Open Access Article
Open Access ArticleP4T-DOTA – a lanthanide chelating tag combining a sterically highly overcrowded backbone with a reductively stable linker†
Daniel
Joss
 and
Daniel
Häussinger
and
Daniel
Häussinger
 *
*
Department of Chemistry, University of Basel, St. Johanns-Ring 19, 4056 Basel, Switzerland. E-mail: daniel.haeussinger@unibas.ch
First published on 9th August 2019
Abstract
Herein we report a DOTA-based lanthanide chelating tag (LCT) with rigidified backbone and a reduction-stable linker. The newly developed tag induces strong pseudocontact shifts suitable for paramagnetic protein nuclear magnetic resonance spectroscopy and the obtained anisotropic susceptibility parameters are in the range of the best performing LCTs.
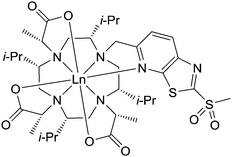
Pseudocontact shifts (PCS) and residual dipolar couplings (RDC) obtained by using lanthanide chelating tags (LCT) yield valuable restraints for investigating protein structures, dynamics and interactions in solution.1–19 The stereo-specifically methyl substituted 1,4,7,10-tetraazacyclododecane-1,4,7,10-tetraacetic acid (DOTA)-based chelators provide a sufficiently rigidified scaffold for observation of significant structural restraints.9,20,21 Interestingly, the methyl substituents on the basic macrocyclic scaffold adopt an equatorial-upper position in the final lanthanide complex.22,23 The methyl substituents thereby arrange in the most suitable way to provide a favourable cavity for the lanthanide ion and minimize the steric repulsion between each other. Furthermore, a crucial factor in the design of the LCTs is to obtain a lanthanide complex that shows only one diastereomer and only one conformation of the pendant arms coordinating to the lanthanide ion.17 The conformationally locked and single diastereomeric LCT provides then only one signal set in 1H–15N HSQC experiments and yields strong paramagnetic effects due to its rigidity and immobilization on the proteins surface.17 As shown by Joss et al., introduction of even more bulkier isopropyl substituents on the backbone of the LCT can significantly enhance the tensor parameters when compared to its methyl substituted predecessor.24 In order to circumvent the inherent instability of disulphide linkers towards a reductive environment, various pyridinesulphone- and iodoacetamide linkers have recently been developed. Therefore, we envisioned to synthesize a lanthanide chelating tag offering the combination of a rigidified backbone and a reductively-stable linker (Fig. 1), that enables convenient and fast protein tagging. The resulting thioether linkage is stable under reductive conditions and has been demonstrated to allow for observation of PCS and RDC in intact eukaryotic cells.2,3
In order to synthesize the newly designed tags, we combined the synthetic approaches by Joss et al. (isopropyl-substituted backbone)24 and Müntener et al. (thiazolo linker)18 and developed them further to yield the target molecule.
To explore the performance, i.e. the range of PCS and RDC as well as the associated Δχ parameters of the Ln-P4T-DOTA tag (P4T-DOTA: (2R,2′R,2′′R)-2,2′,2′′-((2S,5S,8S,11S)-2,5,8,11-tetraisopropyl-10-((2-(methylsulphonyl)thiazolo[5,4-b]pyridine-5-yl)methyl)-1,4,7,10-tetraazacyclododecane-1,4,7-triyl)tripropionate), the Dy-, Tm- and Lu complexes of P4T-DOTA were synthesized and conjugated to ubiquitin S57C, ubiquitin K48C and to selectively 15N leucine labelled human carbonic anhydrase S166C. The newly developed LCT was benchmarked by analysing PCS and RDC (Table 1).
