Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
A facile oxygen-17 NMR method to determine effective viscosity in dilute, molecularly crowded and confined aqueous media†
Nasrollah
Rezaei-Ghaleh
 *ab,
Francesca
Munari
*ab,
Francesca
Munari
 c,
Stefan
Becker
c,
Stefan
Becker
 b,
Michael
Assfalg
b,
Michael
Assfalg
 c and
Christian
Griesinger
c and
Christian
Griesinger
 b
b
aDepartment of Neurology, University Medical Center Göttingen, Göttingen, Germany. E-mail: nrezaei@gwdg.de; nare@nmr.mpibpc.mpg.de
bDepartment for NMR-based Structural Biology, Max Planck Institute for Biophysical Chemistry, Göttingen, Germany
cDepartment of Biotechnology, University of Verona, Verona, Italy
First published on 23rd September 2019
Abstract
We present an NMR method based on natural abundance 17O relaxation of water to determine effective viscosity in biological aqueous samples. The method accurately captures viscosity of dilute and crowded protein solutions and offers a fairly simple way to quantify the internal fluidity of biological condensates formed through phase separation.
Life relies on the spatiotemporal arrangement and coordination of the components and activities of living systems.1 In cells, the spatial arrangement is provided either through compartmentalization via membranous organelles, or alternatively, through a process termed liquid–liquid phase separation (LLPS) which leads to the formation of membrane-less bodies in cells.2 Physical properties of the cellular bodies formed after LLPS exhibit remarkable diversity, ranging from liquid-like droplets to gel-like phases with different degrees of confinement.3 Different micro-rheological and biophysical techniques based on fusion timescale or diffusion analysis are utilized to probe the viscoelastic properties of biological condensates.4,5 Further progress in characterization of biological phase separation requires developments in experimental methods probing the highly crowded and confined environments inside biological condensates.6
Temporal coordination of biological activities depends on the characteristic times of various motional modes of biomolecules,1 influenced potentially by local concentration and the crowding effect.7 Protein dynamics generally occur at multiple time and length scales, which are best characterized through combination of several experimental techniques.8,9 Due to differences in the inherent sensitivity of various experimental techniques, protein samples at different concentrations are often used for protein dynamical studies. It is therefore essential to quantify pure viscosity effects on protein dynamics before any reasonable comparison of experimental data obtained at different protein concentrations can be made.
It has been suggested that water dynamics could be used as a reliable proxy for the local viscosity.10 One of the best methods to study water dynamics is 17O NMR.1117O nucleus has spin quantum number I of 5/2 and natural abundance of 0.037%. The dominant NMR relaxation mechanism of 17O is the anisotropic interaction between the quadrupole moment (Q) of 17O and the electric field gradient present at the site of 17O nuclei.12 Here, we utilize 17O longitudinal relaxation (R1) rate of water, essentially a probe of water structure and dynamics, to determine effective viscosity in biological aqueous samples behaving as Newtonian fluids. Effective viscosity is a local microscopic property of solutions influencing Brownian diffusion of solutes. In non-homogeneous (crowded) solutions, the effective viscosity experienced by the rotationally diffusing solutes may be significantly different from the bulk viscosity.13
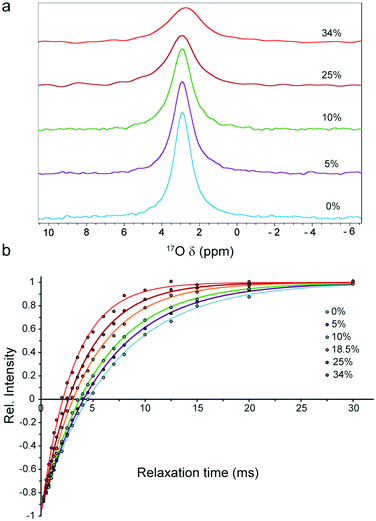
First, we investigated the viscosity dependence of 17O R1 rate of water in glycerol–water mixtures, for which the solution viscosities are well known14 and diffusion obeys the classical Stokes laws.13 The 17O NMR experiments were performed for samples containing 0 to 34% (v/v) glycerol, in which the solution viscosity at 298 K varied from 0.89 to 3.01 cP. In 1D 17O spectra, the intensity of water signal changed in proportion to water content (Fig. S1, ESI†). In addition, the signal linewidths rose from 46.5 ± 0.1 Hz at 0% glycerol to 101.8 ± 0.3 Hz at 34% glycerol, reflecting the increase in viscosity and its resultant signal broadening (Fig. 1a). Similarly, the 17O R1 rates followed an increasing trend from 145 ± 4 s−1 at 0% glycerol to 323 ± 9 s−1 at 34% glycerol (Fig. 1b and Fig. S2, ESI†). The value obtained at 0% glycerol is in excellent agreement with previous reports,15 and implicates a rotational correlation time (τrot) of 1.54 ± 0.04 ps for water molecules in accord with earlier studies.15–17
