A tumor microenvironment model coupled with a mass spectrometry system to probe the metabolism of drug-loaded nanoparticles†
Abstract
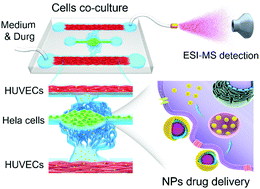
This study describes a novel integrated microfluidic platform with microvessel network channels which are connected to an electrospray ionization-mass spectrometer (ESI-MS), allowing real-time probing of nanoparticle (NP) drug delivery in vascular niche around tumors and understanding the drug delivery efficacy in a realistic tumor microenvironment.


 Please wait while we load your content...
Please wait while we load your content...