Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Bisulfite-free and base-resolution analysis of 5-methylcytidine and 5-hydroxymethylcytidine in RNA with peroxotungstate†
Fang
Yuan‡
ab,
Ying
Bi‡
a,
Paulina
Siejka-Zielinska
a,
Ying-Lin
Zhou
 b,
Xin-Xiang
Zhang
*b and
Chun-Xiao
Song
b,
Xin-Xiang
Zhang
*b and
Chun-Xiao
Song
 *a
*a
aLudwig Institute for Cancer Research and Target Discovery Institute, Nuffield Department of Medicine, University of Oxford, OX3 7FZ, UK. E-mail: chunxiao.song@ludwig.ox.ac.uk
bBeijing National Laboratory for Molecular Sciences (BNLMS), MOE Key Laboratory of Bioorganic Chemistry and Molecular Engineering, College of Chemistry, Peking University, Beijing 100871, China. E-mail: zxx@pku.edu.cn
First published on 30th January 2019
Abstract
5-Methylcytosine (5mC) and 5-hydroxymethylcytosine (5hmC), two of the best-studied DNA modifications, play crucial roles in normal development and disease in mammals. Although 5-methylcytidine (m5C) and 5-hydroxymethylcytidine (hm5C) have also been identified in RNA, their distribution and biological function in RNA remain largely unexplored, due to the lack of suitable sequencing methods. Here, we report a base-resolution sequencing method for hm5C in RNA. We applied the selective oxidation of hm5C to trihydroxylated-thymine (thT) mediated by peroxotungstate. thT was subsequently converted to T during cDNA synthesis using a thermostable group II intron reverse transcriptase (TGIRT). Base-resolution analysis of the hm5C sites in RNA was performed using Sanger sequencing. Furthermore, in combination with the TET enzyme oxidation of m5C to hm5C in RNA, we expand the use of peroxotungstate oxidation to detect m5C in RNA at base-resolution. By using this method, we confirmed three known m5C sites in human tRNA, demonstrating the applicability of our method in analyzing real RNA samples.
Epitranscriptome, which refers to the multitude of RNA chemical modifications, has vital roles in post-transcriptional gene regulation.1–3 5-Methylcytidine (m5C) and 5-hydroxymethylcytidine (hm5C) are two of the major RNA modifications in eukaryotic cells, however, our understanding of them is still in its infancy. M5C is abundant in noncoding RNA, and has the ability to stabilize tRNA secondary structure,4–6 but the knowledge about its distribution and function in mRNA are still very limited due to the inconsistent results obtained from the current sequencing methods.7–9 Hm5C has been shown to be enriched in Drosophila melanogaster mRNA, increase mRNA translation and play a central role in Drosophila brain development.10 Hm5C also exists in mammalian RNA, albeit at low levels,11 and the TET proteins that oxidize 5-methylcytosine (5mC) to 5-hydroxymethylcytosine (5hmC) in DNA can also do so in RNA.10,12 However, the distribution and regulation roles of hm5C in the mammalian transcriptome remain unknown due to the lack of sensitive and robust sequencing methods.
The most common way to sequence m5C in RNA is to adopt bisulfite sequencing, which is widely used to sequence 5mC in DNA. Bisulfite treatment deaminates unmethylated cytosine to uracil in single-strand RNA, while leaving m5C unconverted. Therefore, bisulfite sequencing provides base-resolution information of m5C. Using bisulfite sequencing, widespread m5C sites were identified in both coding and non-coding RNAs.7,8 However, bisulfite treatment employs sequential thermal acidic and alkaline conditions that severely damage the RNA. Further analysis also revealed potential false positives from RNA bisulfite sequencing due to incomplete conversion of unmethylated cytosine in the double-stranded RNA regions and other modifications resistant to bisulfite treatment.13–15 Other methods to sequence m5C in RNA are immunoprecipitation-based that use m5C-specific antibodies or methyltransferases to pull down m5C-containing RNA.16–18 These methods, however, do not have base-resolution and lose the quantitative levels of m5C. Mapping hm5C in RNA is even more challenging. To date, there is no base-resolution sequencing method for hm5C. The only reported method is the antibody-based immunoprecipitation approach.10 This method has been applied to the Drosophila transcriptome, but has yet to be successful in the mammalian transcriptome. Clearly, new RNA-friendly and high-resolution sequencing methods are highly desirable to further study the elusive distribution, localization and biological roles of these two modifications in RNA. Here, we report bisulfite-free and base-resolution sequencing methods for hm5C and m5C based on peroxotungstate oxidation.
