Open Access Article
Open Access ArticleSilica particles with a quercetin–R5 peptide conjugate are taken up into HT-29 cells and translocate into the nucleus†
Giorgia
Del Favero‡
 a,
Friedrich
Bialas‡
b,
Stephanie
Grabher
a,
Anja
Wittig
a,
Birgit
Bräuer
b,
Dagmar
Gerthsen
c,
Cécile
Echalier
a,
Friedrich
Bialas‡
b,
Stephanie
Grabher
a,
Anja
Wittig
a,
Birgit
Bräuer
b,
Dagmar
Gerthsen
c,
Cécile
Echalier
 d,
Meder
Kamalov
d,
Meder
Kamalov
 b,
Doris
Marko
b,
Doris
Marko
 *a and
Christian F. W.
Becker
*a and
Christian F. W.
Becker
 *b
*b
aInstitute of Food Chemistry and Toxicology, Faculty of Chemistry, University of Vienna, Waehringer Strasse 38, 1090 Vienna, Austria. E-mail: doris.marko@univie.ac.at
bInstitute of Biological Chemistry, Faculty of Chemistry, University of Vienna, Waehringer Strasse 38, 1090 Vienna, Austria. E-mail: christian.becker@unvie.ac.at
cLaboratory for Electron Microscopy, Karlsruhe Institute of Technology (KIT), Engesserstrasse 7, 76131 Karlsruhe, Germany
dInstitut des Biomolécules Max Mousseron (IBMM), UMR5247 CNRS, ENSCM, Université de Montpellier, 15 Avenue Charmes Flahault, 34093 Montpellier Cedex 05, France
First published on 17th July 2019
Abstract
Intracellular delivery of bioactive polyphenols is currently evaluated as a protective strategy for cells under pharmaceutical stress. To this end, the 20mer R5 peptide from the marine diatom C. fusiformis was N-terminally modified with a quercetin derivative. This polyphenol–peptide conjugate was used to generate homogeneous silica particles under biomimetic conditions that are efficiently taken up by eukaryotic cells without being cytotoxic. However, not only was accumulation in the cytoplasm of living cells observed via electron and fluorescence microscopy but also translocation into the nucleus. The latter was only seen when the quercetin–peptide conjugate was present within the silica particles and provides a novel targeting option for silica particles to nuclei.
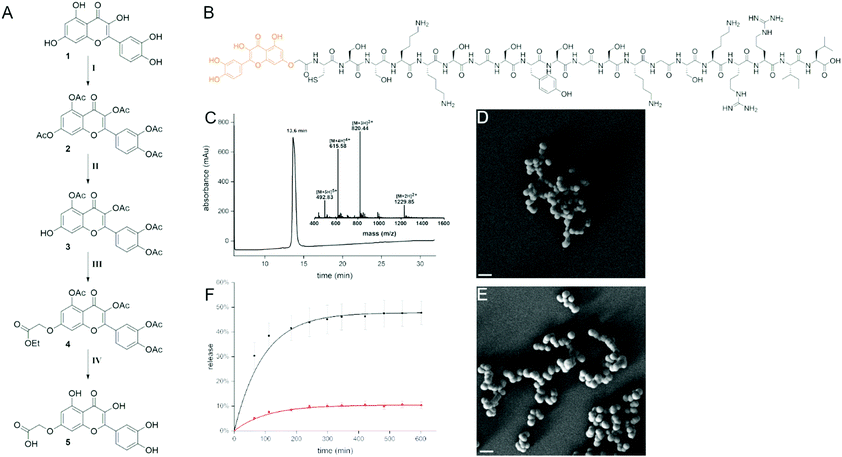
Delivering biological cargo into cells is a complex process that is difficult to control and recent studies have taken advantage of mesoporous silica nanoparticles as a suitable delivery tool.1,2 Here we focused on a biomimetic strategy to generate silica particles that is based on the naturally occurring peptide R5 found in the marine diatom C. fusiformis.3 This 20mer peptide generates, under biomimetic conditions, homogeneous spherical silica particles (SiPs). Alterations to the peptide structure and sequence, e.g. by adding posttranslational modifications or by shuffling the amino acid sequence, can lead to different particle morphologies.4,5 We have previously demonstrated that peptide and protein cargo can be efficiently encapsulated into the resulting silica particles without affecting their function.6,7 Together with our current findings that hydrophobic N-terminal modifications of the R5 sequence support self-assembly of the peptide prior to silica precipitation,8 this has led us to generate a covalent conjugate of a bioactive polyphenol, namely quercetin, with the R5 peptide (Fig. 1A–C). Quercetin is a common dietary polyphenol, also widely used as food supplement.9 Even if well-known and characterized, thanks to its strong biological activity, quercetin is of continuously rising scientific interest.10–12
