Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
4′-Guanidinium-modified siRNA: a molecular tool to control RNAi activity through RISC priming and selective antisense strand loading†
Ganesh N.
Nawale
 a,
Saeed
Bahadorikhalili
a,
Pallabi
Sengupta
b,
Sandeep
Kadekar
a,
Subhrangsu
Chatterjee
b and
Oommen P.
Varghese
a,
Saeed
Bahadorikhalili
a,
Pallabi
Sengupta
b,
Sandeep
Kadekar
a,
Subhrangsu
Chatterjee
b and
Oommen P.
Varghese
 *a
*a
aTranslational Chemical Biology Laboratory, Division of Polymer Chemistry, Department of Chemistry-Ångström, Uppsala University, Uppsala, Sweden. E-mail: oommen.varghese@kemi.uu.se
bBiomolecular NMR and Drug Design Laboratory, Department of Biophysics, Bose Institute, P-1/12 CIT Scheme VII M, Kolkata, India
First published on 5th July 2019
Abstract
We designed novel 4′-C-guanidinocarbohydrazidomethyl-5-methyl uridine (GMU) modified small interfering RNA (siRNA) and evaluated its biophysical and biochemical properties. Incorporation of GMU units significantly increased the thermodynamic stability as well as the enzymatic stability against nucleases in human serum. A gene silencing experiment indicated that GMU modfied siRNA (siRNA6) resulted in ≈4.9-fold more efficient knockdown than unmodified siRNA.
Since the discovery of synthetic siRNA as a potent gene-silencing molecule in mammalian cells, siRNA-based drugs have emerged as a promising technology for the treatment of a variety of diseases.1 RNA interference (RNAi) is an endogenous pathway for post-transcriptional gene silencing that uses sequence-specific knockdown of target mRNA. In this mechanism, short double-stranded RNA (21–23-mer) undergoes a natural antisense or guide strand selection process into the RNA-induced silencing complex (RISC) in the cytosol.2–4 This activated RISC binds mRNA, resulting in gene silencing.5,6 Even though the RNAi based drug development platform looks promising, there are several challenges to overcome. These include challenges to enhance the stability of such drugs against nucleases,7 minimize off-target effects,8 and immune activation,9 and most importantly to enhance in vivo delivery.10 We have recently developed the first non-cationic non-viral siRNA delivery method using hyaluronic acid, a natural biopolymer present in the extracellular matrix.11
In this communication, we present synthesis of a novel 4′-guanidinium modified siRNA and its systematic physicochemical, biochemical, computational and in vitro gene silencing analysis. The GMU modification at the 3′-end is expected to be protonated under physiological conditions,12,13 which could enhance nuclease stability,14–16 and potentially improve immunocompatibility.17 To develop GMU modification, we first synthesized guanidinocarbohydrazide ligand (3) by modifying our recently reported method (Scheme 1).18
The S-methylisothiourea hemisulfate and activated hydroxypropionitrile compound 1 were reacted to obtain compound 2. The S-methyl group was further substituted with carbohydrazide to furnish guanidinocarbohydrazide ligand 3. To prepare the GMU building block, we first synthesized the cyano nucleoside 4 from D-glucose using our previously reported procedure.19 The 2′-acetate group of nucleoside 4 was hydrolysed using sodium methoxide to yield 2′-hydroxy nucleoside 5. Compound 5 was further silylated using tert-butyldimethylsilyl chloride (TBDMS-Cl) to obtain 2′-O-silyl (TBS) nucleoside 6.20 We further reduced the cyano group of 6 to obtain crude aldehyde modified nucleoside 7a which was subsequently converted to hydrazone nucleoside 7.21 The hydrazone derivative was further reduced to afford guanidino nucleoside 8. We further protected the amino group of the hydrazine moiety of 8 using ethyltrifluoro acetate (CF3COOEt) to obtain trifluoro acetate (TFA) protected nucleoside 9. The debenzylation of 9 was performed using 20% Pd(OH)2 to obtain the deprotected diol nucleoside 10. The 5′ hydroxyl of compound 10 was selectively protected using 4,4′-dimethoxytrityl chloride (DMTr-Cl) to furnish 11.22 Finally, phosphitylation of compound 11 was carried out to furnish the phosphoramidite 12.22 The unmodified and GMU modified siRNAs targeting signal transducer and activator of transcription 3 (STAT3) mRNA were synthesized using a solid phase RNA synthesizer (Table 1). The cleavage from the solid support and deprotection of guanidinium and phosphate protecting groups were carried out using 50% piperidine.23 Further nucleobase deprotection, 2′-O-TBDMS group deprotection was carried out and the product was purified by denaturing polyacrylamide gel electrophoresis (PAGE).22 In order to evaluate the effect of chemical modification on the thermal stability of siRNA duplexes, we performed UV-melting studies (Table 1).
