Open Access Article
Open Access ArticleShiga toxin binding alters lipid packing and the domain structure of Gb3-containing membranes: a solid-state NMR study†
Mathias
Bosse
a,
Jeremias
Sibold
b,
Holger A.
Scheidt
a,
Lukas J.
Patalag
 c,
Katharina
Kettelhoit
c,
Annika
Ries
c,
Daniel B.
Werz
c,
Katharina
Kettelhoit
c,
Annika
Ries
c,
Daniel B.
Werz
 c,
Claudia
Steinem
c,
Claudia
Steinem
 bd and
Daniel
Huster
bd and
Daniel
Huster
 *a
*a
aInstitute for Medical Physics and Biophysics, Leipzig University, Härtelstr. 16-18, D-04107 Leipzig, Germany. E-mail: daniel.huster@medizin.uni-leipzig.de
bInstitute for Organic and Biomolecular Chemistry, University of Göttingen, Tammannstr. 2, D-37077 Göttingen, Germany
cTechnische Universität Braunschweig, Institute of Organic Chemistry, Hagenring 30, D-38106 Braunschweig, Germany
dMax-Planck-Institute for Dynamics and Self-Organization, Am Fassberg 11, 37077 Göttingen, Germany
First published on 25th June 2019
Abstract
We studied the influence of globotriaosylceramide (Gb3) lipid molecules on the properties of phospholipid membranes composed of a liquid ordered (lo)/liquid disordered (ld) phase separated 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC)/N-palmitoyl-D-erythro-sphingosylphosphorylcholine (PSM)/cholesterol mixture (40/35/20, mol/mol/mol) supplemented with 5 mol% of either short acyl chain palmitoyl-Gb3 or long acyl chain lignoceryl-Gb3 using 2H solid-state NMR spectroscopy. To this end, both globotriaosylceramides were chemically synthesized featuring a perdeuterated lipid acyl chain. The solid-state 2H NMR spectra support the phase separation into a POPC-rich ld phase and a PSM/cholesterol-rich lo phase. The long chain lignoceryl-Gb3 showed a rather unusual order parameter profile of the acyl chain, which flattens out for the last ∼6 methylene segments. Such an odd chain conformation can be explained by partial chain interdigitation and/or a very fluid midplane region of the membrane. Possibly, the Gb3 molecules may thus preferentially be localized at the lo/ld phase boundary. In contrast, the short chain palmitoyl-Gb3 was well associated with the PSM/cholesterol-rich lo phase. Gb3 molecules act as membrane receptors for the Shiga toxin (STx) produced by Shigella dysenteriae and by enterohemorrhagic strains of Escherichia coli (EHEC). The B-subunits of STx (STxB) forming a pentameric structure were produced recombinantly and incubated with the membrane mixtures leading to alterations in the lipid packing properties and lateral organization of the membranes. Typically, STxB binding led to a decrease in lipid chain order in agreement with partial immersion of protein segments into the lipid–water interface of the membrane. In the presence of STxB, Gb3 preferentially partitioned into the lo membrane phase. In particular the short acyl chain palmitoyl-Gb3 showed very similar chain order parameters to PSM. In the presence of STxB, all lipid species showed isotropic contributions to the 2H NMR powder spectra; this was most pronounced for the Gb3 molecules. Such isotropic contributions are caused by highly curved membrane structures, which have previously been detected as membrane invaginations in fluorescence microscopy. Our analysis estimated that STxB induced highly curved membrane structures with a curvature radius of less than ∼10 nm likely related to the insertion of STxB segments into the lipid–water interface of the membrane.
Introduction
Specific lipid recruitment by membrane-binding proteins is a common theme in protein–membrane interactions.1,2 Particularly electrostatic interactions between polybasic stretches in the protein sequence and negatively charged phospholipids are responsible for increasing the local concentration of acidic lipids in the interaction area with the protein. Often, highly charged phospholipid species such as PIP2 are involved in the very specific lipid–protein interactions.1,3 Furthermore, a specific charge clustering mechanism has also been observed for cationic amphiphilic antimicrobial peptides, which recruit negatively charged phospholipids into highly acidic domains upon peptide binding.4–6 Less well understood are other mechanisms that lead to specific protein–lipid interactions in the membrane.7 It is well observed, however, that the heterogeneous membrane structure which consists of raft-like liquid ordered (lo) and fluid liquid disordered (ld) domains gives rise to specific protein binding and interactions.8–10Shiga toxin is an AB5 protein produced by Shigella dysenteriae and by enterohemorrhagic strains of E. coli (EHEC), binding specifically with its homopentameric B-subunits (STxB) to the receptor lipid globotriaosylceramide (Gb3).11,12 Each B subunit harbors three Gb3 binding sites, which means that the pentamer can specifically bind up to 15 Gb3 molecules.13 There are several lines of evidence that the cytotoxicity of Shiga toxin is strongly associated with Gb3 density and with cholesterol-containing lipid rafts.14,15 Following this hypothesis, it has been demonstrated that STxB binding leads to an increased lipid order in cellular membranes.13 The results obtained from artificial membranes on solid substrates and from pore-spanning membranes further showed that STxB binds to the lo phase in phase-separated membranes and is capable of reorganizing lipids within these membranes concomitant with protein cluster formation.16–20 This process can trigger a protein induced local compaction of particular lipids in one leaflet leading to an asymmetric reduction in the membrane area, which is a prerequisite for membrane invaginations and eventually protein internalization.21
Solid-state 2H NMR has been used to study lipid chain order and packing density in membranes for five decades.22 It is still a very precise tool to investigate these membrane properties and the phase state of complex lipid mixtures.23–25 A substantial amount of 2H NMR work has been performed to characterize the structure and dynamics of simple glycosphingolipids in membranes such as monogalactosyldiacylglycerol and digalactosyldiacylglycerol26 as well as galactosyl ceramides,27 but also more complex glycosphingolipids.28 Most of the previous studies primarily focused on the headgroup orientation of short acyl chain glycerolipids.26,29
As membrane lipids are ubiquitous, highly abundant, and relatively similar in their chemical structure, investigation of specific lipid–protein interactions by biophysical methods rests on an important prerequisite: specific lipid molecules are required that contain functional groups such as (i) fluorescence or EPR labels,30–32 (ii) isotopic enrichment,22,33 or (iii) functional groups for cross linking with other lipids or proteins.34 Here, we have used specifically deuterated Gb3 lipids in interaction studies with STxB by solid-state 2H NMR spectroscopy. Isotopic enrichment is a very well established method allowing highlighting protein interaction with specific lipids. Deuteration of biologically relevant lipid molecules is considered a non-perturbing modification to specifically study their protein interaction and its influence on lipid packing, lateral organization, and the membrane phase structure. We compare the consequences of STxB interaction with complex membrane mixtures containing a natural 24 carbon long saturated chain Gb3 with a Gb3 molecule containing 16 carbons in the chain.
