Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Trehalose-based Siamese twin amphiphiles with tunable self-assembling, DNA nanocomplexing and gene delivery properties†
Ana I.
Carbajo-Gordillo‡
 a,
Julio
Rodríguez-Lavado‡
a,
Julio
Rodríguez-Lavado‡
 b,
José Luis
Jiménez Blanco
b,
José Luis
Jiménez Blanco
 b,
Juan M.
Benito
b,
Juan M.
Benito
 a,
Christophe
Di Giorgio
a,
Christophe
Di Giorgio
 c,
Itziar
Vélaz
c,
Itziar
Vélaz
 d,
Concepción
Tros de Ilarduya
d,
Concepción
Tros de Ilarduya
 *e,
Carmen
Ortiz Mellet
*e,
Carmen
Ortiz Mellet
 *b and
José M.
García Fernández
*b and
José M.
García Fernández
 *a
*a
aInstituto de Investigaciones Químicas (IIQ), CSIC – Universidad de Sevilla, Avda. Américo Vespucio 49, Isla de la Cartuja, 41092 Sevilla, Spain. E-mail: jogarcia@iiq.csic.es
bDepartment of Organic Chemistry, Faculty of Chemistry, University of Seville, C/Profesor García González 1, 41012 Seville, Spain. E-mail: mellet@us.es
cInstitut de Chimie Nice, UMR 7272, Université de Nice Sophia Antipolis, CNRS, 28, Avenue de Valrose, F-06108 Nice, France
dDepartment of Chemistry, Faculty of Sciences, University of Navarra, E-31080, Pamplona, Spain
eDepartment of Pharmaceutical Technology and Chemistry, School of Pharmacy and Nutrition, University of Navarra, 31080 Pamplona, Spain. E-mail: ctros@unav.es
First published on 19th June 2019
Abstract
An original family of multivalent vectors encompassing gemini and facial amphiphilicity, namely cationic Siamese twin surfactants, has been prepared from the disaccharide trehalose; molecular engineering lets us modulate the self-assembling properties and the topology of the nanocomplexes with plasmid DNA for efficient gene delivery in vitro and in vivo.
The approval of several gene therapy treatments by regulatory agencies and the promising results obtained in recent clinical trials in areas like cancer, epilepsies, hemophilia, retinal degeneration, Parkinson's disease or metabolic disorders have strongly nurtured expectations of a new age for personalized medicine.1 The main hurdle for these channels is the lack of delivery vectors capable of efficiently and safely transporting the therapeutic gene material to enforce the desired genetic change in the target cells. Despite great progress, viral systems still raise concerns about immunogenicity, random integration in the host genome, limited DNA packaging capacity and the cost of clinical grade production of large batches.2 Non-viral carriers have the potential to address most of these limitations3 and some are currently being applied in a number of clinical trials.4 However, most of them are intrinsically polydisperse in nature (e.g., cationic polymers), lack conformational definition (e.g., cationic dendrimers) and/or require multicomponent formulation (e.g., cationic lipids), which hampers precise structure–efficiency relationship and optimization studies. Cationic multivalent surfactants with well-defined molecular structures,5 among which cyclodextrin (CD) derivatives are archetypal examples,6 epitomize remarkable exceptions. Their giant amphiphilic surface exposes facially segregated cationic and lipophilic domains that concertedly drive co-assembly with nucleic acids (Fig. 1A); however, keeping diastereomeric purity can be significantly demanding.7 This problem is largely mitigated for the cationic lipid dimers generically termed gemini surfactants,8 the smallest representatives of multivalent amphiphiles (Fig. 1B). Gemini surfactants have proven to be superior to their monomeric counterparts as gene vectors, but unlike their rigid higher-valent facial amphiphilic homologues they generally require a helper lipid to accomplish the task.9 Most importantly, the topological properties and stability of multivalent and gemini-based supramolecular aggregates exhibit a strong dependence on vector structure and local pH, enabling control over cell uptake and cargo release.7–9 Here we report that the disaccharide α,α′-trehalose, a C2-symmetrical glucose dimer, can be engineered through modular “click” strategies to access “Siamese twin” surfactants amalgamating the advantages of gemini and facial amphiphilicity for efficient DNA nanocomplexation and delivery in vitro and in vivo (Fig. 1C).