Besides increasing viscosity and its resultant effect on τrot of water molecules, addition of glycerol may alter the hydrogen-bonded network structure of water and thereby modulate the quadrupole coupling constant (χ) and asymmetry (η) of water 17O nuclei. To our knowledge, there is no report in the literature concerning the quadrupole parameters of water 17O in glycerol–water mixtures, except in the frozen state, where 17O parameters of water are not significantly affected by 60% glycerol.18 Thus, it has been shown that the quadrupole coupling factor, χ2(1 + η2/3), of water 17O changes only minimally over a broad range of temperatures from 274 to 350 K (ca. 6%), where the hydrogen bond network of water undergoes significant alterations.16 Consequently, we assume that the change in quadrupole coupling factor of water 17O over the studied range of glycerol concentrations remains negligibly small for the purpose of current study. Based on this assumption, the τrot of water increases to ∼1.7, 1.9, 2.3, 2.7 and 3.4 ps, respectively at 5, 10, 18.5, 25 and 34% glycerol concentration. It is interesting to note that the change in water dynamics upon addition of glycerol is less prominent when compared to solution viscosity (Fig. S3, ESI†). Contrary to the τrot of water molecules which is supposedly linearly related to 17O R1 rates, the solution viscosity versus17O R1 data showed better fit to a quadratic function when compared to a linear function (p-value < 0.001). The obtained empirical function was:
| η (cp) = 1.719 (±0.194) × 10−5 × (R1)2 + 0.004 (±0.001) × R1 − 0.065 (±0.017) | (1) |
Next, the temperature dependence of 17O R1 rate of water was studied in pure water, for which the temperature dependence of viscosity is well established.19 Upon a decrease from 310 K to 275.4 K, the 17O R1 rate increased from 107 ± 6 s−1 to 385 ± 14 s−1 (Fig. S5a, ESI†). The increase in 17O R1 rates upon cooling is expected because of the longer τrot of water at lower temperatures. This effect is however partially counteracted by the small albeit considerable drop in 17O quadrupole coupling factor of water at low temperatures.16 Taking the temperature dependence of the quadrupole coupling factor into consideration, the τrot of water changes from 1.12 ± 0.06 ps at 310 K to 4.16 ± 0.16 ps at 275.4 K. The relation between viscosity/temperature (η/T) ratio and 17O R1 rates deviated from linearity and was better represented by a quadratic function (p-value < 0.001, Fig. S5b, ESI†). The observed deviations from linearity, both in water–glycerol mixtures and in pure water over the studied temperature range, suggest that the effect of perturbation on solution viscosity cannot be well approximated to a first-order change in the dynamics of individual water molecules.
With the viscosity and temperature dependence of 17O R1 rate of water established, we employed 17O R1 rates to evaluate effective viscosity (henceforth viscosity) in a number of test cases. First, we investigated aqueous solutions containing high protein concentration, such as 4 mM ubiquitin and 3 mM GB3. At 298 K, the 17O R1 rates were 164 ± 3 and 153 ± 4 s−1, respectively, which using eqn (1), corresponded to viscosities of 1.07 ± 0.03 and 0.96 ± 0.03 cP. The ηprotein/ηwater ratio was ∼1.20 ± 0.03 for 4 mM ubiquitin and 1.08 ± 0.04 for 3 mM GB3 solutions, in close agreement with the viscosity ratios of ∼1.17 and 1.08 predicted on the basis of volume occupancies (see ESI†). Then, the rotational correlation time (τc) of ubiquitin and GB3 were determined through 15N cross-correlated relaxation (CCR) rates. At 4 mM concentration, the average CCR rate of ubiquitin was 4.83 ± 0.05 s−1, corresponding to a τc of 4.82 ± 0.04 ns. Compared to the τc of ubiquitin at much lower concentration of 0.37 mM (see below, also ref. 20), the τc of ubiquitin at 4 mM concentration exhibited an increase by a factor of 1.20 ± 0.02. The increase in τc of ubiquitin closely matches the observed viscosity ratio obtained above. This finding is interesting as it rules out significant dimerization of ubiquitin at such a high concentration, as previously suggested.21 For GB3, the average CCR rate was 3.82 ± 0.03 s−1, corresponding to a τc of 3.47 ± 0.04 ns (Fig. S6, ESI†), slightly larger than the value of 3.35 ± 0.03 ns reported at 2 mM GB3 concentration.22 Considering the expected 8% change in viscosity, the τc of GB3 at infinite dilution would be ∼3.23 ns, implying a rather small atomic effective radius (AER) value of ∼2.5 Å for hydrodynamic calculations. Taken together, the 17O R1 measurement of bulk water allowed quantification of viscosity changes at high-concentration protein samples and enabled estimating alterations in the τc of proteins.