For introducing a carboxylic acid linker into quercetin without affecting its function we chose to address the 7-hydroxy group. This was achieved by first completely acetylating quercetin with an excess of acetic anhydride in pyridine at 70 °C. It was then regioselectively deprotected with imidazole and thiophenol in NMP as described by Kim et al. to give the 7-O-monodeacetylated product.13 Subsequently, the carboxylic acid linker was introduced by addition of ethyl iodoacetate. The intermediate 3 was then deprotected using hydrochloric acid in refluxing acetone as described by Mattarei et al. (11% yield over 4 steps, Fig. 1A).14 Coupling to the 20mer R5 peptide was performed on resin prior to cleavage and purification (Fig. 1B). The cleaved conjugate was then purified by RP-HPLC (Fig. 1C). The resulting quercetin–R5 conjugate was dissolved at 1 mg ml−1 concentration in phosphate buffer at pH 7 and freshly generated silicic acid was added to give highly homogeneous silica particles with a peptide loading of >95%. Particles were separated by centrifugation, washed and imaged by scanning electron microscopy (SEM, Fig. 1D). The obtained particles were spherical with a diameter of ∼400 nm. Release of the conjugate from the silica particles in 50 mM potassium phosphate buffer at neutral pH and pH 4 was followed by UV-VIS spectroscopy, based on the absorbance of quercetin at 375 nm. Here, a pH-dependent release could be observed. At pH 4, 45% of the quercetin–R5 conjugate was found in the supernatant after 5 h, whereas at neutral pH only 10% were released after 5 h and no further increase in released conjugate was observed (Fig. 1F).
Therefore, quercetin–R5 containing SiPs were incubated in aqueous buffer at pH 7.4 before incubation with cells for 5 h to remove loosely associated quercetin–R5. To investigate the morphology of the particles after release of a large fraction of the conjugate, a sample of the particles was incubated for 4.5 h in 50 mM phosphate buffer at pH 4. The particles were then imaged by SEM showing that they retained their spherical shape (Fig. 1E). The supernatant of the particles after 10 h incubation at pH 4 was also analysed by LC-MS and clearly indicated that the released conjugate was fully intact (Fig. S1, ESI†).
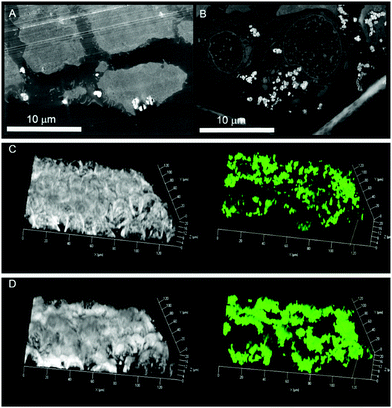
To test uptake of R5-based silica particles, human colon adenocarcinoma HT-29 cells were cultivated and incubated with R5-silica particles not containing quercetin. High-angle annular dark-field scanning transmission electron microscopy (HAADF-STEM) experiments on ultrathin cell sections were performed to analyse intact cells and locate SiPs. Biomimetic SiPs show strong bright contrast in HAADF-STEM images and appear to accumulate at the cellular membrane already after 1 h of incubation with HT-29 cells (Fig. 2A). Accordingly, after 24 h of incubation massive accumulation of SiPs in the intracellular compartment but not in the nucleus was observed (Fig. 2B). Once we verified that biomimetic R5-SiPs can enter the intracellular compartment, a fluorescent variant was used in combination with live cell imaging to follow the kinetic of uptake at the cellular level. The fluorescein-labeled silane triethoxysilyl fluorescein was incorporated into SiPs at a ratio of 1 to 20 to the R5 peptide.15 Similarly, a progressive uptake was observed that led to accumulation of the fluorescent SiPs in the cytoplasm of HT-29 cells after 3 h (Fig. 2C and D). Cytotoxicity measurement revealed no toxicity of biomimetic SiPs in HT-29 cells (Fig. S2, ESI†).16
After demonstrating uptake of R5-SiPs into HT-29 cells without detectable effects on cell viability, we moved on to quercetin–R5-based SiPs. These SiPs behaved similarly to the R5– and fluorescein–R5-SiPs described above. Interestingly, we noticed that the intrinsic fluorescence of the quercetin–R5 peptide, even when incorporated in SiPs, was sufficient for live cell imaging (Fig. S3, ESI†). Concentration-dependent quercetin fluorescence was measured in HT-29 cells to determine the optimal concentration for live cell imaging (Fig. S4, ESI†). Based on these results, in combination with cytotoxicity data for R5 SiP, uptake experiments were performed at concentrations of 70 μM quercetin or quercetin–R5 SiP, respectively.