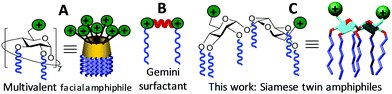 | ||
| Fig. 1 Schematic representations of CD-based multivalent facial amphiphiles (A), gemini surfactants (B) and the novel α,α′-trehalose Siamese twin amphiphiles (C). | ||
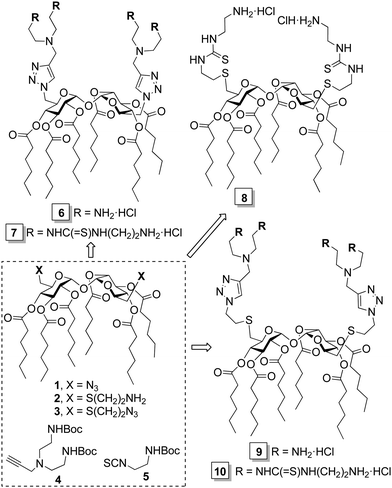
The structure/activity relationship data collected during the last decade for cyclooligosaccharide-based gene delivery systems indicate that facially differentiated constructs consisting of monosaccharide units bearing hexanoyl chains at secondary positions and a bifurcated presentation of amino groups at the primary position are particularly favorable.10 This combination of functional elements provides an appropriate hydrophilic/hydrophobic balance and an optimal density of cationizable centers for efficient interaction with the polyphosphate backbone in the nucleic acid partner.11 We capitalized on this background to develop a set of new trehalose-based Siamese twin prototypes 6–10 (Scheme 1). The rationale is that specific arrangements of flexible spacers (cysteaminyl, ethylene segments), pH buffering groups (triazole heterocycles, tertiary amines), hydrogen bonding modules (thiourea motifs) and peripheral primary amines can translate into distinct self-assembling properties, plasmid DNA (pDNA) complexing abilities and vector/pDNA nanocomplex topologies, thereby impacting transfection capabilities and providing information on the molecular determinants of efficient gene delivery.5
The target gemini facial surfactants 6–10 were synthesized from the 6,6′-diazido-, 6,6′-di-(2-aminoethylthio)- or 6,6′-di-(2-azidoethylthio)-α,α′-trehalose derivatives 1–3 and the alkyne- or isothiocyanate-armed partners 4 and 5 through copper(I)-catalyzed azide–alkyne coupling (CuAAC) and thiourea-forming “click” reactions, respectively (Scheme 1 and ESI†). Their capacity to complex and protect pDNA was initially investigated by electrophoresis mobility shift assay (EMSA) in polyacrylamide gel, using the intercalating agent GelRed® for visualization. Trehalose vector/pDNA formulations were prepared at protonatable nitrogen/anionic phosphate (N/P) ratios of 5, 10 and 20. In all cases, full pDNA complexation, compaction, and protection was achieved, as inferred from the capacity of the compounds to arrest migration of pDNA in the gel, the inability of the fluorescent intercalating agent GelRed® to access the pDNA cargo in the complexes and the recovery of the essentially unaltered pDNA after DNase I/sodium dodecylsulfate (SDS) treatment (Fig. 2B). However, compounds 6 at N/P 5 and 10 and 9 at N/P 5 did not provide stable colloidal suspensions under the conditions used for nanocomplex formulation (Tables S1–S3, ESI†). To ensure a uniform regime, N/P 20 formulations were employed for comparative studies of molecular structure/self-assembly/pDNA nanocomplex topology/gene delivery efficiency relationships.