Second, we studied molecularly crowded aqueous solutions mimicking the environment of interior of cells.7 To address how crowding agents alter the solution viscosity, we examined two commonly used neutral polymeric crowding agents: Ficoll 70 and sucrose. At 200 g L−1 concentration of Ficoll 70, the 17O R1 rate of water was 247 ± 5 s−1, which using eqn (1), corresponded to a viscosity of 2.00 ± 0.07 cP and the ηFicoll/ηwater ratio of 2.23 ± 0.12 (Fig. 2 and Fig. S7, ESI†). Similarly, the 17O R1 rate of water at 200 g L−1 sucrose was 207 ± 7 s−1, corresponding to the viscosity of 1.52 ± 0.08 cP and ηsucrose/ηwater ratio of 1.70 ± 0.11. Assuming that the quadrupole parameters of water 17O remained effectively unperturbed, the τrot of water molecules were 2.62 ± 0.04 ps in Ficoll 70 and 2.19 ± 0.05 ps in sucrose solutions. To further evaluate the 17O R1-based viscosities, we employed 15N relaxation rates R1 and R2, as reported in ref. 20, and determined τc of the 15N-labelled ubiquitin in dilute and crowded samples. The τc of ubiquitin calculated from residue-specific 15N R2/R1 ratios were 3.98 ± 0.30 ns in dilute solution, 9.06 ± 0.84 ns in 200 g L−1 Ficoll 70 and 7.50 ± 0.69 ns in 200 g L−1 sucrose (Fig. S8, ESI†). Interestingly, the τc,Ficoll/τc,water of 2.31 ± 0.05 was in agreement with the viscosity ratio of 2.23 ± 0.12 obtained through 17O R1 rates (Fig. 2). The τc,sucrose/τc,water of 1.88 ± 0.04 was however slightly larger than the 17O R1-based viscosity ratio of 1.70 ± 0.11. Overall, the method based on 17O R1 rates of water provided a fairly accurate value of viscosity in crowded solutions.
Third, we examined confined aqueous media such as biological hydrogels. To investigate whether the 17O R1 of water could be used as a proxy to monitor the confinement level inside biological hydrogels, we first examined gels formed at different agarose concentrations. The 17O R1 rate of water increased from 150 ± 4 s−1 at 0.5% agarose gel to 156 ± 3, 160 ± 4 and 173 ± 4 s−1, respectively, at 1%, 1.5% and 2% agarose gels (Fig. S9, ESI†). The τrot of water molecules is expected to undergo a similar rising trend from 1.59 ± 0.04 ps to 1.65 ± 0.03, 1.69 ± 0.04 and 1.84 ± 0.04 ps, in accord with the increasing level of confinement in dependence of agarose concentration.
Subsequently, we examined a thermo-responsive Ile-Phe dipeptide-based hydrogel.23 Interestingly, the 17O R1 rate of water in the Ile-Phe gel was 184 ± 3 s−1, even bigger than that of 2% agarose (Fig. 3). The large 17O R1 of water in Ile-Phe hydrogel may have its origin in the large level of confinement, or alternatively, be caused by the presence of residual alcohol (HFiP) within the gel and its impact on local viscosity.
Finally, we measured 17O R1 of water in a gel-forming FG-based peptide, part of the nuclear pore complex.24 At 1.6 mM peptide concentration, the 17O R1 rate of water in the formed hydrogel was 147 ± 2 s−1, between the values observed in water and in 0.5% agarose gel. The 17O R1 rate of water exhibited a concentration-dependent increase to 154 ± 2 s−1 at 8 mM and 166 ± 3 s−1 at ∼ 20 mM FG peptide concentration (Fig. 3). The corresponding τrot of water were 1.55 ± 0.02, 1.63 ± 0.02 and 1.76 ± 0.04 ps, reflecting slight restriction of water dynamics within the hydrogel in dependence of FG peptide concentration. The level of mobility restriction in 1.6, 8 and 20 mM FG peptide concentration hydrogels is similar to <0.5%, 1% and 1.5–2% agarose gels, respectively. Taken together, the 17O R1 of water was capable of capturing differences in the confinement level of agarose and peptide-based hydrogels.
