dermatol., 1961, 37, 175–182 CrossRef CAS PubMed.
- D. Toybou, C. Celle, C. Aude-Garcia, T. Rabilloud and J.-P. Simonato, A toxicology-informed, safer by design approach for the fabrication of transparent electrodes based on silver nanowires, Environ. Sci.: Nano, 2019, 6, 684–694 RSC.
- K. S. Song, J. H. Sung, J. H. Ji, J. H. Lee, J. S. Lee, H. R. Ryu, J. K. Lee, Y. H. Chung, H. M. Park, B. S. Shin, H. K. Chang, B. Kelman and I. J. Yu, Recovery from silver-nanoparticle-exposure-induced lung inflammation and lung function changes in Sprague Dawley rats, Nanotoxicology, 2013, 7, 169–180 CrossRef CAS PubMed.
- A. Murphy, A. Casey, G. Byrne, G. Chambers and O. Howe, Silver nanoparticles induce pro-inflammatory gene expression and inflammasome activation in human monocytes, J. Appl. Toxicol., 2016, 36, 1311–1320 CrossRef CAS PubMed.
- Y. Xu, H. Tang, J. H. Liu, H. Wang and Y. Liu, Evaluation of the adjuvant effect of silver nanoparticles both in vitro and in vivo, Toxicol. Lett., 2013, 219, 42–48 CrossRef CAS PubMed.
- C. Aude-Garcia, F. Villiers, V. Collin-Faure, K. Pernet-Gallay, P. H. Jouneau, S. Sorieul, G. Mure, A. Gerdil, N. Herlin-Boime, M. Carriere and T. Rabilloud, Different in vitro exposure regimens of murine primary macrophages to silver nanoparticles induce different fates of nanoparticles and different toxicological and functional consequences, Nanotoxicology, 2016, 10, 586–596 CrossRef CAS PubMed.
- T. Verano-Braga, R. Miethling-Graff, K. Wojdyla, A. Rogowska-Wrzesinska, J. R. Brewer, H. Erdmann and F. Kjeldsen, Insights into the cellular response triggered by silver nanoparticles using quantitative proteomics, ACS Nano, 2014, 8, 2161–2175 CrossRef CAS PubMed.
- R. Miethling-Graff, R. Rumpker, M. Richter, T. Verano-Braga, F. Kjeldsen, J. Brewer, J. Hoyland, H. G. Rubahn and H. Erdmann, Exposure to silver nanoparticles induces size- and dose-dependent oxidative stress and cytotoxicity in human colon carcinoma cells, Toxicol. In Vitro, 2014, 28, 1280–1289 CrossRef CAS PubMed.
- H. G. Drexler, K. Otsuka, G. Gaedicke and J. Minowada, Changes in isoenzyme profiles during induction of differentiation in human myelomonocytic leukemia cell lines, Cancer Res., 1986, 46, 6078–6082 CAS.
- H. G. Welgus, N. L. Connolly and R. M. Senior, 12-o-Tetradecanoyl-phorbol-13-acetate-differentiated U937 cells express a macrophage-like profile of neutral proteinases. High levels of secreted collagenase and collagenase inhibitor accompany low levels of intracellular elastase and cathepsin G, J. Clin. Invest., 1986, 77, 1675–1681 CrossRef CAS PubMed.
- P. Kumarathasan, D. Breznan, D. Das, M. A. Salam, Y. Siddiqui, C. MacKinnon-Roy, J. Guan, N. de Silva, B. Simard and R. Vincent, Cytotoxicity of carbon nanotube variants: a comparative in vitro exposure study with A549 epithelial and J774 macrophage cells, Nanotoxicology, 2015, 9, 148–161 CrossRef CAS PubMed.
- P. Oberbek, T. Bolek, A. Chlanda, S. Hirano, S. Kusnieruk, J. Rogowska-Tylman, G. Nechyporenko, V. Zinchenko, W. Swieszkowski and T. Puzyn, Characterization and influence of hydroxyapatite nanopowders on living cells, Beilstein J. Nanotechnol., 2018, 9, 3079–3094 CrossRef CAS PubMed.