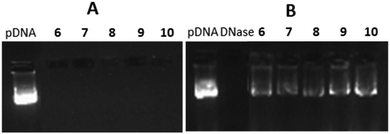 | ||
| Fig. 2 EMSA gels for formulations with 6–10 and pDNA at N/P 20 before (A) and after DNAase I/SDS treatment (B). Virtually identical patterns were obtained at N/P 5 and 10. | ||
Compound 6, featuring a rigid triazole moiety connecting the disaccharide core and the cationic arms, afforded stable homogeneous nanospheres of 15 nm diameter (TEM) in aqueous media, a typical behavior of “conical” surfactants.12 In the presence of pDNA, morula-like nanoplexes of 120–140 nm diameter were formed, suggesting that interaction of the intact vector nanospheres with the polyphosphate chain drives compaction of the plasmid (Fig. 3A and B; left panel). Under identical conditions, the homologous derivative 7 afforded 50 nm vesicles that, upon co-formulation with pDNA, reorganized into spherical nanoparticles (80–100 nm in diameter) exhibiting an alternating arrangement of glycolipid bilayers and plasmid chains (Fig. 3A and B; middle panel). The insertion of a flexible cysteaminyl spacer between the trehalose scaffold and the cationic antennae in compounds 8–10 drastically reduced the predisposition to form defined self-assembled aggregates. In the presence of pDNA, 8 and 9 afforded spherical nanocomplexes similar in size and ultrathin structure to those obtained from 7. Compound 10 exhibited a singular behavior: neat samples showed the presence of bilayer structures characteristic of tubular shaped amphiphiles, whereas in co-formulation with pDNA spherical core–shell nanoparticles (70–80 nm diameter) with a variable level of internal order were formed (Fig. 3A and B; right panel). This scenario strongly suggests a biomimetic two-step mechanism analogous to that characterizing the co-assembly of enveloped viruses:13 first, the “en masse” interaction of individual molecules of 10 with the nucleic acid takes place and then the external unilamellar shell of the amphiphile forms. Nanostructures of such a type have been previously assembled by sophisticated multiformulation strategies and found to be particularly efficient for nucleic acid cargo delivery to target cells.14
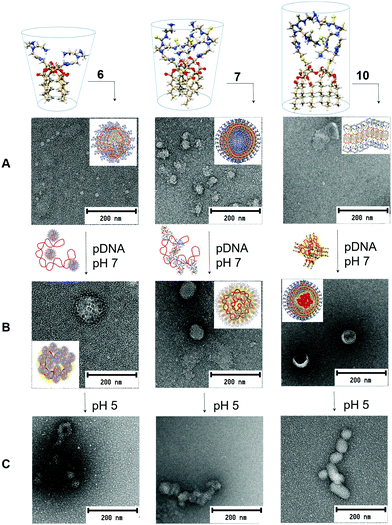 | ||
| Fig. 3 3D molecular models of the gemini facial amphiphiles 6, 7 and 10 and representative TEM micrographs of the corresponding self-assembled (A) and pDNA co-assembled aggregates at pH 7 (B) and pH 5 (C). Schematic representations of the initial interactions with the plasmid and of the final nanoassemblies (pH 7) are provided in the arrows and the inserts, respectively. Control TEM experiments and larger micrographs of the vector/pDNA nanocomplexes are collected in Fig. S12, ESI.† | ||
Dynamic light scattering (DLS) and mixed mode measurement phase analysis light scattering (M3-PALS) provided hydrodynamic diameters slightly higher than those measured by TEM for the above N/P 20 formulations, with polydispersity index values of <0.3 (Table S3, ESI†) and positive ζ-potential values (+21 to +37 mV).
The transfection efficiency of a gene vector depends not only on the stability of the corresponding complexes with nucleic acids in the extracellular milieu (pH ≈ 7) but also on its capability to promote endosomal escape and timely cargo release after cell uptake (pH ≈ 5). Multivalent facial amphiphiles, including gemini surfactants, experience increasing electrostatic repulsions upon acidification, resulting in destabilization of the pDNA co-assemblies.7,8 Individual components can then interact with the endosome membrane, provoking disruption and DNA leakage into the cytoplasm. The new hybrid vectors 6–10 were anticipated to benefit from this mechanism. Indeed, the TEM micrographs recorded at pH 5 (Fig. 3C) revealed aggregates with much less defined internal arrangements. DLS measurements further showed a significant increase in polydispersity and the appearance of higher size nanoparticle populations, altogether supporting decreased stability (Fig. S13, ESI†).
