Catalytic and biophysical investigation of rhodium hydroformylase†
Abstract
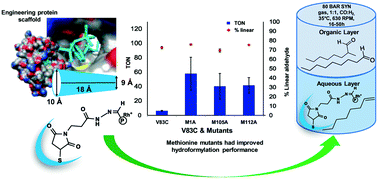
Rh-Containing artificial metalloenzymes based on two mutants of sterol carrier protein_2L (SCP_2L) have been shown to act as hydroformylases, exhibiting significant activity and unexpectedly high selectivity in the hydroformylation of a range of alkenes. Here we report modifications of the catalyst performance by site directed mutagenesis, and studies of the biophysical properties of these catalysts. Catalysts were prepared based on single methionine mutants of SCP_2L for hydroformylation studies. Multinuclear (1H, & 13C), multi-dimensional (2D) solution NMR spectroscopy, EXAFS and XANES were employed to probe the structure. Biophysical studies using 2D [1H 13C] HSQC NMR spectroscopy of 13C-methyl methionine labeled catalysts were used to investigate changes in protein conformation. Hydroformylation studies employing the methionine mutants as catalysts revealed the significant effects of mutation on hydroformylation activity. Among the methionine mutant catalysts M1A of V83C and A100C, and M112A of V83C exhibited significantly higher activity than their parent proteins with improved selectivity. A metal binding role for methionine was suggested by EXAFS and XANES data on selenomethionine variants.
- This article is part of the themed collection: 2019 Catalysis Science & Technology HOT Articles


 Please wait while we load your content...
Please wait while we load your content...