Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Temperature-sensitive protein expression in protocells†
Haiyang Jia ,
Michael Heymann
,
Michael Heymann ,
Tobias Härtel
,
Tobias Härtel ,
Lei Kai
,
Lei Kai and
Petra Schwille
and
Petra Schwille *
*
Dept. Cellular and Molecular Biophysics, Max Planck Institute of Biochemistry, Am Klopferspitz 18, D - 82152 Martinsried, Germany. E-mail: schwille@biochem.mpg.de
First published on 10th May 2019
Abstract
We engineered a synthetic temperature regulation toolbox to enable protocells to sense and respond to heat, utilizing RNA thermometers. The thermo-sensitive protocells were generated by encapsulating temperature feedback transcription/translation machinery in droplets. Based on these temperature-sensing devices, the protocells can be operated with logic AND gates, differentially processing temperature stimuli into biological signals.
The construction of a minimal cell or protocell from the bottom-up may deepen our understanding of the essence of cellular life and its origin on earth.1 It also offers new avenues for applications in biotechnology, medicine, and environmental engineering.2 Recent advances in protocell design also reinvigorated interest and improvements of cell-free protein synthesis (CFPS).3 CFPS reconstitutes the components of the cellular transcription and translation machinery for protein synthesis de novo, thus avoiding purification. Encapsulated into emulsion droplets or lipid vesicles, CFPS provides a versatile platform to specifically utilize protein functionality, ultimately paving the way for the construction of complex living systems from the bottom up.4 A variety of synthetic biological circuits have been adapted for dynamically controlling transcription and translation of protocells, such as negative/positive feedback loops.4,5 These have been used to regulate different components within a protocell, but also to achieve global communication between protocell communities.6–9 Addition of micro-pores, molecule transfer, artificial signalling and differentiation cascades achieved and improved spatiotemporal dynamics in multicellular artificial cell models.10,11 However, one key virtue of living cells and minimal cell models is their capacity to selectively respond to external stimuli and to self-regulate homeostasis,12 which is crucial for surviving adverse and fluctuating environments. With regard to the aspects of resilience and adaptation to the environment, protocells are still much inferior to the standards of nature. In particular, few experimental studies achieved protocells that could convert physical stimuli, such as heat, into biological signals. In living bacteria, e.g., stress response to heat is essential for effective adaptation to environmental fluctuations.13 On the other hand, with regard to biotechnological applications, a temperature-sensitive model system would be ideally poised for efficient control. Thus, implementing a temperature-sensing module for converting temperature stimuli into biological signals would greatly improve the design of protocells, mimicking natural stress responses, or establishing an easy to operate control element. Temperature serves as a well-defined signal that could be used by engineered protocell or bacterial therapeutics to detect and respond to host conditions, or spatially targeted external triggers such as focused ultrasound.14
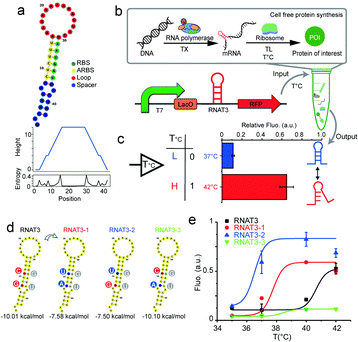
Here we describe the engineering of a protocell that can respond to temperature stimuli, using temperature sensitive non-coding RNA sequences that we refer to as RNA thermometers (RNATs). Natural as well as artificial RNATs have been discovered in vivo15 and designed in vitro.16,17 Most commonly, these RNATs are located in the 5′-untranslated region of the messenger RNA, and fold into secondary structure to control translation by blocking ribosome binding sites (RBS) at low temperature. The RNAT loop unfolds when temperatures exceed a defined threshold, to release the RBS for subsequent gene expression.18
We designed synthetic RNATs based on a simple RNA-melting mechanism.19 Accordingly, we implemented a suite of synthetic RNATs to form a single stem-loop structure, which is composed of three elements: a RBS sequence, a complementary anti-RBS (ARBS) sequence and the loop sequence (Fig. 1a). A reporter gene cassette containing a T7 promoter, RNAT, and red fluorescent protein (RFP) was used to validate various constructs (Fig. 1b). Transcription of mRNA was driven by T7 promoter and its translation was subsequently controlled by the RNA thermometer. We selected the well-established RNAT3 thermometer15 to validate the functionality of our gene cassette using the in vitro PURE reaction20 by quantifying RFP fluorescence at 37 °C and 42 °C. At 37 °C, RNAT3 inhibited the PURE reaction from expressing RFP (“off” state), analogous to previous in vivo results.15 At temperatures of 42 °C, a “jump” in expression could be observed (“on” state). At 42 °C the PURE reaction under RNAT3 regulation yielded 3.3 times higher RFP yields compared to 37 °C. Switching from “off” to “on”, the relative expression ability compared to a control has been increased about sevenfold (Fig. 1c). Ionic strength influences the stability of RNA secondary structure.21 The higher Mg2+ concentration can stabilize the RNAT3 structure; consequently, inhibit its switching in the PURE system (Fig. S1, ESI†).