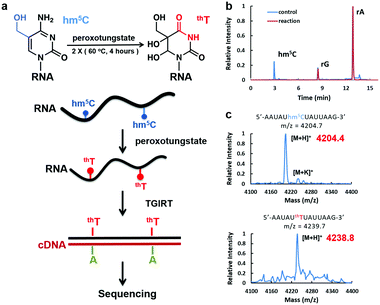
Peroxotungstate oxidation was first developed by the Okamoto group19,20 for selective oxidation of 5hmC in DNA to trihydroxylated-thymine (thT). thT is a thymine derivative, and will induce C-to-T transition in DNA after PCR. However, the peroxotungstate oxidation reaction requires single-strand DNA. The reaction is strongly inhibited in double-strand DNA with a conversion rate of less than 10%, which severely limits its application.20 Although this reaction is not suitable for DNA samples, we hypothesized that it could be ideal to detect hm5C in RNA, which is mostly single-stand (Fig. 1a). We termed this approach peroxotungstate oxidation sequencing (WO-Seq).
We started with optimizing the oxidation conditions of the peroxotungstate against in vitro-transcribed hm5C-containing RNA1. MALDI-TOF MS and HPLC-MS/MS were used to monitor the reaction rate. After two rounds of 4 hours incubation at 60 °C, the hm5C peak in HPLC-MS/MS was undetectable (Fig. 1b), and the MALDI peak of RNA fragments containing one hm5C changed from m/z = 4204.4 to m/z = 4238.8. This is consistent with the calculated m/z change from hm5C-containing RNA to thT-containing RNA (Fig. 1c). Sensitivity of the peroxotungstate treatment for hm5C was also tested (Fig. S1, ESI†). Samples of different combination of hm5C modified RNA and unmodified RNA were treated by peroxotungstate, and then analysed by HPLC-MS/MS. The conversion rates of hm5C were similar in all samples, indicating that the peroxotungstate treatment is suitable for real biological samples which has low hm5C content.
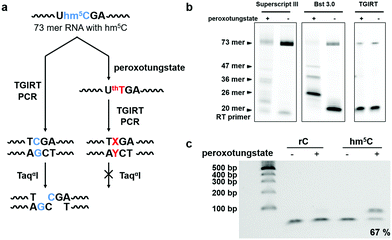
Next, we investigated the potential of the hm5C-to-T transition during cDNA synthesis using the peroxotungstate-oxidized RNA template. We designed and synthesized a 73mer RNA that contained three hm5C sites (RNA2). To enable us to monitor the efficiency of the hm5C-to-T conversion, one hm5C was positioned so that, upon successful hm5C-to-T conversion, a TaqαI restriction enzyme recognition site in the resulting RT-PCR product was destroyed (Fig. 2a). Since thT is not a natural occurring base, we first sought to investigate its behavior during cDNA synthesis. Several commercially available reverse transcriptases were tested on this RNA template. Interestingly, only the thermostable group II intron reverse transcriptase (TGIRT)21,22 could read though all reacted hm5C sites, while Superscript III and Bst 3.0 DNA polymerase induced truncations at the reacted hm5C sites (Fig. 2b). The reaction conditions of TGIRT were further optimized to get the best reverse transcription efficiency, and after subsequent PCR, the DNA products were digested with TaqαI. As shown in Fig. 2c, 67% of the PCR products from the oxidized hm5C-containing RNA2 sample stayed intact after the TaqαI treatment, indicating loss of restriction enzyme cut site and the successful base change induced by the peroxotungstate-oxidized RNA during cDNA synthesis.
We then performed Sanger sequencing of the PCR product from the oxidized RNA samples (Fig. 3). At each hm5C site, a new peak of thymine signal appears, confirming the base change is indeed C-to-T. In order to accurately quantify the C-to-T conversion rate, the PCR product was cloned and sequenced individually. A 62.1% conversion rate was observed from a total of 66 hm5C sites sequenced (Fig. S2, ESI†), consistent with the restriction enzyme digestion result. As a control, PCR products of peroxotungstate-treated normal cytosine (rC)-containing RNA2 and m5C-containing RNA2 were also cloned and sequenced (Fig. S3, ESI†). Results showed that both rC and m5C sites did not change after the treatment, indicating an excellent selectivity of peroxotungstate oxidation on hm5C. Notably, peroxotungstate oxidation is a mild reaction, which showed less damage on RNA compared with bisulfite reaction (Fig. S4, ESI†).