Results
Recombinant expression of STxB
Only little information is available in the literature about the recombinant expression of native STxB.35,36 For our NMR studies, relatively large amounts of unmodified STxB had to be prepared with sufficient purity (>80%). The use of the IMPACT™-system37 for the soluble expression of STxB allows purification and tag removal in one-step, without further modification of the target protein. The pTXB1 vector enables the fusion of the self-cleavable intein tag that contains the chitin binding domain C-terminal to the target protein. The ligase-independent and restriction enzyme-free PIPE method38 was used to clone the codon optimized STxB DNA in the pTXB1 plasmid. The sequence of the construct was confirmed by Sanger sequencing (Eurofins Genomics, Ebersberg, Germany).Although an insoluble expression and refolding of the target protein fused to the IMPACT™-system is in principle possible,39 a soluble expression of the STxB fusion protein avoids essential refolding steps. To favor soluble expression, the temperature was reduced to 24 °C before inducing protein expression. Furthermore, the expression process was terminated after 4–5 h to avoid oxygen restriction to the cells or pH alteration. After cell harvest and lysis, the soluble protein fraction was incubated with chitin beads and loaded on an empty column. To remove all nonbinding proteins, the column was washed excessively with 25 cv of buffer containing Triton-X 100 or a high salt concentration. The STxB fusion protein was cleaved with DTT. The use of 2-mercaptoethanesulfonic acid (MESNA) as a thiol reagent to induce intein-mediated cleavage generates a C-terminal thioester on the target protein, which could be used for further reactions40,41 and facilitates the possibility of generating STxB-drug conjugates. The eluted STxB fraction was rebuffered first in PBS followed by a buffer exchange to HEPES buffer for the NMR measurements. The purity of the expressed STxB was confirmed by Coomassie stained gel electrophoresis (Fig. 1). Finally, 1–2 mg of pure STxB could be isolated from 1 L culture medium. The identity of isolated STxB was confirmed by mass spectrometry (Dr Christian Ihling, Martin Luther University, Halle, Germany).
The STxB preparation of a 2 L cell culture provides enough material for 1–2 NMR samples. Unfortunately, this is not rentable for the production of a sufficient quantity of STxB related to chemical and biotechnological engineering approaches. The use of defined minimal medium42,43 to reach higher optical densities and refolding strategies could increase the final yield.
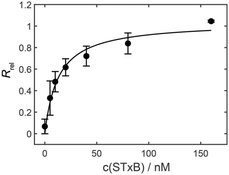
Membrane binding affinity of STxB
STxB is known to bind with high specificity to Gb3 with dissociation constants in the 10 nM range (relative to the STxB monomer).44 To prove the functionality of recombinantly expressed STxB, SPR measurements were performed resulting in the isotherm shown in Fig. 2. Fitting a Langmuir isotherm to the data resulted in a KD = 13 ± 7 nM in agreement with previous findings confirming the binding activity of the recombinant protein.442H NMR studies of POPC/PSM/lignoceryl-Gb3/cholesterol mixtures in the absence and in the presence of STxB
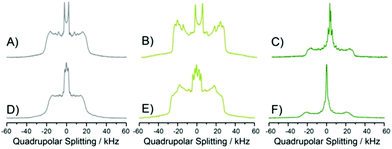
First, a comprehensive set of 2H NMR data of POPC/PSM/lignoceryl-Gb3/cholesterol mixtures at temperatures between 7 and 45 °C in the absence and in the presence of STxB was generated, in which each lipid component was deuterated, respectively, allowing the study of the phase organization, lipid packing and membrane organization of each component in the complex mixture individually.45 Lipid mixtures were investigated in the form of multilamellar vesicles both in the absence and in the presence of STxB. Typical 2H NMR spectra of each deuterated lipid measured at a temperature of 25 °C are shown in Fig. 3. The 2H NMR spectra at all temperatures studied are shown in Fig. S1–S3 (ESI†).In the absence of STxB, each lipid component shows the well-known superposition of 2H NMR Pake spectra indicative of a lipid chain order gradient along the chain at this temperature characteristic of lamellar liquid-crystalline bilayers.22,46 As inferred from the 2H NMR spectra, all lipid molecules undergo rapid axially symmetric rotational diffusion about their long axis. Lignoceryl-Gb3 and PSM exhibited larger residual quadrupolar splittings compared to POPC, suggesting a higher state of order for these saturated lipids than the unsaturated POPC in agreement with previous studies.23–25,47 Furthermore, the 2H NMR spectrum of lignoceryl-Gb3 is characterized by additional intensity in the central region compared to the NMR spectra of the two phospholipids. All 2H NMR spectra showed a single set of quadrupolar splittings for each deuterated segment suggesting that each lipid species is constrained to a single phase state and no demixing of the lipids into micrometer sized domains occurred. However, the significant differences in the observed quadrupolar splittings between individual lipid species indicate lateral fluctuations in the lipid distributions and formation of heterogeneous lipid distributions.
In the presence of STxB, several characteristic alterations of the 2H NMR spectra of each lipid component in the mixture are detected (Fig. 3D–F and Fig. S1–S3, ESI†). First, the width of the 2H NMR spectrum of POPC is reduced indicative of a decrease in lipid chain order upon STxB binding to the membrane. Second, the 2H NMR spectrum of PSM clearly shows two contributions, indicative of PSM molecules residing in two separated phases that are in slow exchange. Third, the 2H NMR spectrum of Gb3 becomes significantly broader in the presence of STxB accompanied by an increase in the line widths of the individual Pake doublets suggestive of a decrease in the correlation times of motions of this lipid component. A fourth notable change in the 2H NMR spectra of STxB-containing membranes is the presence of a pronounced isotropic signal for all three lipid species.
A more quantitative evaluation of the 2H NMR data with respect to the phase state of the membrane and an analysis of the spectral changes of each individual lipid component upon addition of STxB is facilitated by comparing the lipid chain order parameters for each glycerolipid component in the membrane as illustrated in Fig. 4. Order parameter plots for each lipid species are shown for three different temperatures (25, 37, and 45 °C). As expected, order parameters of all lipid components decrease with increasing temperature. In the absence of STxB (Fig. 4A–C), a clear difference in the order parameters of PSM-d31 and POPC-d31 in the mixture is observed. In agreement with previous 2H NMR data on lipid raft mixtures, PSM shows higher order parameters as it preferentially co-localizes with cholesterol in the cholesterol rich lo phase, while POPC remains in the ld phase which is deprived of cholesterol and indicated by low order parameters.23–25 The order parameter profile of lignoceryl-Gb3-d47 in the mixture drastically differs from what is observed for the phospholipids in the mixture. The upper chain segments (C2–C16) show a similar plateau of high order parameters as known for phospholipids in liquid crystalline membranes. The order parameters at this plateau lie between those of the PSM and the POPC in the mixture. Interestingly, the lignoceryl-Gb3 order parameter decreases in the lower half of the chain but this decrease becomes very shallow at segment C20 towards the end of the chain. Such order parameter profiles have also been reported for long chain ceramide species in lipid mixtures mimicking the stratum corneum layer of the skin recently48 and also in older work on glycosphingolipids in model membranes.49–51 It can be inferred from these order parameter plots that the long chain lignoceryl-Gb3 lipids do not incorporate very well in the lipid membrane consisting of 75 mol% of lipids with shorter chains (16 or 18 carbons). It is not clear from these data where the lignoceryl-Gb3 is exactly localized in the demixed membrane; even a localization at the phase boundary as previously found for lipidated proteins seems possible.52 The unusually looking order parameter profiles may also indicate lipid chain interdigitation.50,51 Clearly, the much longer acyl chain of Gb3 creates a packing defect in the membrane that results in a very special chain conformation. For all lipid species, upon an increase in temperature, a decrease in the lipid chain order is recorded as observed for the other lipid species.