- M. Shoeb, V. Kodali, B. Farris, L. M. Bishop, T. Meighan, R. Salmen, T. Eye, J. R. Roberts, P. Zeidler-Erdely, A. Erdely and J. M. Antonini, Evaluation of the molecular mechanisms associated with cytotoxicity and inflammation after pulmonary exposure to different metal-rich welding particles, Nanotoxicology, 2017, 11, 725–736 CAS.
- M. Chen, Y. Li, J. Zhou, Z. Yang, Z. Wang, Y. Yang, H. Zhang, Z. Li and X. Mei, In vitro toxicity assessment of nanocrystals in tissue-type cells and macrophage cells, J. Appl. Toxicol., 2018, 38, 656–664 CrossRef CAS PubMed.
- A. Kroll, C. Dierker, C. Rommel, D. Hahn, W. Wohlleben, C. Schulze-Isfort, C. Gobbert, M. Voetz, F. Hardinghaus and J. Schnekenburger, Cytotoxicity screening of 23 engineered nanomaterials using a test matrix of ten cell lines and three different assays, Part. Fibre Toxicol., 2011, 8, 9 CrossRef CAS PubMed.
- L. Farcal, F. Torres Andon, L. Di Cristo, B. M. Rotoli, O. Bussolati, E. Bergamaschi, A. Mech, N. B. Hartmann, K. Rasmussen, J. Riego-Sintes, J. Ponti, A. Kinsner-Ovaskainen, F. Rossi, A. Oomen, P. Bos, R. Chen, R. Bai, C. Chen, L. Rocks, N. Fulton, B. Ross, G. Hutchison, L. Tran, S. Mues, R. Ossig, J. Schnekenburger, L. Campagnolo, L. Vecchione, A. Pietroiusti and B. Fadeel, Comprehensive In Vitro Toxicity Testing of a Panel of Representative Oxide Nanomaterials: First Steps towards an Intelligent Testing Strategy, PLoS One, 2015, 10, e0127174 CrossRef PubMed.
- D. Breznan, D. D. Das, J. S. O'Brien, C. MacKinnon-Roy, S. Nimesh, N. Q. Vuong, S. Bernatchez, N. DeSilva, M. Hill, P. Kumarathasan and R. Vincent, Differential cytotoxic and inflammatory potency of amorphous silicon dioxide nanoparticles of similar size in multiple cell lines, Nanotoxicology, 2017, 11, 223–235 CrossRef CAS PubMed.
- S. Triboulet, C. Aude-Garcia, M. Carriere, H. Diemer, F. Proamer, A. Habert, M. Chevallet, V. Collin-Faure, J. M. Strub, D. Hanau, A. Van Dorsselaer, N. Herlin-Boime and T. Rabilloud, Molecular responses of mouse macrophages to copper and copper oxide nanoparticles inferred from proteomic analyses, Mol. Cell. Proteomics, 2013, 12, 3108–3122 CrossRef CAS PubMed.
- S. Triboulet, C. Aude-Garcia, L. Armand, A. Gerdil, H. Diemer, F. Proamer, V. Collin-Faure, A. Habert, J. M. Strub, D. Hanau, N. Herlin, M. Carriere, A. Van Dorsselaer and T. Rabilloud, Analysis of cellular responses of macrophages to zinc ions and zinc oxide nanoparticles: a combined targeted and proteomic approach, Nanoscale, 2014, 6, 6102–6114 RSC.
- S. Triboulet, C. Aude-Garcia, L. Armand, V. Collin-Faure, M. Chevallet, H. Diemer, A. Gerdil, F. Proamer, J. M. Strub, A. Habert, N. Herlin, A. Van Dorsselaer, M. Carriere and T. Rabilloud, Comparative proteomic analysis of the molecular responses of mouse macrophages to titanium dioxide and copper oxide nanoparticles unravels some toxic mechanisms for copper oxide nanoparticles in macrophages, PLoS One, 2015, 10, e0124496 CrossRef PubMed.