| Protein mutant | PDB | No. PCS | Ln3+ | Δχax (10−32 m3) | Δχrh (10−32 m3) | X metal (Å) | Y metal (Å) | Z metal (Å) | α (°) | β (°) | γ (°) | Q (%) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Ubiquitin S57C | 1UBI 25 | 50 | Dy | 54.0 | 27.0 | 21.6 | 14.5 | 6.0 | 151.7 | 85.5 | 132.4 | 4.5 |
| 72 | Tm | 39.3 | 14.6 | 21.6 | 14.5 | 6.0 | 60.0 | 34.7 | 2.9 | 4.6 | ||
| Ubiquitin K48C | 32 | Dy | −53.7 | −23.5 | 20.5 | 19.9 | 25.8 | 117.3 | 117.4 | 118.5 | 21.5 | |
| 54 | Tm | 39.6 | 13.4 | 20.5 | 19.9 | 25.8 | 91.4 | 106.5 | 97.3 | 3.6 | ||
| hCA II S166C | 3KS3 26 | 40 | Dy | −46.1 | −30.5 | −15.9 | −3.9 | −10.8 | 24.4 | 43.7 | 38.9 | 11.4 |
| 46 | Tm | 44.3 | 4.8 | −15.9 | −3.9 | −10.8 | 174.7 | 157.0 | 29.0 | 5.8 |
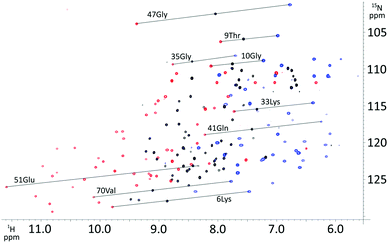
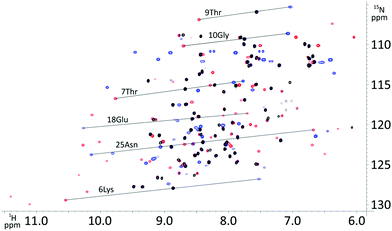
P4T-DOTA delivers large PCS on all tested protein constructs exceeding most current high-performance lanthanide chelating tags (Fig. 2, 3 and 5).3,9,11,15,17,18,28,29
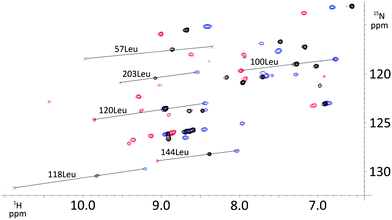 | ||
| Fig. 5 Overlay of 1H–15N HSQC spectra of Dy- (blue), Tm- (red), and Lu-P4T (black) attached to selectively 15N leucine labelled human carbonic anhydrase II S166C. | ||
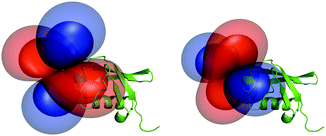
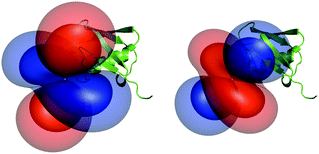
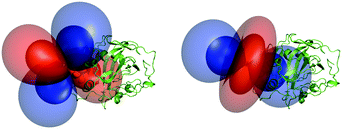
The obtained tensor shapes resemble to the ones found for Ln-DOTA-M8-(4R4S)-SSPy and Ln-P4M4-DOTA, i.e. a significantly less rhombic tensor for the thulium complex when compared to the tensor of the dysprosium complexes (Fig. 4, 6 and 7). The tensors for Tm-P4T-UbS57C/K48C as well as for Tm-P4T attached to selectively 15N leucine labelled hCA II S166C display a much more favourable motional averaging in comparison to Tm-DOTA-M8-(4R4S)-SSPy, and ensure in this way that the magnetic anisotropy of the lanthanide is efficiently transferred to the protein. This feature can be attributed to the rigid and very short thiazolo-linker, that only enables rotation around the Cthiazolo–SCys bond. Furthermore an orientation of the tag is enforced, so that the large axial lobe of the isosurfaces is colinear with the Cthiazolo–SCys bond and therefore, less diminished by rotational averaging (see Fig. 4 and 6–8).18
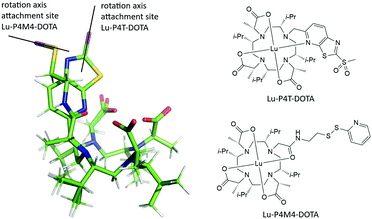 | ||
| Fig. 8 Overlay of DFT structures of Lu-P4T and Lu-P4M4 and their structures. Lu-P4T (top right), Lu-P4M4 (lower right), attachment point of the protein's cysteine residue (magenta). | ||
When compared to the methyl-substituted thiazolo tag described recently by Müntener et al.,18 the isopropyl-substituted thiazolo tag shows an increase in tensor magnitudes due to the sterically more crowded ligand (Table 1).