 | ||
Fig. 4
2H NMR order parameters of the quaternary lipid mixture POPC/PSM/lignoceryl-Gb3/cholesterol (molar ratio: 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 35 35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20) in the absence (A–C) and in the presence of STxB (a molar protein to lipid ratio of 1 20) in the absence (A–C) and in the presence of STxB (a molar protein to lipid ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 120, D–F) at a temperature of 25 °C (top row), 37 °C (middle row), and 45 °C (bottom row) for each glycerolipid compound. Experimental error bars in the determination of the 2H NMR order parameters would be smaller than the symbol size.53 120, D–F) at a temperature of 25 °C (top row), 37 °C (middle row), and 45 °C (bottom row) for each glycerolipid compound. Experimental error bars in the determination of the 2H NMR order parameters would be smaller than the symbol size.53 | ||
In the presence of STxB, interesting alterations in the order parameters are observed (Fig. 4D–F). (i) The upper chain plateau of order parameters of PSM-d31 and lignoceryl-Gb3-d47 become relatively equal, which is caused (ii) by a relatively pronounced 13–15% decrease of the order parameter of PSM-d31 while the order parameters of lignoceryl-Gb3-d47 remain about constant. At 25 °C, STxB addition leads to the appearance of two macroscopically demixed phases for PSM-d31 as already seen from two sets of quadrupolar splittings for each chain methyl and methylene segment in the 2H NMR spectra (Fig. 3). Such 2H NMR spectra cannot be dePaked and therefore no reliable order parameters can be reported in Fig. 4D. At higher temperatures, PSM-d31 is in fast exchange again and chain order parameters can be calculated in the presence of STxB as shown in Fig. 4E and F. (iii) The POPC-d31 order parameters also decrease upon addition of STxB by about 5 to 12%.
These results provide two main conclusions about the restructuring of the mixture upon addition of STxB. First, lignoceryl-Gb3 has a tendency to associate with the lo phase in the presence of STxB and second, the ld phase is deprived of residual lignoceryl-Gb3 and cholesterol indicated by decreased order parameters of the POPC component. This is in agreement with the preferential interaction of Gb3 with STxB reported before.20
2H NMR studies of POPC/PSM/palmitoyl-Gb3/cholesterol mixtures in the absence and in the presence of STxB
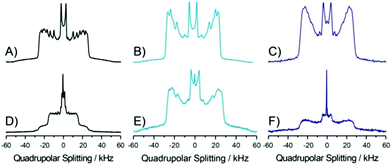
For comparison, we also studied the same lipid mixture in the absence and in the presence of STxB but replaced lignoceryl-Gb3 by the short chain analogue palmitoyl-Gb3. The corresponding 2H NMR spectra of each individual lipid component at a temperature of 25 °C are shown in Fig. 5. The 2H NMR spectra at all temperatures are shown in Fig. S4–S6 (ESI†).In the absence of STxB, the 2H NMR spectra of each lipid component are again well described by the classical superposition of Pake doublets. Very characteristic alterations in the NMR spectra are observed upon addition of STxB. Most strikingly, the POPC lipid component appears to phase separate into two domains with high and low order as seen from the superposition of two sets of 2H NMR spectra of very different width. These domains are large such that the lipids are in slow exchange. This phase separation is observed almost over the entire temperature range investigated here (7–45 °C). Only at 45 °C, the 2H NMR spectrum of the POPC in the mixture consists of a single phase. The changes observed in the 2H NMR spectra of the PSM component of the mixture are relatively minor and also the NMR spectrum of the palmitoyl-Gb3 component does exhibit very large alterations due to the presence of STxB. Again, as in the mixtures containing lignoceryl-Gb3, in the presence of STxB, the 2H NMR spectra of each lipid compound display a (small) isotropic component, which was absent in the absence of the protein.
More quantitatively, these changes are reported in the order parameter profiles, which are shown in Fig. 6. In the absence of STxB (Fig. 6A–C), the differences in the order parameter profiles of the three glycerolipids are much smaller than what was found for the mixture containing lignoceryl-Gb3 (compare Fig. 4). This suggests that the very long chain lignoceryl-Gb3 indeed influences the phase state of the membrane, which at this relatively low cholesterol concentration of 20 mol% has a limited tendency to demix. Nevertheless, at all temperatures, PSM-d31 shows higher order parameters than POPC-d31 in the mixture, which supports the interpretation of lo/ld phase formation. The order parameters of palmitoyl-Gb3 do not follow a clear trend, they are the highest at 25 °C, intermediate at 37 °C, and the lowest at 45 °C compared to the two phospholipids in the mixture. This may confirm a previous finding for lignoceryl-Gb3, which could not be clearly assigned to the lo or the ld phase state.
 | ||
Fig. 6
2H NMR order parameters of the quaternary lipid mixture POPC/PSM/palmitoyl-Gb3/cholesterol (molar ratio: 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40 40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 35 35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20) in the absence (A–C) and in the presence of STxB (a molar protein to lipid ratio of 1 20) in the absence (A–C) and in the presence of STxB (a molar protein to lipid ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 120, D–F) at a temperature of 25 °C (top row), 37 °C (middle row), and 45 °C (bottom row) for each lipid compound. Experimental error bars in the determination of the 2H NMR order parameters would be smaller than the symbol size.53 120, D–F) at a temperature of 25 °C (top row), 37 °C (middle row), and 45 °C (bottom row) for each lipid compound. Experimental error bars in the determination of the 2H NMR order parameters would be smaller than the symbol size.53 | ||
The alterations in the phase state of the mixture that are induced by addition of STxB as illustrated in Fig. 6D and E are very clear. The formation of two phases in the mixture at all observed temperatures is very obvious: a lo phase formed by PSM/palmitoyl-Gb3/cholesterol characterized by high order parameters, and a cholesterol- and palmitoyl-Gb3-deprived ld phase enriched in POPC. This is indicated by very drastic alterations in the chain order parameters of POPC upon addition of STxB: POPC order parameters decrease by 33–39%. In contrast, the order parameter of PSM and palmitoyl-Gb3 only moderately change to become more equal for the two lipids: at 25 °C, palmitoyl-Gb3 order decreases by 10% while PSM order increases by 3%; at 37 °C, palmitoyl-Gb3 order decreases by 3% while PSM remains constant; and at 45 °C, palmitoyl-Gb3 order increases by 4% while PSM order decreases by 3%. These pronounced order alterations lead to a clear phase separation of the system.