- E. Eymard-Vernain, C. Lelong, A. E. Pradas Del Real, R. Soulas, S. Bureau, V. Tardillo Suarez, B. Gallet, O. Proux, H. Castillo-Michel and G. Sarret, Impact of a Model Soil Microorganism and of Its Secretome on the Fate of Silver Nanoparticles, Environ. Sci. Technol., 2018, 52, 71–78 CrossRef CAS PubMed.
- W. E. Hathaway, L. A. Newby and J. H. Githens, The Acridine Orange Viability Test Applied to Bone Marrow Cells. I. Correlation with Trypan Blue and Eosin Dye Exclusion and Tissue Culture Transformation, Blood, 1964, 23, 517–525 CAS.
- A. Moore, C. J. Donahue, K. D. Bauer and J. P. Mather, Simultaneous measurement of cell cycle and apoptotic cell death, Methods Cell Biol., 1998, 57, 265–278 CrossRef CAS PubMed.
- S. W. Perry, J. P. Norman, J. Barbieri, E. B. Brown and H. A. Gelbard, Mitochondrial membrane potential probes and the proton gradient: a practical usage guide, BioTechniques, 2011, 50, 98–115 CrossRef CAS PubMed.
- T. Rabilloud, Optimization of the cydex blue assay: A one-step colorimetric protein assay using cyclodextrins and compatible with detergents and reducers, PLoS One, 2018, 13, e0195755 CrossRef PubMed.
- K. M. Mayer and F. H. Arnold, A colorimetric assay to quantify dehydrogenase activity in crude cell lysates, J. Biomol. Screening, 2002, 7, 135–140 CrossRef CAS PubMed.
- B. Plaut and J. R. Knowles, pH-dependence of the triose phosphate isomerase reaction, Biochem. J., 1972, 129, 311–320 CrossRef CAS PubMed.
- P. Fossati, Phosphate determination by enzymatic colorimetric assay, Anal. Biochem., 1985, 149, 62–65 CrossRef CAS PubMed.
- V. B. Ritov and D. E. Kelley, Hexokinase isozyme distribution in human skeletal muscle, Diabetes, 2001, 50, 1253–1262 CrossRef CAS PubMed.
- D. E. Baranano, M. Rao, C. D. Ferris and S. H. Snyder, Biliverdin reductase: a major physiologic cytoprotectant, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 16093–16098 CrossRef CAS PubMed.
- J. A. Kerry, M. Rohde and F. Kwok, Brain pyridoxal kinase. Purification and characterization, Eur. J. Biochem., 1986, 158, 581–585 CrossRef CAS PubMed.
- V. E. Anderson, P. M. Weiss and W. W. Cleland, Reaction intermediate analogues for enolase, Biochemistry, 1984, 23, 2779–2786 CrossRef CAS PubMed.
- W. R. Wright, J. C. Rainwater and L. D. Tolle, Glucose assay systems: evaluation of a colorimetric hexokinase procedure, Clin. Chem., 1971, 17, 1010–1015 CAS.
- T. C. Wagner and M. D. Scott, Single extraction method for the spectrophotometric quantification of oxidized and reduced pyridine nucleotides in erythrocytes, Anal. Biochem., 1994, 222, 417–426 CrossRef CAS PubMed.
- K. Wosikowski, K. Mattern, I. Schemainda, M. Hasmann, B. Rattel and R. Loser, WK175, a novel antitumor agent, decreases the intracellular nicotinamide adenine dinucleotide concentration and induces the apoptotic cascade in human leukemia cells, Cancer Res., 2002, 62, 1057–1062 CAS.
- C. Aude-Garcia, B. Dalzon, J. L. Ravanat, V. Collin-Faure, H. Diemer, J. M. Strub, S. Cianferani, A. Van Dorsselaer, M. Carriere and T. Rabilloud, A combined proteomic and targeted analysis unravels new toxic mechanisms for zinc oxide nanoparticles in macrophages, J. Proteomics, 2016, 134, 174–185 CrossRef CAS PubMed.
- E. Gianazza, F. Celentano, S. Magenes, C. Ettori and P. G. Righetti, Formulations for immobilized pH gradients including pH extremes, Electrophoresis, 1989, 10, 806–808 CrossRef CAS PubMed.