| Name | Passenger (5′–3′) and guide strand (3′–5′) | T m | ΔTmb/mod. |
|---|---|---|---|
| a T m represents melting temperatures for unmodified and GMU modified siRNA duplexes (bold text indicates modification) in °C. b ΔTm represents the [Tm (RNA mod.) − Tm (RNA unmod.)]. The Tm values were determined using 1 μM of siRNA in buffer containing 50 mM NaCl, 10 mM Na2PO4, pH 7.4. All experiments were triplicated, and the Tm values have reported an average of 3 measurements with the estimated standard deviation. | |||
| siRNA1 | GGAAGCUGCAGAAAGAUACTTTTCCUUCGACGUCUUUCUAUG | 66.9 ± 0.1 | |
| siRNA2 | GGAAGCUGCAGAAAGAUACTTTTCCUUCGACGUCUUUCUAUG | 67.8 ± 0.3 | +0.9 |
| siRNA3 | GGAAGCUGCAGAAAGAUACTTTTCCUUCGACGUCUUUCUAUG | 69.8 ± 0.1 | +2.9 |
| siRNA4 | GGAAGCUGCAGAAAGAUACTTTTCCUUCGACGUCUUUCUAUG | 69.5 ± 0.2 | +2.6 |
| siRNA5 | GGAAGCUGCAGAAAGAUACTTTTCCUUCGACGUCUUUCUAUG | 69.7 ± 0.3 | +2.8 |
| siRNA6 | GGAAGCUGCAGAAAGAUACTTTTCCUUCGACGUCUUUCUAUG | 70.3 ± 0.2 | +3.4 |
Incorporation of single GMU modification at the 3′-terminal of the passenger strand of siRNA (siRNA2) showed a change in melting temperature (ΔTm) of +0.9 °C per modification as compared to the unmodified duplex. Incorporation of GMU modification on both the overhang nucleotides of siRNA in either the passenger (siRNA3) or guide (siRNA4) strand demonstrated a similar increase in the melting temperature (ΔTm = 2.6–2.9 °C). When both the strands of siRNA were modified together, an increase in ΔTm of 2.8–3.4 °C was observed (siRNA5 and siRNA6). In general, GMU modifications resulted in a significant thermal stabilization of all the modified siRNA duplexes (Table 1).
To elucidate the role of guanidinium modifications in thermal stability of siRNA duplexes, we performed in silico molecular modelling and simulation of the modified and unmodified siRNA duplexes (Fig. S1 and S2, ESI†). These studies implied that the guanidinium linkers near 3′-ribonucleotides (C4′ of ribose sugar) and penultimate thymidine residues (deoxyribose sugar) at siRNA6 termini conferred additional non-covalent interactions to protect the fraying ends of the siRNA duplex and impart thermodynamic stability. In siRNA6, dual GMU modifications at the 3′-overhang amplified the network of electrostatic interactions with adjacent residues (Fig. S1B, ESI†). We further analyzed the energetics of siRNA conformers, by performing MM-PBSA calculations, which decompose the interaction energies for the modified residues by considering molecular mechanics and solvation energies (Table S1, ESI†).