Discussion
Incorporation of Gb3 into POPC/PSM/cholesterol mixed membranes
Lateral organization of the lipids in bilayers is governed by the interactions between all membrane components. The interaction energies between lipids include all segments of the molecules, but very critically depend on hydrophobic chain length matching between neighboring lipids. Gb3 is typically found in nature with long fatty acid chains (C20–C24), which can also be unsaturated. Thus, the membrane incorporation of natural Gb3 is crucially dependent on the length of its acyl chain. Such long chains as found in Gb3 molecules create a significant packing defect in the host membrane, which typically contain lipids with C16–C18 chains.54 Interdigitation of the chains may occur to accommodate such long chains in order to avoid the exposure of the hydrophobic methylene segments to the aqueous environment.50,55 Acyl chain unsaturation, which is found in some of the Gb3 species in nature, may partially relieve the membrane packing defects induced by the very long acyl chains of the Gb3 molecules. Studying the response of complex lipid mixtures to protein binding by experimental techniques represents a challenging task since phospholipids share a rather common chemical structure providing few spectroscopically unique features.7,56,57 This challenge is typically overcome by specific lipids carrying probe molecules30,58 or isotopic labelling. 2H NMR spectroscopy is particularly well suited as phospholipids with individually 2H-labeled acyl chains can readily be synthesized allowing the study of the response of each individual deuterated lipid species to membrane binding ions, peptides, proteins or other molecules.59–61In the current study, we investigated in detail the lipid chain packing properties of both the Gb3 and the phospholipids from the host membrane (POPC and PSM). To this end, isotopic enrichment of Gb3 with deuterated long (24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0) or short (16
0) or short (16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0) saturated chains was accomplished by chemical synthesis. Using 2H NMR, the individual chain order parameters of each lipid component were measured and evaluated with respect to lipid packing. From the NMR data, we could calculate the average chain extent of each lipid component of the mixture using the mean torque model.62 For instance, at 37 °C, the chain extent of POPC and PSM in the quaternary lipid mixtures is 13.6 and 15.4 Å, respectively (Tables S1–S4, ESI†). This agrees with the formation of the well-known ld and lo phases as also observed in the absence of Gb3.23–25 In contrast to the two phospholipids, the acyl chain of lignoceryl-Gb3 extents to 19.5 Å, creating a pronounced chain length mismatch relative to the host membrane. This large discrepancy in acyl chain lengths suggests that lignoceryl-Gb3 is not well incorporated into the membrane, may undergo interdigitation or may show a very unusual chain conformation.50,51 Such a response of the system seems typical for long chain lipids in classical membrane systems as seen for long chain ceramides in stratum corneum lipid mixtures48 and also in previous work on glycosphingolipids.49–51 It has been suggested before that the longer chains of Gb3 may better present the carbohydrate moiety for protein binding,63 but such a scenario would expose hydrophobic chain segments to the aqueous environment, which would be energetically very unfavorable.
0) saturated chains was accomplished by chemical synthesis. Using 2H NMR, the individual chain order parameters of each lipid component were measured and evaluated with respect to lipid packing. From the NMR data, we could calculate the average chain extent of each lipid component of the mixture using the mean torque model.62 For instance, at 37 °C, the chain extent of POPC and PSM in the quaternary lipid mixtures is 13.6 and 15.4 Å, respectively (Tables S1–S4, ESI†). This agrees with the formation of the well-known ld and lo phases as also observed in the absence of Gb3.23–25 In contrast to the two phospholipids, the acyl chain of lignoceryl-Gb3 extents to 19.5 Å, creating a pronounced chain length mismatch relative to the host membrane. This large discrepancy in acyl chain lengths suggests that lignoceryl-Gb3 is not well incorporated into the membrane, may undergo interdigitation or may show a very unusual chain conformation.50,51 Such a response of the system seems typical for long chain lipids in classical membrane systems as seen for long chain ceramides in stratum corneum lipid mixtures48 and also in previous work on glycosphingolipids.49–51 It has been suggested before that the longer chains of Gb3 may better present the carbohydrate moiety for protein binding,63 but such a scenario would expose hydrophobic chain segments to the aqueous environment, which would be energetically very unfavorable.
Generally, the length of a lipid chain in the membrane is determined by the number of gauche conformers. Each gauche defect decreases the chain length by 1.1 Å.64 At 37 °C, we can estimate from the 2H NMR data that the 24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 chain of Gb3 within the quaternary mixture already contains 8 gauche defects. In order to match the hydrophobic thickness of the host membrane, 3–4 additional gauche defects would have to be created, which is apparently energetically more costlier than a backfolded chain conformation or lipid chain interdigitation. The latter possibility has been looked at before in great detail.50 In this seminal study, galactosyl ceramide with a 24
0 chain of Gb3 within the quaternary mixture already contains 8 gauche defects. In order to match the hydrophobic thickness of the host membrane, 3–4 additional gauche defects would have to be created, which is apparently energetically more costlier than a backfolded chain conformation or lipid chain interdigitation. The latter possibility has been looked at before in great detail.50 In this seminal study, galactosyl ceramide with a 24![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 acyl chain was incorporated into SOPC membranes providing very similar order parameter plots to that reported in the current work. Morrow et al. concluded that the long chain can partially extend the bilayer midplane and penetrate the other membrane leaflet. The extra chain length may also form a highly flexible layer in the bilayer midplane penetrating the methyl groups of the opposing monolayer.50 In light of the current membrane models characterized by highly mobile lipid chains in membranes, these two scenarios may not fully explain the behavior of this elongated chain end, it seems to be clear that the long Gb3 chain is in a highly dynamic state.
0 acyl chain was incorporated into SOPC membranes providing very similar order parameter plots to that reported in the current work. Morrow et al. concluded that the long chain can partially extend the bilayer midplane and penetrate the other membrane leaflet. The extra chain length may also form a highly flexible layer in the bilayer midplane penetrating the methyl groups of the opposing monolayer.50 In light of the current membrane models characterized by highly mobile lipid chains in membranes, these two scenarios may not fully explain the behavior of this elongated chain end, it seems to be clear that the long Gb3 chain is in a highly dynamic state.
The very different chain order parameters of PSM and POPC in the mixture are in agreement with an lo/ld phase separation of the membrane. These domains are small and likely transient representing time-dependent fluctuations in the lateral lipid distribution. On the basis of the 2H NMR experiments, it is not entirely clear where the Gb3 molecules are localized in the complex mixtures, even localization at the interface between lo and ld phases is possible.52 From the distinctive shape of the chain order parameter profile of lignoceryl-Gb3 it is likely that chain interdigitation occurred. It seems clear, though, that Gb3 is not uniformly distributed in the membrane as previously assumed.65 Previous AFM and fluorescence microscopy work clearly showed that Gb3 is associated with the lo phase of the membrane after STxB binding.19 Moreover, the fatty acid of Gb3 appears to influence the partition of Gb3 and probably also sphingomyelin in the phase separated membranes.20 We note, however, that a chain length mismatch between the PSM in the lo phase and the chain of Gb3 of 4.1 Å exists at 37 °C.