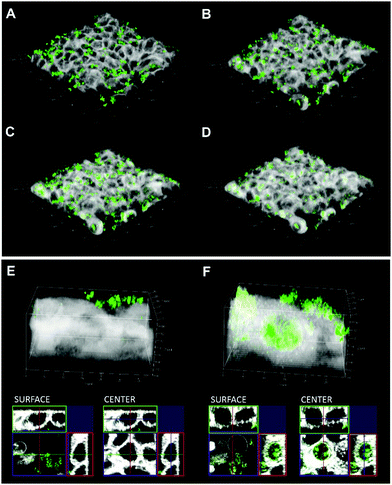
The intrinsic fluorescence of the quercetin–R5 conjugate allowed us to study uptake without any additional labelling, which also excludes undesired effects of labels on the uptake and targeting mechanisms. Here, uptake into HT-29 cells was followed by live cell imaging and proved that already after 3 h a major fraction of quercetin–R5 SiPs was found in cells (Fig. 3A–D). However, uptake of quercetin–R5 SiPs starts immediately after the application but it takes at least 90 min to reach the nucleus/peri-nuclear region (Fig. S5, ESI†). In order to verify if the uptake of quercetin–R5-SiPs was an active process or a consequence of an alteration of the permeability of the cell membrane, respective live cell imaging experiments were performed (Fig. S6, ESI†). Confocal live imaging microscopy allowed us to follow the uptake of quercetin–R5 SiPs and subsequent interaction with the plasma membrane (Fig. S6, ESI†). Uptake of the quercetin–R5 SiPs occurred through active engulfment of the particles followed by recovery of the membrane surface to its original integrity, thus suggesting the SiPs uptake to be an active, energy-dependent process and not a secondary effect of the particles sustained by a loss of membrane integrity. These findings are supported by uptake experiments at 4 °C and in the presence of wortmannin as an inhibitor of phagocytosis/micropinocytosis17 and after treatment with methyl-β-cyclodextrin (mβCD) to deplete cholesterol and induce alterations of membrane organization and fluidity (Fig. S7, ESI†).18 The uptake of quercetin–R5 SiPs was severely reduced at 4 °C and SiPs are mainly found on the cell surface after 3 h. However, as the wortmannin and mβCD treatment did not lead to significant changes in uptake of quercetin–R5 SiPs, we can currently only exclude the related uptake mechanisms of phagocytosis/micropinocytosis and caveolin-mediated endocytosis for HT-29 cells. Moreover, the quercetin-modified SiPs accumulated inside the nucleus (Fig. 3E and F and cross sections of the cell surface/central section), a behaviour not observed for any other SiPs investigated here and rarely for silica (nano-) particles tested in other studies.19,20
To control if this targeting effect was accompanied by a biological effect, comet assay (single cell gel electrophoresis) experiments were performed with the quercetin–R5 SiPs in comparison to the corresponding concentration of quercetin (70 μM), quercetin–R5 and R5 SiPs. Interestingly, binding of quercetin to SiPs abolished the genotoxic effect of the unconjugated compound (Fig. S8, ESI†).
Overall, we describe a new strategy to target biomimetic silica particles to the nucleus of eukaryotic cells without any detectable effects on cell viability. Covalent conjugation of the bioactive food constituent quercetin to a similarly safe silaffin-based peptide that is used to generate biomimetic silica, provides an almost ideal cellular shuttle. To attach cargo to this shuttle the cysteine residue offers itself as an attachment point that could be used to release cargo via redox-mediated processes on its way through the cytoplasm or via more stable linkages within the nucleus.