pH-Responsiveness was further checked by titration experiments (Fig. S14, ESI†). Compounds 7 and 10, displaying aminoethylthiourea peripheral branches, exhibited much stronger buffering capabilities, determined as the meq. of NaOH required to switch a 50 mM solution (referring to the total number of protonatable amino groups in the vector) from pH 5 to pH 7, as compared with their homologues 6 and 9 (181.6 and 195.0 versus 61.2 and 85.6 meq., respectively). The buffering potentials of 7 and 10 are even higher than that of branched polyethyleneimine (bPEI 25 kDa; 134.4 meq.),11,15 a cationic polymer used as gold standard for nonviral gene delivery across multiple studies.16 A proton sponge mechanism, resulting in swelling and disruption on progressing from the early to the late endosome state, has been postulated for PEI polyplexes and likely also facilitates endosomal escape in the present case.17
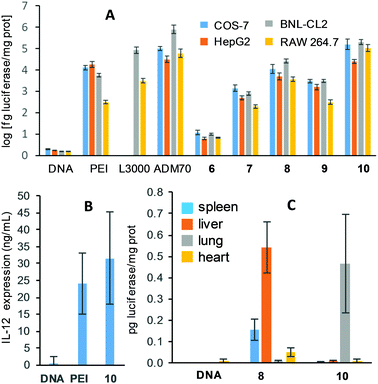
The transfection efficiency of the Siamese twin vectors 6–10 was initially assessed in COS-7 primate epithelial kidney cells and HepG2 human hepatocarcinoma cells. PEI and the CD-based vector ADM70 (Fig. S15, ESI†), one of the most efficient multivalent molecular vectors reported to date,18 as well as naked pDNA (luciferase encoding), were used as positive and negative controls, respectively (Fig. 4A). Formulations prepared from 7 or 10 at N/P 20 were clearly superior to those elaborated from 6 or 9, having identical numbers of cationic centers, in both cell lineages, spotlighting the aminoethylthioureido motif as a preferred functional element in multivalent gene delivery system prototypes. The 10/pDNA nanocomplexes were the most efficient in these experimental settings, promoting 10- and 1.2-fold higher levels of luciferase expression as compared to bPEI in COS-7 and HepG2 cells, respectively. They also rivalled the transfection proficiency of ADM70/pDNA complexes. Compound 10 further compared favorably with PEI and Lipofectamine 3000® (a commercial cationic lipid formulation) in murine embryonic hepatocyte BNL-CL2 and macrophage RAW 264.7 cells, which are notably more resistant to transfection. Only ADM70 in BNL-CL2 cells was superior. Importantly, irrespective of the trehalose-based vector structure, all the pDNA formulations showed low cytotoxicities (Alamar blue® and MTT cell viabilities >80% and >70%, respectively, except for 6; Fig. S16A and B, ESI†). The best performing vector 10 was next assessed for its ability to complex the plasmid pCMVIL12, which encodes the potent antitumoral cytokine interleukin-12 (IL-12),19 and deliver it into HepG2 cells. The corresponding nanocomplexes (N/P 20) performed about 30% better than bPEI in this assay (Fig. 4B).
The possibility of molding the topological properties at the nanoscale level by acting on the molecular structure of the vector offers unique opportunities to affect the tropism of the nanoparticles in vivo. To explore this issue, nanocomplexes formulated with 8 (spherical multilayered) or 10 (spherical core–shell) at N/P 20 were next injected systemically into mice, and their activity was compared with PBS and the naked DNA as negative controls. The results, based on the luciferase reporter gene expression, indicated that 24 h after the intravenous administration, transfection occurred mainly in the liver in the case of 8, which is consistent with previous data for CD-based vectors affording nanocomplexes with similar topology.8 In sharp contrast, compound 10 promoted transfection in the lung (Fig. 4B), an important target in gene therapy,20 with negligible luminescence detected in other organs, highlighting the potential of nanocomplex topology control through molecular vector tailoring for tissue-specific gene delivery.