We tuned RNATs activation temperatures by utilizing a cell-free transcription/translation system in the test tube (Fig. S2, ESI†). A series of RNATs with different minimum free energies was designed through the RNAfold Web Server and inserted into the reporter gene cassette. New RNATs contained the introduced base pair mismatches into the stem. Their melting temperatures were verified through coupled cell-free transcription and translation at different temperatures, while monitoring RFP synthesis by a fluorescence plate reader assay. By avoiding the need for cell culturing, this screening approach can be at least three days shorter in comparison to conventional in vivo approaches. This significantly accelerates the design and validation process.
The screen obtained three excellent candidates derived from the original RNAT3 (Fig. 1d and e). While RNAT3 dehybridized at 40 °C15, the RNAT3-1 mutant sequence had a threshold temperature of 37 °C, while the final RFP expression yield was unaffected. RNAT3-2 had a transition temperature of 35 °C. The switching behaviour of RNAT3-3 was similar to RNAT3, but with overall reduced expression. Considering the sequence of the RNATs, since the bond strength of C–G pair is stronger than U–G, mutation from C to U in the mismatch of the RBS region would make the stem-loop easier to open. Therefore, the switching temperature of RNAT3-2 was reduced, while the expression was enhanced. Conversely, the RNAT3-3 mutation of (G to A) stabilized the loop structure, resulting in the lowest observed expression level. RNAT3-1 contained both mutations, thus featured with all the factors of RNAT3, RNAT3-2 and RNAT3-3. Owing to the balance effects of the two sites, RNAT3-1 could maintain similar expression ability with the original RNAT3; meanwhile it switched on at lower temperature and yielded 30% weaker expression level compared with RNAT3-2. This toolbox of different RNTAs provides us with a variety of different expression control modules in dependence of temperature (temperature sensors). In vitro designed RNATs could also work in vivo, and showed similar “on” and “off” manners (Fig. S3, ESI†).
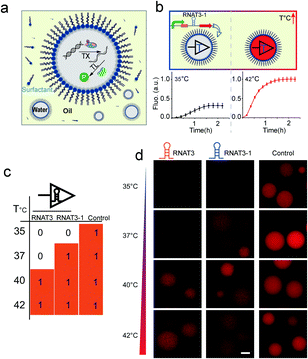
Based on these results, we then prototyped a temperature sensitive protocell model by co-encapsulating the thermometer controlled gene circuits and the PURE transcription/translation machinery in picoliter emulsion droplet compartments (Fig. 2a). Emulsion droplets were formed through mechanical agitation, to disperse the aqueous phase into HFE7500 fluorinated oil with a PFPE-PEG-PFE biocompatible surfactant.22 The resulting micro-emulsion droplets are stable against coalescence for days and can tolerate temperature fluctuations. We encapsulated the RNAT3-1 controlled cell-free protein synthesis system into water-in-oil droplets (Fig. 2b). Protocells programmed with RNAT3 expressed RFP (red) signal at 40 °C, but were insensitive to 35 and 37 °C (Fig. 2d). In turn, RNAT3-1 protocells were activated by lower temperature around 37 °C. Both did not express RFP (“off”) at 35 °C incubation. Thus, consistent with our previous bulk experiments, RNATs laden protocells responded specifically to different temperatures. The results confirm that synthetic RNATs endowed the protocells with the ability to convert ambient temperature into a specific biological response, demonstrating a direct connection between the protocell and environmental temperature cues.