For the palmitoyl-Gb3 in contrast, the situation is very clear and a relatively similar chain extension for the lo phase forming lipids is observed (15.3 Å for palmitoyl-Gb3 and 15.6 Å for PSM). This indicates that palmitoyl-Gb3 is well incorporated into the host membrane and partitions into the lo phase of the host membrane.
Membrane restructuring induced by STxB binding
STxB binding to the membrane causes an important restructuring of the lateral membrane organization and lipid re-distribution16–20 as also seen in the changes in the 2H NMR spectra recorded here. This is suggested from the previous observation that one STxB pentamer can bind up to 15 Gb3 molecules.66 Our data are in agreement with the previous finding that STxB interaction with the membrane occurs predominantly with the lo phase domains.19 However, we do not see a consistent increase in Gb3 lipid chain order upon interaction with STxB as seen in previous molecular dynamics simulations.67 At 37 °C, all lipids of the lignoceryl-Gb3-containing mixture actually slightly decrease their chain order parameters upon STxB binding, which implies slight thinning of the membrane. This could be caused by insertion of protein segments into the lipid–water interface of the membrane.Interestingly, the differences in chain extension of the individual lipids that was observed in the absence of the toxin still remained after STxB binding. The 2H NMR order parameters provide very detailed insights into chain packing alterations upon STxB binding. For instance, the upper half of the acyl chain of lignoceryl-Gb3 (C2–12) shows very similar order parameters to PSM (Fig. 4E and F), which supports the notion that Gb3 is associated with PSM/cholesterol-rich lo domains of the bilayer in agreement with previous results.19 Such a conclusion is much clearer for the mixture containing palmitoyl-Gb3, where the order parameters and consequently the chain extension of the Gb3 chain and the palmitoyl chain of PSM are indeed very similar as expected for well mixed lipids (Fig. 6D–F). The POPC-rich ld phase must be clearly segregated from this well ordered lo phase, rich in PSM, Gb3, and cholesterol.
It has been reported previously that STxB binding induces membrane invaginations and membrane structures with high curvature when binding to cells13 and artificial membranes.44 While the situation in live cells is much more complicated compared to the conditions in the artificial membranes studied here, we note pronounced isotropic spectral intensity in the 2H NMR spectra of all lipid molecules as a consequence of STxB binding, but most pronounced for the Gb3 lipids. This strongly suggests that highly curved membrane structures are formed in response to STxB binding. Considering that the molecular motions that would lead to the full averaging of the 2H NMR spectra to produce isotropic signals have to be much faster than ∼50 μs and assuming an effective lipid diffusion constant of 3 × 10−12 m2 s−1,68 the curvature radius of such structures should be smaller than ∼10 nm to observe the isotropic spectral components. While all lipid species of the mixture show such isotropic components in the presence of STxB, this peak is most pronounced for the Gb3 lipids. This confirms that STxB binding to the mixed membrane induces highly curved lipid structures, which involve predominantly the Gb3 lipids along with some of the other membrane components. While tubule formation has been shown before to predominantly occur in membranes containing monounsaturated Gb3 molecules,13 our current data show that STxB-induced membrane curvature apparently also has an effect on membranes containing saturated Gb3. Computer simulations also showed that STxB can induce small local curvature effects in membranes in the presence of saturated Gb3 as used in our study.67 But the small curvature of membranes as detected by 2H NMR does not prove that microscopic invaginations may occur in the systems in the presence of saturated Gb3. This is also suggested as similar effects were observed in the presence of both long chain lignoceryl-Gb3 and short chain palmitoyl-Gb3.
Experimental
Materials
Synthetic DNA for STxB and primers for cloning were purchased from Thermo Scientific (Waltham, MA, USA). The pTXB1 vector, ER2566 cells and chitin beads were obtained from New England Biolabs (Frankfurt am Main, Germany). All other chemicals were purchased from Sigma Aldrich (Taufkirchen, Germany) or Carl Roth (Karlsruhe, Germany). The lipids 1,2-dioleoyl-sn-glycero-3-phosphocholine (DOPC), 1-palmitoyl-2-oleoyl-sn-glycero-3-phosphocholine (POPC), and N-palmitoyl-D-erythro-sphingosylphosphorylcholine (PSM) as well as the derivatives of these molecules with a deuterated palmitoyl chain in the sn-1 position (POPC-d31, PSM-d31), and cholesterol were purchased from Avanti Polar Lipids, Inc. (Alabaster, Al). Porcine Gb3 used for the SPR study was purchased from Matreya (State College, PA). All chemicals were used without further purification.Chemical synthesis of lignoceryl-Gb3 and palmitoyl-Gb3
The chemical synthesis of the four Gb3 derivatives was conducted starting from literature-known azide 1, whose synthesis was previously reported by us (Scheme 1).19 Reduction of the azide under Staudinger conditions utilizing PPh3 and water in benzene led to a free amine. HATU-mediated coupling with the respective fatty acids afforded four different protected glycosphingolipids. To finalize the synthesis all protecting groups were removed under Zemplén conditions and deuterated lignoceryl-Gb32(D), palmitoyl-Gb33(D) and the protonated congeners 2(H)19 and 3(H)69 were obtained. A thin layer chromatogram of all Gb3 lipids is reproduced in Fig. S7 (ESI†).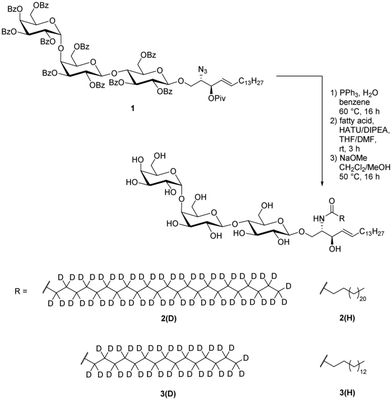 | ||
| Scheme 1 Chemical synthesis of lignoceryl-Gb3-d472(D), palmitoyl-Gb3-d313(D) and protonated congeners 2(H) and 3(H) from literature-known azide 1.19 | ||
Recombinant xpression of the shiga toxin subunit B
The DNA encoding the wild type Shiga toxin subunit B (STxB) was amplified by PCR and cloned in frame with the Mxe intein and chitin binding domain by using the PIPE method38 to generate the STxB_pTXB1 plasmid.STxB was expressed as Mxe intein and chitin binding domain (CBD) fusion protein to fully exploit the IMPACT™-system (New England Biolabs, Germany)37 in Escherichia coli ER2566 cells. After growing the cells at 37 °C in LB medium, the temperature was reduced to 24 °C and protein expression was induced at an OD600 of ∼1 with 0.5 mM IPTG for 4–5 h. All following steps were carried out at 4 °C or one ice. Cells were harvested by centrifugation and resuspended in lysis buffer (20 mM Tris–HCl, 500 mM NaCl, 1 mM EDTA, 1% (v/v) Triton X-100, pH 8.5), incubated for 30 min and lysed by a homogenizer (APV-2000). After 60 min incubation with DNaseI and 3 mM MgCl2 and centrifugation, the supernatant was diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 with column buffer (CB) (20 mM Tris–HCl, 500 mM NaCl, 1 mM EDTA, pH 8.5) and incubated in a batch with the chitin beads (equilibrated with CB) overnight. The batch was transferred to an empty column and washed stepwise with CB, CB +0.2% (v/v) Triton X-100, CB, CB +1 M NaCl and CB (5 column volumes (cv) for each step). Cleavage was induced by incubation for 48 h with CB containing 50 mM dithiothreitol (DTT). After elution, the STxB sample was rebuffered via dialysis in phosphate buffered saline (PBS, 2.7 mM KCl, 136 mM NaCl, 1.5 mM KH2PO4, 8.1 mM Na2HPO4, pH 7.4) for binding studies and subsequently for the NMR studies in HEPES buffer (100 mM HEPES, 10 mM NaCl, pH 7.4) respectively. Finally, the protein was concentrated to ∼1 mg ml−1 before incubation with the membrane vesicles.