Polyplexes obtained with bPEI exhibited 100% mortality in this assay, whereas all mice survived treatment with formulations containing the trehalose-based Siamese twin amphiphiles. Taken together, the results provide a testable prototype to guide future molecular vector design.
The authors thank MINECO/MCIU (SAF2016-76083-R, CTQ2015-64425-C2-1-R, CTQ2015-64425-C2-2-R, RTI2018-097609-B-C21 and RTI2018-097609-B-C22), the Junta de Andalucía (FQM2012-1467) and the European Regional Development Funds (FEDER and FSE) for financial support. We acknowledge the CSIC (URICI) for supporting open access publication and the CITIUS (Univ. Seville) for technical support.
Conflicts of interest
There are no conflicts to declare.Notes and references
- (a) E. Bender, Nature, 2016, 537, S57 CrossRef CAS PubMed; (b) B. Blits and H. Petry, Front. Neuroanat., 2017, 10, 128 Search PubMed; (c) S. L. Ginn, A. K. Amaya, I. E. Alexander, M. Edelstein and M. R. Abedi, J. Gene Med., 2018, 10, 128 Search PubMed; (d) N. Matharu, S. Rattanasopha, S. Tamura, L. Maliskova, Y. Wang, A. Bernard, A. Hardin, W. L. Eckalbar, C. Vaisse and N. Ahituv, Science, 2019, 363, eaau0629 CrossRef PubMed.
- P. Colella, G. Ronzitti and F. Mingozzi, Mol. Ther.–Methods Clin. Dev., 2018, 8, 87 CrossRef CAS PubMed.
- (a) M. Zeng, D. Zhou, F. Alshehri, I. Lara-Sáez, Y. Lyu, J. Creagh-Flynn, Q. Xu, A. Sigen, J. Zhang and W. Wang, Nano Lett., 2019, 19, 381 CrossRef CAS PubMed; (b) F. Wang, J. Gao, J. Xiao and J. Du., Nano Lett., 2018, 18, 5562 CrossRef CAS PubMed; (c) S. Liu, D. Zhou, J. Yang, H. Zhou, J. Chen and T. Guo, J. Am. Chem. Soc., 2017, 139, 5102 CrossRef CAS PubMed.
- (a) I. Lostalé-Seijo and J. Montenegro, Nat. Rev. Chem., 2018, 2, 258 CrossRef; (b) C. Chen, Z. Yang and X. Tang, Med. Res. Rev., 2018, 38, 829 CrossRef PubMed.
- (a) E. Junquera and E. Aicart, Adv. Colloid Interface Sci., 2016, 233, 161 CrossRef CAS PubMed; (b) J. L. Jiménez Blanco, J. M. Benito, C. Ortiz Mellet and J. M. García Fernández, J. Drug Delivery Sci. Technol., 2017, 42, 18 CrossRef; (c) J. M. García Fernández, J. M. Benito and C. Ortiz Mellet, Pure Appl. Chem., 2013, 85, 1825 Search PubMed.
- (a) C. Ortiz Mellet, J. M. García Fernández and J. M. Benito, Chem. Soc. Rev., 2011, 40, 1586 RSC; (b) D. Manzanares, I. Araya-Durán, L. Gallego-Yerga, P. Játiva, V. Márquez-Miranda, J. Canan, J. L. Jiménez Blanco, C. Ortiz Mellet, F. D. González-Nilo, J. M. García Fernández and V. Ceña, Nanomedicine, 2017, 12, 1607 CrossRef CAS PubMed; (c) X. Zhou, P. Pathak and J. Jayawickramarajah, Chem. Commun., 2018, 54, 11668 RSC; (d) R. P. Singh, T. Hidalgo, P.-A. Cazade, R. Darcy, M. F. Cronin, I. Dorin, C. M. O’Driscoll and D. Thompson, Mol. Pharmaceutics, 2019, 16, 1358 CrossRef CAS PubMed.
- L. Gallego-Yerga, J. M. Benito, L. Blanco-Fernández, M. Martínez-Negro, I. Vélaz, E. Aicart, E. Junquera, C. Ortiz Mellet, C. Tros de Ilarduya and J. M. García Fernández, Chem. – Eur. J., 2018, 24, 3825 CrossRef CAS PubMed.