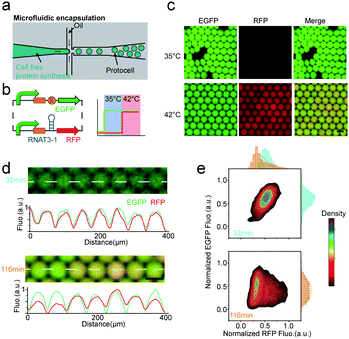
Dynamic logic circuits capable of controlling multiple gene functions at different temperatures are required to further integrate thermal protocell responses with other metabolic cues. Such genetic logic circuits would enable a synthetic cell to initiate different biological functions in a defined workflow to ultimately connect a functional biological network to specific thermal cues in vitro. In particular, differential expression of proteins in response to defined environmental settings is an elegant way of titrating intracellular reactions. Towards this goal, we engineered a multiplexed protocell capable of reporting metabolic activity through EGFP fluorescence, while simultaneously reporting temperature through RFP fluorescence. Expression levels of enhanced green fluorescent protein (EGFP) and RFP in picoliter droplets under constitutively active T7 promoters were identical for all investigated temperatures (Fig. S4, ESI†). We then combined constitutively active EGFP expression with RFP expression under RNAT3-1 control (Fig. 3b). As expected, only EGFP was detected for low temperature of 35 °C. Although the stretch of DNA transcribed into an RNA molecule, the translation of RFP was blocked by RNAT3-1. When the temperature was shifted to 42 °C (above the 37 °C threshold), co-expression of RFP and EGFP inside of microcompartments was observed (Fig. 3c and Fig. S4, ESI†), demonstrating that the thermal genetic multiplexer can enable the protocells to selectively transform expression of different genes in response to a changing environment.
Controlled microfluidic encapsulation into identically sized protocell droplets was then carried out in order to access response fluctuations. We measured the fluorescence intensity of EGFP (pCoofy1-T7-RBS-EGFP) and RFP (pCoofy1-T7-RNAT3-1-E1010) per droplet for a population of >3000 droplets every 10 min, allowing us to track the behaviour of protocell populations with thermal multiplexer. Intriguingly, in the initial half hour, the protocell population showed almost homogeneous expression for both independent genes, egfp and rfp (Fig. 3d). Afterwards, the protocell population started to differentiate over time, especially RFP expression noise accumulated over time (Fig. 3d, e and Fig. S5a, ESI†). This may be due to stochastic on/off switching of the RNATs that leads to slight differences in expression rates (Fig. S5b, ESI†). Such stochasticity is inherent to biochemical gene expression,23 and may further be amplified through inhomogeneous distributions of materials among different bioreactors.23,24 The long-time moderate heat stress (42 °C) may reduce the maturation rate and decrease fluorescence,25 which may also be reason of the overtime change.
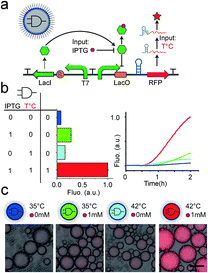
In a next step, we extended our protocell expression control architecture to implement a temperature sensitive AND gate, enabling protocells sensing both chemical and thermal signals (Fig. 4). Different published logic gates have been implanted as gene circuits.26,27 We design our thermo-responsive AND gate to function on both, the transcription and translation level (Fig. 4a). The AND gate utilized chemical inducer and temperature as the two independent inputs. A T7 promoter with a lac operon as regulator was used to control transcription of the RFP, to respond to the chemical inducer (IPTG). LacI repressor was constitutively controlled by another T7 promoter (without lac operon). Therefore, the transcription was kept off in the absence of IPTG. IPTG mediated inhibition of the LacI repressor in turn activated mRNA synthesis. After mRNA expression, our RNA thermometer inhibits translation at low temperature. This combined transcriptional and translational based AND gate could produce different reporter outputs according to different input combinations: the two input signals temperature and IPTG (Fig. 4b). We validated the function of this circuit in a bulk assay in test tube. Compared with the highest background, the expression was increased about fourfold (Fig. 4b). We then encapsulated the mini-AND into our water-in-oil emulsion droplets to generate protocells that can integrate the different signals. The compartmentalized protocells can perform logic as well as the AND gate in the bulk reaction, by responding to the chemical environment and temperature (Fig. 4c). This minimal AND gate only required one inducible promoter, one constitutive promoter, one repressor and one RNA thermometer. Compared with other transcription based genetic AND gates,28 our approach reduced the amount of required gene parts by half. The mini-AND gates would thus be more flexible for the application of protocell engineering.