1 with column buffer (CB) (20 mM Tris–HCl, 500 mM NaCl, 1 mM EDTA, pH 8.5) and incubated in a batch with the chitin beads (equilibrated with CB) overnight. The batch was transferred to an empty column and washed stepwise with CB, CB +0.2% (v/v) Triton X-100, CB, CB +1 M NaCl and CB (5 column volumes (cv) for each step). Cleavage was induced by incubation for 48 h with CB containing 50 mM dithiothreitol (DTT). After elution, the STxB sample was rebuffered via dialysis in phosphate buffered saline (PBS, 2.7 mM KCl, 136 mM NaCl, 1.5 mM KH2PO4, 8.1 mM Na2HPO4, pH 7.4) for binding studies and subsequently for the NMR studies in HEPES buffer (100 mM HEPES, 10 mM NaCl, pH 7.4) respectively. Finally, the protein was concentrated to ∼1 mg ml−1 before incubation with the membrane vesicles.
Functionality assay for STxB
Functionality of recombinantly expressed STxB, i.e., its binding affinity to the receptor lipid Gb3, was analyzed using the surface plasmon resonance (SPR) technique. Gold-covered chips (SPR sensor chips AU, Xantex bioanalytics, Düsseldorf, Germany) were incubated with a 16 mM ethanolic 1-octanethiol solution overnight. After mounting the chip into the SPR system (SR700DC spectrometer, Reichert technologies life sciences, Buffalo, NY), the chip was rinsed with PBS. Small unilamellar vesicles (SUVs) composed of DOPC (reference channel) or DOPC/porcine Gb3 (95![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5) (measurement channel) were prepared by ultrasonication. DOPC/porcine Gb3 SUVs were flushed through the measurement channel to spread a monolayer on the 1-octanethiol functionalized chip, followed by DOPC SUVs added to the measurement and reference channel. To remove non-spread SUVs the system was rinsed 5 times for 2 min with 50 nM NaOH solution.44 The system was equilibrated with PBS for 5 min. STxB at different concentrations was added for 20 min followed by rinsing with PBS for 20 min. The change in reflectivity was plotted vs. the STxB concentration in solution and a Langmuir adsorption isotherm was fit to the data to obtain the dissociation constant.
5) (measurement channel) were prepared by ultrasonication. DOPC/porcine Gb3 SUVs were flushed through the measurement channel to spread a monolayer on the 1-octanethiol functionalized chip, followed by DOPC SUVs added to the measurement and reference channel. To remove non-spread SUVs the system was rinsed 5 times for 2 min with 50 nM NaOH solution.44 The system was equilibrated with PBS for 5 min. STxB at different concentrations was added for 20 min followed by rinsing with PBS for 20 min. The change in reflectivity was plotted vs. the STxB concentration in solution and a Langmuir adsorption isotherm was fit to the data to obtain the dissociation constant.
Sample preparation for 2H NMR measurements
For 2H NMR measurements, either lignoceryl-Gb3 or palmitoyl-Gb3 was mixed with POPC, PSM, and cholesterol at a molar ratio of 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 40
40![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 35
35![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 20 in a chloroform/methanol mixture. In each sample, either chain deuterated lignoceryl-Gb3-d47 or palmitoyl-Gb3-d31, chain deuterated POPC-d31, or chain deuterated PSM-d31 was used, respectively. After evaporating the solvent, the lipid film was re-suspended in buffer solution (100 mM HEPES, 10 mM NaCl, pH 7.4). From this suspension, large unilamellar vesicles (LUVs) were prepared by extrusion through 100 nm polycarbonate membranes70 and STxB was added at a molar ratio of 1 STxB pentamer per 20 Gb3 molecules. After overnight incubation at room temperature, the suspension was ultra-centrifuged for 5 h at ∼225
20 in a chloroform/methanol mixture. In each sample, either chain deuterated lignoceryl-Gb3-d47 or palmitoyl-Gb3-d31, chain deuterated POPC-d31, or chain deuterated PSM-d31 was used, respectively. After evaporating the solvent, the lipid film was re-suspended in buffer solution (100 mM HEPES, 10 mM NaCl, pH 7.4). From this suspension, large unilamellar vesicles (LUVs) were prepared by extrusion through 100 nm polycarbonate membranes70 and STxB was added at a molar ratio of 1 STxB pentamer per 20 Gb3 molecules. After overnight incubation at room temperature, the suspension was ultra-centrifuged for 5 h at ∼225![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g. The lipid pellet was transferred into 5 mm glass vials and tightly sealed for NMR measurements. Centrifugation typically results in the production of μm sized multilamellar vesicles (MLV), which enable detection of the typical 2H NMR spectra as observed.
000 × g. The lipid pellet was transferred into 5 mm glass vials and tightly sealed for NMR measurements. Centrifugation typically results in the production of μm sized multilamellar vesicles (MLV), which enable detection of the typical 2H NMR spectra as observed.