- B. E. Brycki, I. H. Kowalczyk, A. Szulc, O. Kaczerewska and M. Pakiet. Application and Characterization of Surfactants, ed. R. Najjar, IntechOpen, 2017, ch. 4, p. 97 Search PubMed.
- (a) M. Damen, A. J. J. Groenen, S. F. M. van Dongen, R. J. M. Nolte, B. J. Scholteb and M. C. Feiters, Med. Chem. Commun., 2018, 9, 1404 RSC; (b) M. Martínez-Negro, K. Kumar, A. L. Barrán-Berdón, S. Datta, P. Kondaiah, E. Junquera, S. Bhattacharya and E. Aicart, ACS Appl. Mater. Interfaces, 2016, 8, 22113 CrossRef PubMed; (c) M. Martínez-Negro, L. Blanco-Fernández, P. M. Tentori, L. Pérez, A. Pinazo, C. Tros de Ilarduya, E. Aicart and E. Junquera, Nanomaterials, 2018, 8, 1061 CrossRef.
- L. Gallego-Yerga, M. Lomazzi, V. Franceschi, F. Sansone, C. Ortiz Mellet, G. Donofrio, A. Casnati and J. M. García Fernández, Org. Biomol. Chem., 2015, 13, 1708 RSC.
- I. Pflueger, C. Charrat, C. Ortiz Mellet, J. M. García Fernández, C. Di Giorgio and J. M. Benito, Org. Biomol. Chem., 2016, 14, 10037 RSC.
- M. Al-Dulaymi, D. Michel, J. M. Chitanda, A. El-Aneed, R. E. Verrall, P. Grochulski and I. Badea, Bioconjugate Chem., 2018, 29, 3293 CrossRef CAS.
- R. Ni, J. Zhou, N. Hossain and Y. Chau, Adv. Drug Delivery Rev., 2016, 106, 3 CrossRef CAS.
- L. Zhang, P. Wang, Q. Feng, N. Wang, Z. Chen, Y. Huang, W. Zheng and X. Jiang, NPG Asia Mater., 2017, 9, e441 CrossRef CAS.
- R. V. Benjaminsen, M. A. Mattebjerg, J. R. Henriksen, S. M. Moghimi and T. L. Andresen, Mol. Ther., 2013, 21, 149 CrossRef CAS PubMed.
- (a) B. Abdallah, A. Hassan, C. Benoist, D. Goula, J. P. Behr and B. A. Demeneix, Hum. Gene Ther., 1996, 7, 1947 CrossRef CAS PubMed; (b) Y. Wang, M. Ye, R. Xie and S. Gong, Bioconjugate Chem., 2019, 30, 325 CrossRef.
- T. Bus, A. Traeger and U. S. Schubert, J. Mater. Chem. B, 2018, 6, 6904 RSC.
- M. Martínez-Negro, G. Caracciolo, S. Palchetti, D. Pozzi, A. L. Capriotti, C. Cavaliere, A. Laganà, C. Ortiz Mellet, J. M. Benito, J. M. García Fernández, E. Aicart and E. Junquera, Biochim. Biophys. Acta, Gen. Subj., 2017, 1861, 1737 CrossRef.
- X. Liu, X. Gao, S. Zheng, B. Wang, Y. Li, C. Zhao, Y. Muftuoglu, S. Chen, Y. Li, H. Yao, H. Sun, Q. Mao, C. You, G. Guo and Y. Wei, Nanomedicine, 2017, 13, 1993 CrossRef CAS.
- K. Minami, K. Okamoto, K. Doi, K. Harano, E. Noiri and E. Nakamura, Sci. Rep., 2014, 4, 4916 CrossRef CAS.
Footnotes |
| † Electronic supplementary information (ESI) available: Experimental details (synthesis, nanoparticle characterization, in vitro and in vivo transfection) and copies of the NMR spectra of the new compounds. See DOI: 10.1039/c9cc04489b |
| ‡ These authors contribute equally. |
| This journal is © The Royal Society of Chemistry 2019 |