Design of a liquid cell toward three-dimensional imaging of unidirectionally-aligned particles in solution using X-ray free-electron lasers
Abstract
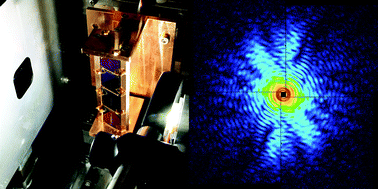
X-ray free-electron lasers (XFELs) opened up a possibility for molecular-scale single particle imaging (SPI) without the need for crystallization. In SPI experiments, the orientation of each particle has to be determined from the measured diffraction pattern. Preparing unidirectionally-aligned particles can facilitate the determination of the sample orientation. Here, we show the design principles of a liquid cell for three-dimensional imaging of unidirectionally-aligned particles in solution with XFELs. The liquid cell was designed so that neither incident X-rays nor diffracted X-rays are blocked by the substrate of the liquid cell even at high tilt angles. As a feasibility evaluation, we performed coherent diffraction measurements using the cells with a 1 μm focused XFEL beam. We successfully measured coherent diffraction patterns of a nano-fabricated metal pattern at 70° tilt angle and obtained the reconstructed image by applying iterative phase retrieval. The liquid cell will be usefully applied to molecular-scale SPI by using more tightly focused XFELs. In particular, imaging of membrane proteins embedded in lipid membranes is expected to have an enormous impact on life science and medicine.
- This article is part of the themed collections: XFELs: cutting edge X-ray light for chemical and material sciences and 2019 PCCP HOT Articles


 Please wait while we load your content...
Please wait while we load your content...