- T. Rabilloud, C. Valette and J. J. Lawrence, Sample application by in-gel rehydration improves the resolution of two-dimensional electrophoresis with immobilized pH gradients in the first dimension, Electrophoresis, 1994, 15, 1552–1558 CrossRef CAS PubMed.
- S. Luche, H. Diemer, C. Tastet, M. Chevallet, A. Van Dorsselaer, E. Leize-Wagner and T. Rabilloud, About thiol derivatization and resolution of basic proteins in two-dimensional electrophoresis, Proteomics, 2004, 4, 551–561 CrossRef CAS PubMed.
- A. Gorg, W. Postel, J. Weser, S. Gunther, J. R. Strahler, S. M. Hanash and L. Somerlot, Elimination of Point Streaking on Silver Stained Two-Dimensional Gels by Addition of Iodoacetamide to the Equilibration Buffer, Electrophoresis, 1987, 8, 122–124 CrossRef.
- C. Tastet, P. Lescuyer, H. Diemer, S. Luche, A. van Dorsselaer and T. Rabilloud, A versatile electrophoresis system for the analysis of high- and low-molecular-weight proteins, Electrophoresis, 2003, 24, 1787–1794 CrossRef CAS PubMed.
- P. Sinha, J. Poland, M. Schnolzer and T. Rabilloud, A new silver staining apparatus and procedure for matrix-assisted laser desorption/ionization-time of flight analysis of proteins after two-dimensional electrophoresis, Proteomics, 2001, 1, 835–840 CrossRef CAS PubMed.
- A. G. Herrmann, J. L. Searcy, T. Le Bihan, J. McCulloch and R. F. Deighton, Total variance should drive data handling strategies in third generation proteomic studies, Proteomics, 2013, 13, 3251–3255 CrossRef CAS PubMed.
- D. Yekutieli and Y. Benjamini, Resampling-based false discovery rate controlling multiple test procedures for correlated test statistics, J. Stat. Plan. Inference, 1999, 82, 171–196 CrossRef.
- A. Carvajal-Rodriguez and J. de Una-Alvarez, Assessing significance in high-throughput experiments by sequential goodness of fit and q-value estimation, PLoS One, 2011, 6, e24700 CrossRef CAS PubMed.
- A. P. Diz, A. Carvajal-Rodriguez and D. O. Skibinski, Multiple hypothesis testing in proteomics: a strategy for experimental work, Mol. Cell. Proteomics, 2011, 10, M110 004374 CrossRef PubMed.
- Ø. Hammer, D. A. T. Harper and P. D. Ryan, Paleontological statistics software package for education and data analysis, Palaeontol. Electronica, 2001, 4, XIX–XX Search PubMed.
- F. Gharahdaghi, C. R. Weinberg, D. A. Meagher, B. S. Imai and S. M. Mische, Mass spectrometric identification of proteins from silver-stained polyacrylamide gel: A method for the removal of silver ions to enhance sensitivity, Electrophoresis, 1999, 20, 601–605 CrossRef CAS PubMed.
- S. Richert, S. Luche, M. Chevallet, A. Van Dorsselaer, E. Leize-Wagner and T. Rabilloud, About the mechanism of interference of silver staining with peptide mass spectrometry, Proteomics, 2004, 4, 909–916 CrossRef CAS PubMed.
- P. Skehan, R. Storeng, D. Scudiero, A. Monks, J. McMahon, D. Vistica, J. T. Warren, H. Bokesch, S. Kenney and M. R. Boyd, New colorimetric cytotoxicity assay for anticancer-drug screening, J. Natl. Cancer Inst., 1990, 82, 1107–1112 CrossRef CAS PubMed.
- V. Vichai and K. Kirtikara, Sulforhodamine B colorimetric assay for cytotoxicity screening, Nat. Protoc., 2006, 1, 1112–1116 CrossRef CAS PubMed.
- D. W. Huang, B. T. Sherman and R. A. Lempicki, Bioinformatics enrichment tools: paths toward the comprehensive functional analysis of large gene lists, Nucleic Acids Res., 2009, 37, 1–13 CrossRef CAS PubMed.
- D. W. Huang, B. T. Sherman and R. A. Lempicki, Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources, Nat. Protoc., 2009, 4, 44–57 CrossRef CAS PubMed.