We gratefully acknowledge help from Carolin Lechner, Helge Gehrke, Georg Aichinger, Svenja Hankele and Holger Blank with sample preparation and data collection. This project has received funding from the European Union's Horizon 2020 research and innovation programme under the Marie Sklodowska-Curie grant agreement no. 675007.
Conflicts of interest
There are no conflicts to declare.Notes and references
- J. Lee, D. G. Jo, D. Park, H. Y. Chung and M. P. Mattson, Pharmacol. Rev., 2014, 66, 815–868 CrossRef PubMed.
- A. Watermann and J. Brieger, Nanomaterials, 2017, 7, 189–205 CrossRef PubMed.
- R. D. Nils Kröger and M. Sumper, Science, 1999, 286, 1129–1132 CrossRef PubMed.
- C. C. Lechner and C. F. W. Becker, Chem. Sci., 2012, 3, 3500 RSC.
- C. C. Lechner and C. F. Becker, J. Pept. Sci., 2014, 20, 152–158 CrossRef CAS PubMed.
- C. C. Lechner and C. F. Becker, Bioorg. Med. Chem., 2013, 21, 3533–3541 CrossRef CAS PubMed.
- C. C. Lechner and C. F. Becker, Biomater. Sci., 2015, 3, 288–297 RSC.
- M. Kamalov, A. Hajradini, C. Rentenberger and C. F. W. Becker, Mater. Lett., 2018, 212, 114–117 CrossRef CAS.
- C. Chalet, J. Rubbens, J. Tack, G. S. Duchateau and P. Augustijns, J. Pharm. Pharmacol., 2018, 70, 1002–1008 CrossRef CAS PubMed.
- Q. Wu, P. A. Kroon, H. Shao, P. W. Needs and X. Yang, J. Agric. Food Chem., 2018, 66, 7181–7189 CrossRef CAS PubMed.
- Y. Jin, Z.-l. Huang, L. Li, Y. Yang, C.-h. Wang, Z.-t. Wang and L.-l. Ji, Acta Pharmacol. Sin., 2018, 40, 75–85 CrossRef PubMed.
- F. Aghapour, A. A. Moghadamnia, A. Nicolini, S. N. M. Kani, L. Barari, P. Morakabati, L. Rezazadeh and S. Kazemi, Biochem. Biophys. Res. Commun., 2018, 500, 860–865 CrossRef CAS PubMed.
- M. K. Kim, H. Choo and Y. Chong, J. Med. Chem., 2014, 57, 7216–7233 CrossRef CAS PubMed.
- A. Mattarei, L. Biasutto, F. Rastrelli, S. Garbisa, E. Marotta, M. Zoratti and C. Paradisi, Molecules, 2010, 15, 4722–4736 CrossRef CAS PubMed.
- J. Ciccione, T. Jia, J.-L. Coll, K. Parra, M. Amblard, S. Jebors, J. Martinez, A. Mehdi and G. Subra, Chem. Mater., 2016, 28, 885–889 CrossRef CAS.
- A. Wittig, H. Gehrke, G. Del Favero, E. M. Fritz, M. Al-Rawi, S. Diabate, C. Weiss, H. Sami, M. Ogris and D. Marko, Nanomaterials, 2017, 7, 18 CrossRef PubMed.
- H. Herd, N. Daum, A. T. Jones, H. Huwer, H. Ghandehari and C. M. Lehr, ACS Nano, 2013, 7, 1961–1973 CrossRef CAS PubMed.
- X. Zhang, J. Hurng, D. L. Rateri, A. Daugherty, G. W. Schmid-Schonbein and H. Y. Shin, Am. J. Physiol.: Cell Physiol., 2011, 301, C451–C460 CrossRef CAS PubMed.
- A. S.-S. K. AbouAitah, A. A. Farghali, J. Wojnarowicz, A. Stefanek, S. Gierlotka, A. Opalinska, A. K. Allayeh, T. Ciach and W. Lojkowski, Oncotarget, 2018, 9, 26466–26490 CrossRef PubMed.
- L. Xiong, X. Du, F. Kleitz and S. Z. Qiao, Small, 2015, 11, 5919–5926 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Experimental procedures, supplementary figures and tables. See DOI: 10.1039/c9cc02215e |
| ‡ Authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2019 |