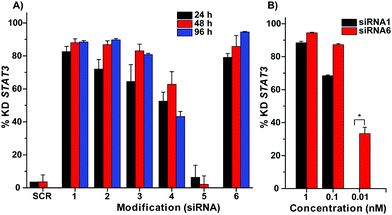
The modified siRNAs were further evaluated for their RNAi efficacy in a human colon cancer cell line (HCT116). HCT116 cells were transfected with different concentrations of siRNA, and the expression levels of STAT3 mRNA were analyzed after 24 h, 48 h or 96 h (Fig. 1). The scrambled siRNA sequence (SCR) was used as a negative control in these experiments. The gene knockdown (KD) results of unmodified and GMU modified siRNAs are summarized in Fig. 1. These results indicate that the modifications of overhang are well tolerated in the passenger strand as well as guide strand of siRNA (Fig. 1A). The modifications in the passenger strand of siRNA duplexes such as siRNA2 and siRNA3 have shown no loss of RNAi activity when compared to the unmodified siRNA1 (in 48 or 96 h 2–5% less KD). The effect of modifications in the guide strand (siRNA4), however, was found to be sensitive with 20–40% less KD efficiency as compared to siRNA1. Surprisingly, when both the strands of siRNA were modified in an asymmetric manner with a single modification on the passenger and two modifications on the guide strand (siRNA5), there was a complete loss of RNAi activity. Interestingly, in the case of siRNA6 where both the overhangs were modified with GMU in a symmetrical manner, the RNAi activity was restored and was in fact superior (7% more KD for 96 h) to the unmodified siRNA (Fig. 1A). We further performed a concentration-dependent study to validate the actual difference in RNAi activity between siRNA1 and siRNA6. Gratifyingly, these experiments suggested that siRNA6 with completely modified overhangs was ≈4.9 fold more efficient as compared to the siRNA1 control (IC50 for siRNA1 ≈311 pM, while for siRNA6 ≈63 pM, Fig. S3, ESI†). Specifically, siRNA6 demonstrated 20–40% higher activity as compared to the native siRNA after 96 h (Fig. 1B). One of the key aspects of the siRNA duplex that dictates its activity is RISC priming, which enables correct orientation of the duplex for specific loading of the antisense strand.24 The 3′-overhang plays an important role in this step as it binds to the PAZ domain of the Argonaute-2 protein within the RISC.25 Intrigued by the difference in bioactivity between siRNA5 and siRNA6, we were prompted to investigate the differences in actual loading of sense and antisense strands. For this purpose, we performed a recruitment assay using stem-loop qPCR.26 This experiment clearly indicated enhanced recruitment of the guide strand in the case of modified siRNA6 (≈1.8-fold enhancement as compared to the siRNA1; Fig. 2A) while the recruitment of the guide strand in siRNA5 was not observed (Fig. 2A). Interestingly, we observed exclusive recruitment of the sense strand in the case of siRNA5 (≈234-fold enhancement as compared to the siRNA1) and significantly less in siRNA6 (0.5 fold less compared to the siRNA1 (Fig. 2B)).
To further investigate the role of guanidinium modifications in guide strand recruitment, we carried out molecular docking studies between Argonaute protein and siRNA duplexes (siRNA1 and siRNA6) and compared the intermolecular interactions. The PAZ domain of Argonaute, which is regarded as the principal RNA binding module to recognize the 3′ overhang of siRNA is observed to make stable non-covalent interactions with siRNA6, consistent with our experimental evidence (Fig. 2C). Symmetric guanidinium modifications at both ends favoured spatial positioning of siRNA into the binding pocket of the PAZ domain of the RISC complex. Thr 361, Ile-353, and Lys-355 allow the passage of siRNA1 and siRNA6 into the RISC complex (Fig. 3D and Fig. S4B, ESI†). In siRNA6, Cys-352 and Thr-368 additionally play important roles in constituting electrostatic interactions with A10, G8 and U7 and enable improved strand recruitment into the RISC complex (Fig. 2D). Structural dynamics of siRNA1 evidenced stable Watson–Crick base pairing in the central domain, which spanned the binding pocket of PAZ. We observed strong pairing between G11:C9, stabilized by neighbouring G8, which projected a π–π stacking interaction with C9 in the same strand. This sequestered the hydrogen-bond interaction between Lys 355 and C9, which might play a critical role to compromise its recruitment into the RISC complex (Fig. S4A and B, ESI†). Further structural investigations confirmed that the sugar puckering of siRNA6 remained in C3′-endo (N-type sugar) sugar conformation (Fig. S5, ESI†), which is essential to maintain the integrity of A-type RNA.