Besides the favourable Δχ-tensor properties, the reductively stable linker offers new possibilities, as e.g. applications in in-cell NMR, when compared to the Ln-P4M4-DOTA tag, that can only be employed under non-reductive conditions. The strong paramagnetic relaxation enhancement (PRE), e.g. generated for the Dy-P4T-UbK48C construct leads to relatively few detectable signals. However, the results obtained for the thulium tag when attached to ubiquitin S57C and hCA II S166C, i.e. the very large pseudocontact shifts combined with moderate PRE, renders the thulium P4T tag as an ideal solution to investigate not only small and medium sized proteins up to 29 kDa but shows potential for future applications to proteins with significantly larger size.
In order to further characterize the LCT presented in this study, geometries of the Lu-, Tm- and Dy complexes were calculated using density functional theory (DFT) calculations. For the calculations, BP86 was used as functional, SARC-TZVP as basis set for the ligands, while SARC2-QZVP was used as basis set for the lanthanide metal. The calculations were performed using the relativistic ZORA approximation, as well as the RI approximation to speed up the calculations. To model the water solvent, CPCM solvent model was implemented into the calculations. The corresponding references are included in the SI.
Ln-DOTA-M8-(8S)-SSPy complexes show two conformational isomers depending on the ionic radius of the coordinated lanthanide, whereas the (4R4S) stereoisomer shows exclusively a Λ(δδδδ) conformation.17,22 Based on the performed calculations with and without implicit solvent model, a clear stabilization of all investigated complexes towards a Λ(δδδδ) geometry is observed, a result that matches the outcomes for related tags (Table 2).17,18 The obtained stabilization energies for the square antiprism (SAP) conformation of the lanthanide complexes in an implicit water solvent of 17.1 (lutetium), 10.6 (thulium) and 19.3 kJ mol−1 (dysprosium) correspond to equilibrium constants of 999, 73 and 2388 towards the favoured SAP conformation. Due to the higher steric demand of the thiazolo ligand close to the ninth coordination site when compared to the amide ligand of DOTA-M8-(4R4S)-SSPy and the experimental evidence by Strickland et al. that for the Yb-DOTA-M8-(8S)-SSPy with a significantly less hindered apical coordination site there is no coordination of a water molecule,30 no calculations with an explicit water molecule on the ninth coordination site were performed.
| Calculated energies of the different complexes Ln(L) given in Hartree, L = P4T-DOTA | Stabilization Λ(δδδδ) (SAP) over Δ(δδδδ) (TSAP) (kJ mol−1) | |
|---|---|---|
| Lu(L) TSAP | Lu(L) SAP | |
| −17023.78390 | −17023.78679 | −7.6 |
| Lu(L) TSAP solv. | Lu(L) SAP solv. | |
| −17023.87054 | −17023.87706 | −17.1 |
| Tm(L) TSAP | Tm(L) SAP | |
| −16112.77041 | −16112.77262 | −5.8 |
| Tm(L) TSAP solv. | Tm(L) SAP solv. | |
| −16112.85311 | −16112.85716 | −10.6 |
| Dy(L) TSAP | Dy(L) SAP | |
| −14814.69023 | −14814.69733 | −18.7 |
| Dy(L) TSAP solv. | Dy(L) SAP solv. | |
| −14814.77349 | −14814.78084 | −19.3 |
Interestingly, from an overlay of the DFT structures of Lu-P4M4 and Lu-P4T, the angle of the LCT to the protein can be estimated (Fig. 8). Two striking differences can be observed: (i) the linker in Ln-P4T, which is rigidified by the non-flexible aromatic system, is significantly shorter and more rigid than the corresponding linker in Ln-P4M4, (ii) while Ln-P4T is attached in a favourable angle to the protein in terms of motional averaging, Ln-P4M4 is more prone to averaging of the magnetic anisotropy.