- S. M. Zhao, W. Xu, W. Q. Jiang, W. Yu, Y. Lin, T. F. Zhang, J. Yao, L. Zhou, Y. X. Zeng, H. Li, Y. X. Li, J. Shi, W. L. An, S. M. Hancock, F. C. He, L. X. Qin, J. Chin, P. Y. Yang, X. Chen, Q. Y. Lei, Y. Xiong and K. L. Guan, Regulation of Cellular Metabolism by Protein Lysine Acetylation, Science, 2010, 327, 1000–1004 CrossRef CAS PubMed.
- Y. Chen, H. G. Shertzer, S. N. Schneider, D. W. Nebert and T. P. Dalton, Glutamate cysteine ligase catalysis: dependence on ATP and modifier subunit for regulation of tissue glutathione levels, J. Biol. Chem., 2005, 280, 33766–33774 CrossRef CAS PubMed.
- G. Veronesi, C. Aude-Garcia, I. Kieffer, T. Gallon, P. Delangle, N. Herlin-Boime, T. Rabilloud and M. Carriere, Exposure-dependent Ag+ release from silver nanoparticles and its complexation in AgS2 sites in primary murine macrophages, Nanoscale, 2015, 7, 7323–7330 RSC.
- N. Yang, O. Higuchi, K. Ohashi, K. Nagata, A. Wada, K. Kangawa, E. Nishida and K. Mizuno, Cofilin phosphorylation by LIM-kinase 1 and its role in Rac-mediated actin reorganization, Nature, 1998, 393, 809–812 CrossRef CAS PubMed.
- K. Mizuno, Signaling mechanisms and functional roles of cofilin phosphorylation and dephosphorylation, Cell. Signalling, 2013, 25, 457–469 CrossRef CAS PubMed.
- C. DerMardirossian, A. Schnelzer and G. M. Bokoch, Phosphorylation of RhoGDI by Pak1 mediates dissociation of Rac GTPase, Mol. Cell, 2004, 15, 117–127 CrossRef CAS PubMed.
- C. DerMardirossian, G. Rocklin, J. Y. Seo and G. M. Bokoch, Phosphorylation of RhoGDI by Src regulates Rho GTPase binding and cytosol-membrane cycling, Mol. Biol. Cell, 2006, 17, 4760–4768 CrossRef CAS PubMed.
- L. Kvitek, A. Panacek, J. Soukupova, M. Kolar, R. Vecerova, R. Prucek, M. Holecova and R. Zboril, Effect of surfactants and polymers on stability and antibacterial activity of silver nanoparticles (NPs), J. Phys. Chem. C, 2008, 112, 5825–5834 CrossRef CAS.
- M. A. H. Khondoker, S. C. Mun and J. Kim, Synthesis and characterization of conductive silver ink for electrode printing on cellulose film, Appl. Phys. A: Mater. Sci. Process., 2013, 112, 411–418 CrossRef CAS.
- M. Ismail and R. Jabra, Investigation the parameters affecting on the synthesis of silver nanoparticles by chemical reduction method and printing a conductive pattern, J. Mater. Environ. Sci., 2017, 8, 4152–4159 CAS.
- D. Mau Chien, D. T. M. Dung and E. Fribourg-Blanc, Silver nanoparticles ink synthesis for conductive patterns fabrication using inkjet printing technology, Adv. Nat. Sci.: Nanosci. Nanotechnol., 2015, 6 DOI:10.1088/2043-6262/6/1/015003.
- D. R. Raj, S. Prasanth, T. V. Vineeshkumar and C. Sudarsanakumar, Ammonia sensing properties of tapered plastic optical fiber coated with silver nanoparticles/PVP/PVA hybrid, Opt. Commun., 2015, 340, 86–92 CrossRef.
- Q. Liu, L. Seveyrat, F. Belhora and D. Guyomar, Investigation of polymer-coated nano silver/polyurethane nanocomposites for electromechanical applications, J. Polym. Res., 2013, 20 DOI:10.1007/s10965-013-0309-z.