One of the major challenges of the nucleic acid drug is the stability towards endo and exonucleases present in blood plasma and within the cytosol.7 We have previously shown that a single guanidinium modified locked nucleoside at the 3′-terminal can impart significant enzymatic stability for over 24 h in human serum.18 This prompted us to evaluate the stability of the GMU modified siRNAs in human serum. Incubation of modified and unmodified siRNAs with 70% human serum at various time points followed by PAGE analysis of the nuclease digested product indicated that all modified sequences (siRNA2–6) showed a significant improvement in nuclease stability (Fig. 3A).
The siRNA duplex with single or double GMU modifications at the 3′-end of the passenger or guide strand showed stability for over 4 h in human serum. The dual modification on both the guide and passenger strands (siRNA6) further enhanced nuclease stability with a nearly intact duplex after 12 h and almost 50% intact siRNA after 72 h (h*, Fig. 3A). The unmodified siRNA duplex (siRNA1) on the other hand was completely degraded within one hour of incubation time. We reasoned that the stabilized non-covalent interactions by GMU modifications at the 3′-termini sequester siRNA molecules from the catalytic pockets of RNase A-type endonucleases, which enhance nuclease resistance of the siRNA molecules in the sera. Structural analyses of in silico RNase A-siRNA complexes further underscored that the RNase A catalytic triad (His12, His119, and Lys41) invades siRNA1 to promote RNA cleavage (Fig. 3B and C). However, the Watson–Crick pairing in siRNA6 is perturbed due to the end modifications rendering reduced efficiency of the RNase A catalytic triad to remain in the spatial proximity of the ribonucleotides, required for the nucleophilic attack towards 2′-OH of the ribose sugars. Therefore, siRNA6 is not as vulnerable as siRNA1 and is able to evade nuclease cleavage (Fig. 3D and E).
In summary, we have synthesized novel 4′-guanidinium-modified amidite to obtain several chemically modified siRNAs. These modified siRNAs not only have increased thermal stability but also significantly improved stability in human serum. The incorporation of GMU nucleotides at the two terminal 3′-overhangs (siRNA6) also resulted in sustained gene silencing activity at significantly lower concentration (picomolar) after 96 h of transfection. The stem-loop qPCR experiments confirmed that such sustained activity was due to enhanced guide strand recruitment within the RISC complex. Structural investigations of the Argonaute protein also confirmed altered spatial positioning of siRNA into the PAZ domain resulting in successful recruitment of the desired guide strand into the RISC complex and enhanced activity. We believe that the 4′-guanidino modifications at the 3′-overhangs of siRNA will allow development of the next generation of RNAi drugs with enhanced potency and minimal sense-strand mediated off-target effects. Since such modifications are incorporated at the overhang region, it could be adapted to other siRNA sequences, without losing its bioactivity.
Conflicts of interest
There are no conflicts to declare.Notes and references
- R. Kanasty, J. R. Dorkin, A. Vegas and D. Anderson, Nat. Mater., 2013, 12, 967–977 CrossRef CAS.
- P. D. Zamore, T. Tuschl, P. A. Sharp and D. P. Bartel, Cell, 2000, 101, 25–33 CrossRef CAS.
- A. Fire, S. Xu, M. K. Montgomery, S. A. Kostas, S. E. Driver and C. C. Mello, Nature, 1998, 391, 806–811 CrossRef CAS.
- S. M. Elbashir, J. Harborth, W. Lendeckel, A. Yalcin, K. Weber and T. Tuschl, Nature, 2001, 411, 494–498 CrossRef CAS.
- W. Filipowicz, Cell, 2005, 122, 17–20 CrossRef CAS.
- D. M. Dykxhoorn, C. D. Novina and P. A. Sharp, Nat. Rev. Mol. Cell Biol., 2003, 4, 457–467 CrossRef CAS.
- J. J. Turner, S. W. Jones, S. A. Moschos, M. A. Lindsay and M. J. Gait, Mol. BioSyst., 2007, 3, 43–50 RSC.