2H NMR measurements
2H NMR spectra were measured on a Bruker Avance 750 MHz NMR spectrometer (Bruker Biospin GmbH, Rheinstetten, Germany) operating at a resonance frequency of 115.1 MHz for 2H. A single-channel solid probe equipped with a 5 mm solenoid coil was used. A phase-cycled quadrupolar echo sequence71 was applied. The length of a 90° pulse was 2.3–2.5 μs and a relaxation delay of 1 s was applied. The measurements were conducted at various temperatures from 7 to 45 °C. 2H NMR spectra were processed as described in detail in Ref. 59 Briefly, 2H NMR powder spectra were dePaked using the algorithm of McCabe and Wassall.72 If spectral quality is allowed, smooth chain order parameters were calculated according to the literature.73Conclusions
2H isotopic enrichment of short and long chain Gb3 was key to individually study the lipid chain packing and phase organization of mixed membranes forming small heterogeneities in the lipid distribution. Short chain Gb3 preferentially associates with the lo membrane phase and the long chain variant of the molecules may interdigitate, show a very fluid lower chain and might be localized at the lo/ld phase boundary. STxB binding leads to a slight disordering of all lipids in the mixture possibly due to insertion of protein segments into the lipid–water interface of the membrane along with a very preferential partitioning of Gb3 into the lo phase. STxB binding also induces highly curved membrane structures, which preferentially involves the Gb3 species.Conflicts of interest
There are no conflicts to declare.Acknowledgements
MB and DH thank Dr Christian Ihling (Martin Luther University Halle) for mass spectrometry measurements. CS and DBW thank the DFG (SFB 803, project A05) for financial support. The project was also supported by the DFG project HU 720/18-1.Notes and references
- S. McLaughlin and D. Murray, Nature, 2005, 438, 605 CrossRef CAS PubMed.
- L. Brunsveld, H. Waldmann and D. Huster, Biochim. Biophys. Acta, 2009, 1788, 273 CrossRef CAS PubMed.
- U. Golebiewska, A. Gambhir, G. Hangyas-Mihalyne, I. Zaitseva, J. Radler and S. McLaughlin, Biophys. J., 2006, 91, 588 CrossRef CAS PubMed.
- R. F. Epand, W. L. Maloy, A. Ramamoorthy and R. M. Epand, Biochemistry, 2010, 49, 4076 CrossRef CAS PubMed.
- F. Jean-Francois, S. Castano, B. Desbat, B. Odaert, M. Roux, M. H. Metz-Boutigue and E. J. Dufourc, Biochemistry, 2008, 47, 6394 CrossRef CAS PubMed.
- A. Arouri, M. Dathe and A. Blume, Biochim. Biophys. Acta, 2009, 1788, 650 CrossRef CAS PubMed.
- D. Huster, Biochim. Biophys. Acta, 2014, 1841, 1146 CrossRef CAS PubMed.
- K. Simons and E. Ikonen, Nature, 1997, 387, 569 CrossRef CAS PubMed.
- G. Vereb, J. Szollosi, J. Matko, P. Nagy, T. Farkas, L. Vigh, L. Matyus, T. A. Waldmann and S. Damjanovich, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 8053 CrossRef CAS PubMed.
- M. Heinrich, A. Tian, C. Esposito and T. Baumgart, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 7208 CrossRef CAS PubMed.
- T. Waddell, A. Cohen and C. A. Lingwood, Proc. Natl. Acad. Sci. U. S. A., 1990, 87, 7898 CrossRef CAS PubMed.
- M. S. Jacewicz, M. Mobassaleh, S. K. Gross, K. A. Balasubramanian, P. F. Daniel, S. Raghavan, R. H. McCluer and G. T. Keusch, J. Infect. Dis., 1994, 169, 538 CrossRef CAS PubMed.
- W. Römer, L. Berland, V. Chambon, K. Gaus, B. Windschiegl, D. Tenza, M. R. Aly, V. Fraisier, J. C. Florent, D. Perrais, C. Lamaze, G. Raposo, C. Steinem, P. Sens, P. Bassereau and L. Johannes, Nature, 2007, 450, 670 CrossRef PubMed.
- O. Kovbasnjuk, M. Edidin and M. Donowitz, J. Cell Sci., 2001, 114, 4025 CAS.
- T. Hanashima, M. Miyake, K. Yahiro, Y. Iwamaru, A. Ando, N. Morinaga and M. Noda, Microb. Pathog., 2008, 45, 124 CrossRef CAS PubMed.
- N. Teske, J. Sibold, J. Schumacher, N. K. Teiwes, M. Gleisner, I. Mey and C. Steinem, Langmuir, 2017, 33, 14175 CrossRef CAS PubMed.
- M. Safouane, L. Berland, A. Callan-Jones, B. Sorre, W. Römer, L. Johannes, G. E. Toombes and P. Bassereau, Traffic, 2010, 11, 1519 CrossRef CAS PubMed.
- A. Orth, L. Johannes, W. Römer and C. Steinem, ChemPhysChem, 2012, 13, 108 CrossRef CAS PubMed.
- O. M. Schütte, A. Ries, A. Orth, L. J. Patalag, W. Römer, C. Steinem and D. W. Werz, Chem. Sci., 2014, 5, 3104 RSC.
- B. Windschiegl, A. Orth, W. Römer, L. Berland, B. Stechmann, P. Bassereau, L. Johannes and C. Steinem, PLoS One, 2009, 4, e6238 CrossRef PubMed.
- L. Johannes and S. Mayor, Cell, 2010, 142, 507 CrossRef CAS PubMed.
- J. Seelig, Q. Rev. Biophys., 1977, 10, 353 CrossRef CAS PubMed.
- S. L. Veatch, O. Soubias, S. L. Keller and K. Gawrisch, Proc. Natl. Acad. Sci. U. S. A., 2007, 104, 17650 CrossRef CAS PubMed.
- T. Bartels, R. S. Lankalapalli, R. Bittman, K. Beyer and M. F. Brown, J. Am. Chem. Soc., 2008, 130, 14521 CrossRef CAS PubMed.
- A. Bunge, P. Müller, M. Stöckl, A. Herrmann and D. Huster, Biophys. J., 2008, 94, 2680 CrossRef CAS PubMed.
- B. Bechinger, P. M. Macdonald and J. Seelig, Biochim. Biophys. Acta, 1988, 943, 381 CrossRef CAS.
- E. F. Smeets, P. Comfurius, E. M. Bevers and R. F. Zwaal, Biochim. Biophys. Acta, 1994, 1195, 281 CrossRef.
- D. M. Singh, X. Shan, J. H. Davis, D. H. Jones and C. W. Grant, Biochemistry, 1995, 34, 451 CrossRef CAS PubMed.
- M. R. Morrow, D. M. Singh and C. W. Grant, Biophys. J., 1995, 69, 955 CrossRef CAS.
- T. K. M. Nyholm, S. Jaikishan, O. Engberg, V. Hautala and J. P. Slotte, Biophys. J., 2019, 116, 296 CrossRef CAS PubMed.
- O. Engberg, T. Yasuda, V. Hautala, N. Matsumori, T. K. M. Nyholm, M. Murata and J. P. Slotte, Biophys. J., 2016, 110, 1563 CrossRef CAS PubMed.
- L. J. Patalag, J. Sibold, O. M. Schütte, C. Steinem and D. B. Werz, ChemBioChem, 2017, 18, 2171 CrossRef CAS PubMed.
- M. R. Elkins, I. V. Sergeyev and M. Hong, J. Am. Chem. Soc., 2018, 140, 15437 CrossRef CAS PubMed.
- C. Wang, Y. Yu and S. L. Regen, Angew. Chem., Int. Ed., 2017, 56, 1639 CrossRef CAS PubMed.
- S. Gao, L. Kang and J. Wang, J. Life Sci., 2010, 4, 26 Search PubMed.