Artificial cells or protocells, which can be built by top-down or bottom-up methods, have attracted much attention as substitutes for natural cells mimicking some of their essential properties. However, despite the impressive progress to date, there is still a wide gap between artificial cells and biological cells. To fill this gap, several issues remain to be solved. A key issue is the efficient regulation of “cells” by their environment, and the respective communication with it.2 Our study addressed this problem by developing a synthetic temperature feedback regulation toolbox to render protocells responsive to heat. We validated our RNA thermometer in vitro and developed an in vitro strategy for tuning the thermal response rapidly and simply. As a translation level regulator, RNA thermometers can minimize the transcriptional redundancy of the genetic circuits to make them easier to be implanted in a minimal cell model.
We generated protocells from the bottom-up by encapsulating a temperature feedback transcription/translation machinery into picoliter emulsion droplets. With these temperature sensitive devices, the protocells can process different inputs in a deterministic way. We demonstrated a thermally driven gene multiplexer and a logic AND gate, capable of processing both thermal and chemical signals. Such tunable temperature sensitive protocells will enable a variety of applications in biotechnology, medicine, and industry. For example, by their ability to detect temperature changes, respectively optimized medical protocells may at some point replace microbial therapy as a much safer way to produce and deliver pharmaceuticals by sensing the fever of host. Alternatively, drug synthesis and release from protocells may be controlled by focused ultrasound heating, potentially targeting drugs directly to the specific sites in the host. With respect to fundamental research, temperature dependent differential regulation of protein expression, as shown here, will enable us to establish reaction schemes that require a delicate concentration ratio, such as the self-organization of MinDE proteins,29 in cell free expression schemes. Undoubtedly, further development of environment sensitive logics as shown here within artificial cells or protocells will bring attractive opportunities to many fields.
This research has been supported by the GRK2062 funded by Deutsche Forschungsgemeinschaft (DFG); and MaxSynBio consortium funded by the Federal Ministry of Education and Research of Germany and the Max Planck Society.
Open Access funding provided by the Max Planck Society.
Conflicts of interest
There are no conflicts to declare.References
- P. Schwille and S. Diez, Crit. Rev. Biochem. Mol. Biol., 2009, 44, 223–242 CrossRef CAS PubMed.
- C. Xu, S. Hu and X. Chen, Mater. Today, 2016, 19, 516–532 CrossRef CAS PubMed.
- V. Noireaux, R. Bar-Ziv and A. Libchaber, Proc. Natl. Acad. Sci. U. S. A., 2003, 100, 12672–12677 CrossRef CAS PubMed.
- J. Shin and V. Noireaux, ACS Synth. Biol., 2012, 1, 29–41 CrossRef CAS.
- H. Niederholtmeyer, Z. Z. Sun, Y. Hori, E. Yeung, A. Verpoorte, R. M. Murray and S. J. Maerkl, eLife, 2015, 4, e09771 CrossRef PubMed.
- P. M. Gardner, K. Winzer and B. G. Davis, Nat. Chem., 2009, 1, 377–383 CrossRef CAS PubMed.
- Y. Qiao, M. Li, R. Booth and S. Mann, Nat. Chem., 2017, 9, 110 CrossRef CAS PubMed.