The 17O R1-based method presented above offers a simple and quick way for determination of viscosity in biological aqueous samples. Despite the very low natural abundance of 17O and its low gyromagnetic ratio, the presented method is not particularly demanding in terms of sample requirements or magnetic fields. At 9.4 T magnetic field corresponding to proton Larmor frequency of 400.13 MHz, the 17O R1 experiment of a typical sample required 3–4 hours of measurement. This is because of the high concentration of water and fairly high mobility of water molecules, even in environments as confined as 2% agarose gel studied above. Compared to the alternative methods of viscosity determination in crowded cellular or confined media, such as NMR line width analysis,25 or 19F NMR- or fluorescence-based analysis of rotational and translational diffusion,5,26 the method based on natural abundance 17O relaxation of water is advantageous as it does not require additional labelling or external probes, and is relatively immune to transient protein–protein interactions ubiquitous in cellular media.
The 17O relaxation rate of water is sensitive not only to the τrot of water molecules, but also to the quadrupole coupling factor of H217O.16 As a result, the use of 17O R1 rates of water in viscosity determination is in principle limited to the cases in which changes in water structure and consequently in the quadrupole coupling factor of H217O are negligibly small. Theoretical and experimental investigations suggest that the above condition is indeed not very stringent and can be met over a wide range of temperatures and possible hydrogen-bonded network structures of water.16 Another potential limitation arises when 17O relaxation of water is altered by paramagnetic relaxation effects. Consequently, the presented method cannot be applied to samples in which the presence of a paramagnetic component (such as Cu2+ or Mn2+) cannot be avoided. Moreover, to eliminate unwanted paramagnetic ions and dissolved molecular oxygen, it is essential to use deionized water and degas the samples before NMR experiments.
NMR spin relaxation is a powerful technique to probe protein dynamics at atomistic resolution. NMR-based dynamical studies are frequently performed at high protein concentrations, where concentration-dependent changes in viscosity influence the timescale of different motional modes of proteins. In particular, low-field proton relaxometry, a powerful technique monitoring slow reorientational dynamics in disordered proteins,9 requires millimolar protein concentrations. As exemplified by the ubiquitin and GB3 data above, the viscosity determination through 17O R1 of water enables direct evaluation of the viscosity effects on protein dynamics and allows disentangling pure viscosity effects from potential concentration-dependent phenomena such as protein self-association events. In addition, NMR experiments in non-standard conditions such as crowded media, super-cooled temperatures and high hydrostatic pressures can benefit from direct viscosity determination.27
The traffic between cell nucleus and cytoplasm is controlled via gel forming FG-rich domains of nucleoporins.24 For the FG peptide used in this study, the hydrogel formed at low peptide concentration showed 17O R1 of water only slightly larger than the bulk water, suggesting that the dynamics of water within the channels of the FG hydrogel were largely unaffected. On the other hand, the 17O R1 of water in the high-concentration FG hydrogel was comparable to 1.5–2% agarose gel, indicating a considerable level of water mobility restriction. We suggest that the 17O R1 of water can be used as a proxy to monitor changes in the local environment of biological hydrogels.
In summary, a facile 17O NMR method to determine effective viscosity in biological samples is presented. The method captures effective viscosity in dependence of protein concentration and crowding, and is capable of reporting the confinement level in biological hydrogels, therefore has the potential to be utilized in quantitative characterization of biological condensates in the rapidly growing fields of phase separation and peptide-based pharmaceutical hydrogels.
N. R.-G. acknowledges German Research Foundation (DFG) for a research grant (RE 3655/2-1). The FG peptide was kindly provided by Dirk Görlich. Open Access funding provided by the Max Planck Society.
Conflicts of interest
There are no conflicts to declare.References
- P. Machamer, L. Darden and C. F. Craver, Philos. Sci., 2000, 67, 1–25 CrossRef.
- S. F. Banani, H. O. Lee, A. A. Hyman and M. K. Rosen, Nat. Rev. Mol. Cell Biol., 2017, 18, 285–298 CrossRef CAS PubMed.