- V. Luginbuehl, N. Meier, K. Kovar and J. Rohrer, Biotechnol. Adv., 2018, 36, 613 CrossRef CAS PubMed.
- S. Chong, F. B. Mersha, D. G. Comb, M. E. Scott, D. Landry, L. M. Vence, F. B. Perler, J. Benner, R. B. Kucera, C. A. Hirvonen, J. J. Pelletier, H. Paulus and M. Q. Xu, Gene, 1997, 192, 271 CrossRef CAS PubMed.
- H. E. Klock and S. A. Lesley, Methods Mol. Biol., 2009, 498, 91 CrossRef CAS PubMed.
- A. Pichert, D. Schlorke, S. Franz and J. Arnhold, Biomatter, 2012, 2, 142 CrossRef PubMed.
- R. David, Z. Machova and A. G. Beck-Sickinger, Biol. Chem., 2003, 384, 1619 CAS.
- K. Nordsieck, L. Baumann, V. Hintze, M. T. Pisabarro, M. Schnabelrauch, A. G. Beck-Sickinger and S. A. Samsonov, Biopolymers, 2018, 109, e23103 CrossRef.
- C. Berger, C. Montag, S. Berndt and D. Huster, Protein Expression Purif., 2011, 76, 25 CrossRef CAS PubMed.
- P. Schmidt, B. J. Bender, A. Kaiser, K. Gulati, H. A. Scheidt, H. E. Hamm, J. Meiler, A. G. Beck-Sickinger and D. Huster, Front. Mol. Biosci., 2018, 4, 100 CrossRef PubMed.
- O. M. Schütte, L. J. Patalag, L. M. Weber, A. Ries, W. Römer, D. B. Werz and C. Steinem, Biophys. J., 2015, 108, 2775 CrossRef PubMed.
- S. Stahlberg, B. Skolova, P. K. Madhu, A. Vogel, K. Vavrova and D. Huster, Langmuir, 2015, 31, 4906 CrossRef CAS.
- J. H. Davis, Biophys. J., 1979, 27, 339 CrossRef CAS.
- S. P. Soni, J. A. Ward, S. E. Sen, S. E. Feller and S. R. Wassall, Biochemistry, 2009, 48, 11097 CrossRef CAS PubMed.
- A. Kovacik, A. Vogel, J. Adler, P. Pullmannova, K. Vavrova and D. Huster, Biochim. Biophys. Acta, 2018, 1860, 1162 CrossRef CAS PubMed.
- M. R. Morrow, D. Singh and C. W. Grant, Biochim. Biophys. Acta, 1995, 1235, 239 CrossRef.
- M. R. Morrow, D. Singh, D. Lu and C. W. Grant, Biophys. J., 1993, 64, 654 CrossRef CAS PubMed.
- R. N. Lewis, R. N. McElhaney, M. A. Monck and P. R. Cullis, Biophys. J., 1994, 67, 197 CrossRef CAS PubMed.
- A. Vogel, G. Reuther, K. Weise, G. Triola, J. Nikolaus, K. T. Tan, C. Nowak, A. Herrmann, H. Waldmann, R. Winter and D. Huster, Angew. Chem., Int. Ed., 2009, 48, 8784 CrossRef CAS PubMed.
- L. L. Holte, F. Separovic and K. Gawrisch, Lipids, 1996, 31, S-199 CrossRef CAS.
- M. M. Manni, J. Sot, E. Arretxe, R. Gil-Redondo, J. M. Falcon-Perez, D. Balgoma, C. Alonso, F. M. Goni and A. Alonso, Chem. Phys. Lipids, 2018, 217, 29 CrossRef CAS.
- H. Ohtaka, Y. Kawasaki and M. Kodama, J. Therm. Anal. Calorim., 2013, 113, 1593 CrossRef CAS.
- G. Jaipuria, T. Ukmar-Godec and M. Zweckstetter, Cell. Mol. Life Sci., 2018, 75, 2137 CrossRef CAS.
- M. A. Sani and F. Separovic, Acc. Chem. Res., 2016, 49, 1130 CrossRef CAS PubMed.
- H. A. Scheidt, P. Müller, A. Herrmann and D. Huster, J. Biol. Chem., 2003, 278, 45563 CrossRef CAS PubMed.
- D. Huster, K. Arnold and K. Gawrisch, Biochemistry, 1998, 37, 17299 CrossRef CAS PubMed.
- D. Huster, K. Arnold and K. Gawrisch, Biophys. J., 2000, 78, 3011 CrossRef CAS.
- O. Soubias, W. E. Teague, Jr., K. G. Hines and K. Gawrisch, Biophys. J., 2015, 108, 1125 CrossRef CAS PubMed.
- A. Vogel, C. P. Katzka, H. Waldmann, K. Arnold, M. F. Brown and D. Huster, J. Am. Chem. Soc., 2005, 127, 12263 CrossRef CAS.
- E. B. Watkins, H. Gao, A. J. C. Dennison, N. Chopin, B. Struth, T. Arnold, J. C. Florent and L. Johannes, Biophys. J., 2014, 107, 1146 CrossRef CAS PubMed.
- S. E. Feller, K. Gawrisch and A. D. MacKerell, Jr., J. Am. Chem. Soc., 2002, 124, 318 CrossRef CAS.
- V. Solovyeva, L. Johannes and A. C. Simonsen, Soft Matter, 2015, 11, 186 RSC.
- H. Ling, A. Boodhoo, B. Hazes, M. D. Cummings, G. D. Armstrong, J. L. Brunton and R. J. Read, Biochemistry, 1998, 37, 1777 CrossRef CAS PubMed.
- W. Pezeshkian, A. G. Hansen, L. Johannes, H. Khandelia, J. C. Shillcock, P. B. Kumar and J. H. Ipsen, Soft Matter, 2016, 12, 5164 RSC.
- H. A. Scheidt, D. Huster and K. Gawrisch, Biophys. J., 2005, 89, 2504 CrossRef CAS PubMed.
- P. Wisse, H. Gold, M. Mirzaian, M. J. Ferraz, G. Lutteke, R. J. B. H. N. van den Berg, H. van den Elst, J. Lugtenbuft, G. A. van der Marel, J. M. F. G. Aerts, J. D. C. Codée and H. S. Overkleeft, Eur. J. Org. Chem., 2015, 2661 CrossRef CAS.
- M. J. Hope, M. B. Bally, G. Webb and P. R. Cullis, Biochim. Biophys. Acta, 1985, 812, 55 CrossRef CAS.
- J. H. Davis, K. R. Jeffrey, M. Bloom, M. I. Valic and T. P. Higgs, Chem. Phys. Lett., 1976, 42, 390 CrossRef CAS.
- M. A. McCabe and S. R. Wassall, J. Magn. Reson., Ser. B, 1995, 106, 80 CrossRef CAS.
- M. Lafleur, B. Fine, E. Sternin, P. R. Cullis and M. Bloom, Biophys. J., 1989, 56, 1037 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9cp02501d |
| This journal is © the Owner Societies 2019 |