To conclude, a new, strongly paramagnetic lanthanide chelating tag is presented that yields pseudocontact shifts in a very high range, exhibits large anisotropy tensors for both employed lanthanide ions and forms a reductively stable linkage to the target protein. The newly developed tag was benchmarked on three different protein constructs, ubiquitin S57C, ubiquitin K48C and hCA II S166C. When compared to its predecessors, the presented LCT yields strongly enhanced pseudocontact shifts due to the very rigid and short linker in combination with a highly sterically crowded backbone. In order to enable further studies on large proteins, protein complexes and other biomacromolecules by PCS NMR spectroscopy, the development of high-performance LCTs will be continued.
The Chemistry Department of the University of Basel and the Swiss National Science Foundation grant 200021_130263 are acknowledged for financial support. Biological structures were generated using the open source software PyMOL (http://www.pymol.org/). Calculations were performed at sciCORE (http://scicore.unibas.ch/) scientific computing core facility at University of Basel. C. E. Housecroft, E. C. Constable, T. Müntener and R. Vogel are acknowledged for helpful discussions.
Conflicts of interest
There are no conflicts of interest to declare.Notes and references
- C. Nitsche and G. Otting, Prog. Nucl. Magn. Reson. Spectrosc., 2017, 98–99, 20–49 CrossRef CAS PubMed.
- B.-B. Pan, F. Yang, Y. Ye, Q. Wu, C. Li, T. Huber and X.-C. Su, Chem. Commun., 2016, 52, 10237–10240 RSC.
- T. Müntener, D. Häussinger, P. Selenko and F.-X. Theillet, J. Phys. Chem. Lett., 2016, 7, 2821–2825 CrossRef PubMed.
- Y. Hikone, G. Hirai, M. Mishima, K. Inomata, T. Ikeya, S. Arai, M. Shirakawa, M. Sodeoka and Y. Ito, J. Biomol. NMR, 2016, 66, 99–110 CrossRef CAS PubMed.
- W.-M. Liu, M. Overhand and M. Ubbink, Coord. Chem. Rev., 2014, 273–274, 2–12 CrossRef CAS.
- C. T. Loh, K. Ozawa, K. L. Tuck, N. Barlow, T. Huber, G. Otting and B. Graham, Bioconjugate Chem., 2013, 24, 260–268 CrossRef CAS PubMed.
- G. Otting, Annu. Rev. Biophys., 2010, 39, 387–405 CrossRef CAS PubMed.
- X.-C. Su, H. Liang, K. V. Loscha and G. Otting, J. Am. Chem. Soc., 2009, 131, 10352–10353 CrossRef CAS PubMed.
- D. Häussinger, J.-R. Huang and S. Grzesiek, J. Am. Chem. Soc., 2009, 131, 14761–14767 CrossRef PubMed.
- X.-C. Su, B. Man, S. Beeren, H. Liang, S. Simonsen, C. Schmitz, T. Huber, B. A. Messerle and G. Otting, J. Am. Chem. Soc., 2008, 130, 10486–10487 CrossRef CAS PubMed.
- P. H. J. Keizers, A. Saragliadis, Y. Hiruma, M. Overhand and M. Ubbink, J. Am. Chem. Soc., 2008, 130, 14802–14812 CrossRef CAS PubMed.
- G. Pintacuda, A. Y. Park, M. A. Keniry, N. E. Dixon and G. Otting, J. Am. Chem. Soc., 2006, 128, 3696–3702 CrossRef CAS PubMed.