- S. Alberti, Curr. Biol., 2017, 27, R1097–R1102 CrossRef CAS PubMed.
- S. Elbaum-Garfinkle, Y. Kim, K. Szczepaniak, C. C. Chen, C. R. Eckmann, S. Myong and C. P. Brangwynne, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 7189–7194 CrossRef CAS PubMed.
- J. Wang, J. M. Choi, A. S. Holehouse, H. O. Lee, X. Zhang, M. Jahnel, S. Maharana, R. Lemaitre, A. Pozniakovsky, D. Drechsel, I. Poser, R. V. Pappu, S. Alberti and A. A. Hyman, Cell, 2018, 174(688–699), e616 Search PubMed.
- S. Alberti, A. Gladfelter and T. Mittag, Cell, 2019, 176, 419–434 CrossRef CAS PubMed.
- G. Rivas and A. P. Minton, Trends Biochem. Sci., 2016, 41, 970–981 CrossRef CAS PubMed.
- N. Salvi, A. Abyzov and M. Blackledge, J. Phys. Chem. Lett., 2016, 2483–2489 CrossRef CAS PubMed.
- N. Rezaei-Ghaleh, G. Parigi, A. Soranno, A. Holla, S. Becker, B. Schuler, C. Luchinat and M. Zweckstetter, Angew. Chem., Int. Ed., 2018, 57, 15262–15266 CrossRef CAS PubMed.
- B. Halle and M. Davidovic, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 12135–12140 CrossRef CAS PubMed.
- J. Qvist, E. Persson, C. Mattea and B. Halle, Faraday Discuss., 2009, 141, 131–144 RSC ; discussion 175–207.
- I. P. Gerothanassis, Prog. Nucl. Magn. Reson. Spectrosc., 2010, 56, 95–197 CrossRef CAS PubMed.
- C. Li, Y. Wang and G. J. Pielak, J. Phys. Chem. B, 2009, 113, 13390–13392 CrossRef CAS PubMed.
- A. Volk and C. J. Kahler, Exp. Fluids, 2018, 59, 75 CrossRef.
- D. Lankohrst, J. Schriever and J. C. Leyte, Ber. Bunsenges. Phys. Chem., 1982, 86, 215–221 CrossRef.
- J. Ropp, C. Lawrence, T. C. Farrar and J. L. Skinner, J. Am. Chem. Soc., 2001, 123, 8047–8052 CrossRef CAS PubMed.
- K. Modig and B. Halle, J. Am. Chem. Soc., 2002, 124, 12031–12041 CrossRef CAS PubMed.
- V. K. Michaelis, B. Corzilius, A. A. Smith and R. G. Griffin, J. Phys. Chem. B, 2013, 117, 14894–14906 CrossRef CAS PubMed.
- R. C. Weast, Handbook of Chemistry and Physics, CRC Press, West Palm Beach, FL, 1978 Search PubMed.
- F. Munari, A. Bortot, S. Zanzoni, M. D'Onofrio, D. Fushman and M. Assfalg, FEBS Lett., 2017, 591, 979–990 CrossRef CAS PubMed.
- Z. Liu, W. P. Zhang, Q. Xing, X. Ren, M. Liu and C. Tang, Angew. Chem., Int. Ed., 2012, 51, 469–472 CrossRef CAS PubMed.
- J. B. Hall and D. Fushman, J. Biomol. NMR, 2003, 27, 261–275 CrossRef CAS PubMed.
- N. S. de Groot, T. Parella, F. X. Aviles, J. Vendrell and S. Ventura, Biophys. J., 2007, 92, 1732–1741 CrossRef PubMed.
- B. B. Hulsmann, A. A. Labokha and D. Gorlich, Cell, 2012, 150, 738–751 CrossRef PubMed.
- Q. Wang, A. Zhuravleva and L. M. Gierasch, Biochemistry, 2011, 50, 9225–9236 CrossRef CAS PubMed.
- Y. Ye, X. Liu, Z. Zhang, Q. Wu, B. Jiang, L. Jiang, X. Zhang, M. Liu, G. J. Pielak and C. Li, Chem. – Eur. J., 2013, 19, 12705–12710 CrossRef CAS PubMed.
- L. P. Singh, B. Issenmann and F. Caupin, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 4312–4317 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental methods and supplementary figures. See DOI: 10.1039/c9cc06124j |
| This journal is © The Royal Society of Chemistry 2019 |