- M. Tejamaya, I. Romer, R. C. Merrifield and J. R. Lead, Stability of citrate, PVP, and PEG coated silver nanoparticles in ecotoxicology media, Environ. Sci. Technol., 2012, 46, 7011–7017 CrossRef CAS PubMed.
- A. R. Gliga, S. Skoglund, I. O. Wallinder, B. Fadeel and H. L. Karlsson, Size-dependent cytotoxicity of silver nanoparticles in human lung cells: the role of cellular uptake, agglomeration and Ag release, Part. Fibre Toxicol., 2014, 11, 11 CrossRef PubMed.
- B. Dalzon, C. Aude-Garcia, V. Collin-Faure, H. Diemer, D. Beal, F. Dussert, D. Fenel, G. Schoehn, S. Cianferani, M. Carriere and T. Rabilloud, Differential proteomics highlights macrophage-specific responses to amorphous silica nanoparticles, Nanoscale, 2017, 9, 9641–9658 RSC.
- H. Shao, C. Wu and A. Wells, Phosphorylation of alpha-actinin 4 upon epidermal growth factor exposure regulates its interaction with actin, J. Biol. Chem., 2010, 285, 2591–2600 CrossRef CAS PubMed.
- Z. Zhang, G. Izaguirre, S. Y. Lin, H. Y. Lee, E. Schaefer and B. Haimovich, The phosphorylation of vinculin on tyrosine residues 100 and 1065, mediated by SRC kinases, affects cell spreading, Mol. Biol. Cell, 2004, 15, 4234–4247 CrossRef CAS PubMed.
- G. Pearce, T. Audzevich and R. Jessberger, SYK regulates B-cell migration by phosphorylation of the F-actin interacting protein SWAP-70, Blood, 2011, 117, 1574–1584 CrossRef CAS PubMed.
- R. Silverman-Gavrila, L. Silverman-Gavrila, G. Hou, M. Zhang, M. Charlton and M. P. Bendeck, Rear polarization of the microtubule-organizing center in neointimal smooth muscle cells depends on PKCalpha, ARPC5, and RHAMM, Am. J. Pathol., 2011, 178, 895–910 CrossRef CAS PubMed.
- P. Singaravelu, W. L. Lee, S. Wee, U. Ghoshdastider, K. Ding, J. Gunaratne, J. M. Grimes, K. Swaminathan and R. C. Robinson, Yersinia effector protein (YopO)-mediated phosphorylation of host gelsolin causes calcium-independent activation leading to disruption of actin dynamics, J. Biol. Chem., 2017, 292, 8092–8100 CrossRef CAS PubMed.
- L. L. LeClaire, M. Baumgartner, J. H. Iwasa, R. D. Mullins and D. L. Barber, Phosphorylation of the Arp2/3 complex is necessary to nucleate actin filaments, J. Cell Biol., 2008, 182, 647 CrossRef CAS PubMed.
- R. Prudent, N. Demoncheaux, H. Diemer, V. Collin-Faure, R. Kapur, F. Paublant, L. Lafanechere, S. Cianferani and T. Rabilloud, A quantitative proteomic analysis of cofilin phosphorylation in myeloid cells and its modulation using the LIM kinase inhibitor Pyr1, PLoS One, 2018, 13, e0208979 CrossRef PubMed.
- J. Murphy, R. Summer, A. A. Wilson, D. N. Kotton and A. Fine, The prolonged life-span of alveolar macrophages, Am. J. Respir. Cell Mol. Biol., 2008, 38, 380–385 CrossRef CAS PubMed.
- S. Yona, K. W. Kim, Y. Wolf, A. Mildner, D. Varol, M. Breker, D. Strauss-Ayali, S. Viukov, M. Guilliams, A. Misharin, D. A. Hume, H. Perlman, B. Malissen, E. Zelzer and S. Jung, Fate mapping reveals origins and dynamics of monocytes and tissue macrophages under homeostasis, Immunity, 2013, 38, 79–91 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9en00408d |
| ‡ These authors contributed equally to this work. |
| § Deceased: Nov. 21st, 2018. This paper is dedicated to her memory. |
| This journal is © The Royal Society of Chemistry 2019 |