- K. D. Brewer, T. Bacaj, A. Cavalli, C. Camilloni, J. D. Swarbrick, J. Liu, A. Zhou, P. Zhou, N. Barlow, J. Xu, A. B. Seven, E. A. Prinslow, R. Voleti, D. Häussinger, A. M. J. J. Bonvin, D. R. Tomchick, M. Vendruscolo, B. Graham, T. C. Südhof and J. Rizo, Nat. Struct. Mol. Biol., 2015, 22, 555 CrossRef CAS PubMed.
- V. Gaponenko, A. S. Altieri, J. Li and R. A. Byrd, J. Biomol. NMR, 2002, 24, 143–148 CrossRef CAS PubMed.
- F. Peters, M. Maestre-Martinez, A. Leonov, L. Kovacic, S. Becker, R. Boelens and C. Griesinger, J. Biomol. NMR, 2011, 51, 329–337 CrossRef CAS PubMed.
- J. Wöhnert, K. J. Franz, M. Nitz, B. Imperiali and H. Schwalbe, J. Am. Chem. Soc., 2003, 125, 13338–13339 CrossRef PubMed.
- D. Joss, R. M. Walliser, K. Zimmermann and D. Häussinger, J. Biomol. NMR, 2018, 29–38 CrossRef CAS PubMed.
- T. Müntener, J. Kottelat, A. Huber and D. Häussinger, Bioconjugate Chem., 2018, 29, 3344–3351 CrossRef PubMed.
- K. Zimmermann, D. Joss, T. Müntener, E. S. Nogueira, L. Knörr, M. Schäfer, F. W. Monnard and D. Häussinger, Chem. Sci., 2019, 10, 5064–5072 RSC.
- R. S. Ranganathan, N. Raju, H. Fan, X. Zhang, M. F. Tweedle, J. F. Desreux and V. Jacques, Inorg. Chem., 2002, 41, 6856–6866 CrossRef CAS PubMed.
- R. S. Ranganathan, R. K. Pillai, N. Raju, H. Fan, H. Nguyen, M. F. Tweedle, J. F. Desreux and V. Jacques, Inorg. Chem., 2002, 41, 6846–6855 CrossRef CAS PubMed.
- A. C. L. Opina, M. Strickland, Y.-S. Lee, N. Tjandra, R. A. Byrd, R. E. Swenson and O. Vasalatiy, Dalton Trans., 2016, 45, 4673–4687 RSC.
- D. Parker, R. S. Dickins, H. Puschmann, C. Crossland and J. A. K. Howard, Chem. Rev., 2002, 102, 1977–2010 CrossRef CAS PubMed.
- D. Joss, M.-S. Bertrams and D. Häussinger, Chem. – Eur. J., 2019 DOI:10.1002/chem.201901692.
- R. Ramage, J. Green, T. W. Muir, O. M. Ogunjobi, S. Love and K. Shaw, Biochem. J., 1994, 299(pt 1), 151–158 CrossRef CAS PubMed.
- B. S. Avvaru, C. U. Kim, K. H. Sippel, S. M. Gruner, M. Agbandje-McKenna, D. N. Silverman and R. McKenna, Biochemistry, 2010, 49, 249–251 CrossRef CAS PubMed.
- A. S. Maltsev, A. Grishaev, J. Roche, M. Zasloff and A. Bax, J. Am. Chem. Soc., 2014, 136, 3752–3755 CrossRef CAS PubMed.
- B. Graham, C. T. Loh, J. D. Swarbrick, P. Ung, J. Shin, H. Yagi, X. Jia, S. Chhabra, N. Barlow, G. Pintacuda, T. Huber and G. Otting, Bioconjugate Chem., 2011, 22, 2118–2125 CrossRef CAS PubMed.
- F. Yang, X. Wang, B.-B. Pan and X.-C. Su, Chem. Commun., 2016, 52, 11535–11538 RSC.
- M. Strickland, C. D. Schwieters, C. Göbl, A. C. L. Opina, M.-P. Strub, R. E. Swenson, O. Vasalatiy and N. Tjandra, J. Biomol. NMR, 2016, 66, 125–139 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9cc04676c |
| This journal is © The Royal Society of Chemistry 2019 |