- A. L. Jackson and P. S. Linsley, Nat. Rev. Drug Discovery, 2010, 9, 57–67 CrossRef CAS.
- G. Hartmann, J. Clin. Invest., 2009, 119, 438–441 CrossRef CAS.
- S. F. Dowdy, Nat. Biotechnol., 2017, 35, 222–229 CrossRef CAS.
- M. Paidikondala, V. K. Rangasami, G. N. Nawale, T. Casalini, G. Perale, S. Kadekar, G. Mohanty, T. Salminen, O. P. Oommen and O. P. Varghese, Angew. Chem., Int. Ed., 2019, 58, 2815–2819 CrossRef CAS.
- D. Francoise, A. Said, D. Gaelle, M. M. Hong, C. Philippe, J. G. Michael, V. Jean-Jacques and L. Bernard, Curr. Top. Med. Chem., 2007, 7, 727–737 CrossRef.
- T. Kubo, K. Yanagihara, Y. Takei, K. Mihara, Y. Sato and T. Seyama, Biochem. Biophys. Res. Commun., 2012, 426, 571–577 CrossRef CAS.
- R. H. Griffey, B. P. Monia, L. L. Cummins, S. Freier, M. J. Greig, C. J. Guinosso, E. Lesnik, S. M. Manalili, V. Mohan, S. Owens, B. R. Ross, H. Sasmor, E. Wancewicz, K. Weiler, P. D. Wheeler and P. D. Cook, J. Med. Chem., 1996, 39, 5100–5109 CrossRef CAS.
- M. Kanazaki, Y. Ueno, S. Shuto and A. Matsuda, J. Am. Chem. Soc., 2000, 122, 2422–2432 CrossRef CAS.
- X. Luo, T. Sugiura, R. Nakashima, Y. Kitamura and Y. Kitade, Bioorg. Med. Chem. Lett., 2013, 23, 4157–4161 CrossRef CAS.
- M. D. Marimani, A. Ely, M. C. Buff, S. Bernhardt, J. W. Engels, D. Scherman, V. Escriou and P. Arbuthnot, J. Controlled Release, 2015, 209, 198–206 CrossRef CAS.
- J. Barman, D. Gurav, O. P. Oommen and O. P. Varghese, RSC Adv., 2015, 5, 12257–12260 RSC.
- O. P. Varghese, J. Barman, W. Pathmasiri, O. Plashkevych, D. Honcharenko and J. Chattopadhyaya, J. Am. Chem. Soc., 2006, 128, 15173–15187 CrossRef CAS PubMed.
- K. K. Ogilvie, S. L. Beaucage, A. L. Schifman, N. Y. Theriault and K. L. Sadana, Can. J. Chem., 1978, 56, 2768–2780 CrossRef CAS.
- Y. Liu, J. Xu, M. Karimiahmadabadi, C. Zhou and J. Chattopadhyaya, J. Org. Chem., 2010, 75, 7112–7128 CrossRef CAS.
- K. R. Gore, G. N. Nawale, S. Harikrishna, V. G. Chittoor, S. K. Pandey, C. Höbartner, S. Patankar and P. I. Pradeepkumar, J. Org. Chem., 2012, 77, 3233–3245 CrossRef CAS.
- T. P. Prakash, A. Puschl, E. Lesnik, V. Mohan, V. Tereshko, M. Egli and M. Manoharan, Org. Lett., 2004, 6, 1971–1974 CrossRef CAS.
- C. L. Noland, E. Ma and Jennifer A. Doudna, Mol. Cell, 2011, 43, 110–121 CrossRef CAS.
- A. Alagia, A. F. Jorge, A. Aviñó, T. F. G. G. Cova, R. Crehuet, S. Grijalvo, A. A. C. C. Pais and R. Eritja, Chem. Sci., 2018, 9, 2074–2086 RSC.
- A. Cheng, A. V. Vlassov and S. Magdaleno, Methods Mol. Biol., 2011, 764, 183–197 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental details, and spectroscopic and analytical data. See DOI: 10.1039/c9cc04141a |
| This journal is © The Royal Society of Chemistry 2019 |