- G. Rampioni, F. D’Angelo, M. Messina, A. Zennaro, Y. Kuruma, D. Tofani, L. Leoni and P. Stano, Chem. Commun., 2018, 54, 2090–2093 RSC.
- T. D. Tang, D. Cecchi, G. Fracasso, D. Accardi, A. Coutable-Pennarun, S. S. Mansy, A. W. Perriman, J. R. Anderson and S. Mann, ACS Synth. Biol., 2017, 7, 339–346 CrossRef.
- G. Villar, A. D. Graham and H. Bayley, Science, 2013, 340, 48–52 CrossRef CAS PubMed.
- A. Dupin and F. C. Simmel, Nat. Chem., 2019, 11, 32 CrossRef CAS PubMed.
- H. Jia, M. Heymann, F. Bernhard, P. Schwille and L. Kai, Nat. Biotechnol., 2017, 39, 199–205 CAS.
- G. Segal and E. Z. Ron, Ann. N. Y. Acad. Sci., 1998, 851, 147–151 CrossRef CAS.
- D. I. Piraner, M. H. Abedi, B. A. Moser, A. Lee-Gosselin and M. G. Shapiro, Nat. Chem. Biol., 2017, 13, 75 CrossRef CAS PubMed.
- H. Jia, X. Sun, H. Sun, C. Li, Y. Wang, X. Feng and C. Li, ACS Synth. Biol., 2016, 5, 312–320 CrossRef CAS PubMed.
- S. Sen, D. Apurva, R. Satija, D. Siegal and R. M. Murray, ACS Synth. Biol., 2017, 6, 1461–1470 CrossRef CAS.
- F. W. Sadler, I. Dodevski and C. A. Sarkar, ACS Synth. Biol., 2017, 7, 292–296 CrossRef.
- J. Kortmann and F. Narberhaus, Nat. Rev. Microbiol., 2012, 10, 255–265 CrossRef CAS PubMed.
- A. S. Mironov, I. Gusarov, R. Rafikov, L. E. Lopez, K. Shatalin, R. A. Kreneva, D. A. Perumov and E. Nudler, Cell, 2002, 111, 747–756 CrossRef CAS PubMed.
- Y. Shimizu, A. Inoue, Y. Tomari, T. Suzuki, T. Yokogawa, K. Nishikawa and T. Ueda, Nat. Biotechnol., 2001, 19, 751–755 CrossRef CAS PubMed.
- D. E. Draper, RNA, 2004, 10, 335–343 CrossRef CAS.
- C. Holtze, A. Rowat, J. Agresti, J. Hutchison, F. Angile, C. Schmitz, S. Köster, H. Duan, K. Humphry and R. Scanga, Lab Chip, 2008, 8, 1632–1639 RSC.
- M. B. Elowitz, A. J. Levine, E. D. Siggia and P. S. Swain, Science, 2002, 297, 1183–1186 CrossRef CAS PubMed.
- M. M. Hansen, L. H. Meijer, E. Spruijt, R. J. Maas, M. V. Rosquelles, J. Groen, H. A. Heus and W. T. Huck, Nat. Nanotechnol., 2016, 11, 191 CrossRef CAS PubMed.
- K. Deepankumar, S. P. Nadarajan, D.-H. Bae, K.-H. Baek, K.-Y. Choi and H. Yun, Biotechnol. Bioprocess Eng., 2015, 20, 67–72 CrossRef CAS.
- B. Wang, R. I. Kitney, N. Joly and M. Buck, Nat. Commun., 2011, 2, 508 CrossRef PubMed.
- T. S. Moon, C. Lou, A. Tamsir, B. C. Stanton and C. A. Voigt, Nature, 2012, 491, 249–253 CrossRef CAS PubMed.
- J. Bonnet, P. Yin, M. E. Ortiz, P. Subsoontorn and D. Endy, Science, 2013, 340, 599–603 CrossRef CAS PubMed.
- P. Glock, B. Ramm, T. Heermann, S. Kretschmer, J. Schweizer, J. Mücksch, G. K. Alagöz and P. Schwille, ACS Synth. Biol., 2018, 8, 148–157 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9cc02734c |
| This journal is © The Royal Society of Chemistry 2019 |