Understanding chemical interaction between phosphonate-derivative molecules and a silver surface cluster in SERS: a combined experimental and computational approach†
Abstract
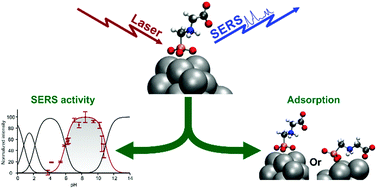
The interaction between phosphonate functions and a silver surface cluster is investigated using Surface-Enhanced Raman Spectroscopy (SERS). Changing the functional group (methylphosphonic acid based molecule) by studying the effect of protonation, methylation and substitution of the side chain with amine and carboxylate functions enabled us to modulate the chemical interactions between the different functions and the metal cluster. We find that the adsorption energy of the methylphosphonic acid decreases with the protonation, the methylation processes and the substitution of the side chain. In all cases, only the deprotonated phosphonate forms are SERS active. To understand how the molecules interact with the nanoparticle, the electronic structure, adsorption energies and Raman spectra were computed for molecules adsorbed on a 20 atom silver cluster representing a nanoparticle surface. The qualitative agreement between computed static Raman spectra and experimental SERS spectra makes it possible to determine stable geometries of the analyte–silver cluster complexes and to characterize the adsorption modes. The findings presented here provide a framework for designing analytical developments based on SERS for simultaneous detection of phosphonated molecules, including pesticides such as glyphosate, creating practical opportunities in key areas such as environmental and water resource in situ monitoring.


 Please wait while we load your content...
Please wait while we load your content...