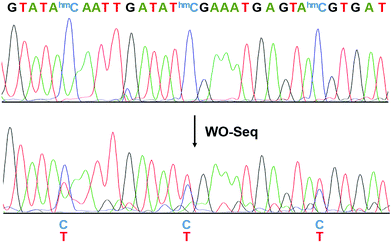 | ||
| Fig. 3 Electropherograms of Sanger sequencing results before and after WO-Seq. The conversion of C-to-T happened at each hm5C site. | ||
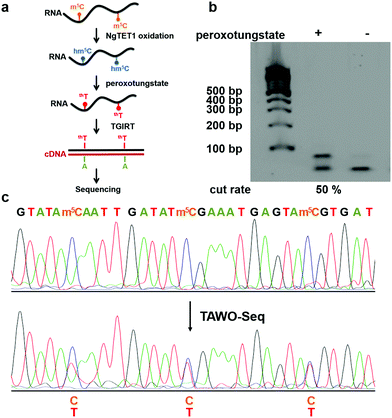
After demonstrating WO-Seq for hm5C sequencing, we next sought to expand its use for m5C sequencing in RNA. In DNA, 5hmC is generated by the oxidation of 5mC mediated by the TET enzyme. Recently, the mammalian TET enzyme was reported to have the ability of oxidizing m5C to hm5C in RNA.12 We tested Naeglaria Tet-like oxygenase (NgTET1)23 and showed it can also oxidize m5C to hm5C on m5C-containing RNA1 by both MALDI-MS and HPLC-MS/MS (Fig. S5, ESI†). Based on this, we further aimed to combine the peroxotungstate oxidation with NgTET1 oxidation to detect m5C in a procedure we termed TET-Assisted WO-Seq (TAWO-Seq) (Fig. 4a). The results of both oxidation reactions were verified by HPLC-MS/MS (Fig. S6, ESI†). Restriction enzyme digestion assays and Sanger sequencing were performed (Fig. 4b and c). Sanger sequencing results showed the C-to-T transition at each m5C site. The m5C-to-T conversion rate was 50% estimated by restriction enzyme analysis, lower than that of hm5C, due to incomplete m5C to hm5C oxidation by NgTET1 (Fig. S6, ESI†). We also cloned and sequenced individual PCR product for the m5C sample. As shown in Fig. S7 (ESI†), 33.3% of the total m5C sites were successfully detected. Commercially available mouse Tet1 (mTet1) was also tested for the TAWO-Seq, which gives similar results with NgTET1 (Fig. S8a, ESI†). We further demonstrated that β-glucosyltransferase (βGT) can label hm5C with glucose and thereby protect it from peroxotungstate oxidation (Fig. S9, ESI†). Combining βGT protection with TAWO-Seq could therefore enable it to detect m5C specifically.
To further demonstrate the utility of TAWO-Seq on real RNA sample, we applied it to the endogenous tRNAAsp(GUC) in 293T cells. The tRNAAsp(GUC) contains three known m5C sites at structural positions 38, 47 and 48 (Fig. S10a, ESI†).24–26 Both NgTET1 and mTet1 were used to oxidize the tRNA and the products were then treated with peroxotungstate. The RT-PCR product of treated tRNAAsp(GUC) was cloned and sequenced. As shown in Fig. S10b and c (ESI†), 35.2% of the m5C sites were successfully detected with NgTET1 assisted WO-Seq, and 37.5% of the m5C sites were detected using mTet1 assisted WO-Seq, which demonstrated the applicability of TAWO-Seq to real RNA samples. Among the three m5C sites in tRNAAsp(GUC), we found that m5C at position 48 has the highest C-to-T conversion rate (68.4% by NgTET1 assisted WO-Seq, 77.8% by mTet1 assisted WO-Seq). According to the tRNAAsp(GUC) structure, this site is in a double-stranded CpG context, which is an ideal substrate of TET enzyme.23 It is likely that the different m5C-to-T conversion rates of three m5C sites are caused by the sequence preference of the TET proteins.
In conclusion, we have described WO-Seq as an RNA friendly, chemical oxidation-based, base-resolution method to sequence hm5C in RNA. We demonstrate the specific hm5C-to-T transition using peroxotungstate to oxidize the RNA followed by cDNA synthesis with the TGIRT enzyme, and Sanger sequencing results have proved the base-resolution sequencing ability of this method. We further demonstrate the ability of TAWO-Seq to detect m5C by combining WO-Seq with the prior NgTET1 or mTet1 oxidation of m5C to hm5C. The successful detection of m5C sites in human tRNA demonstrates our method is applicable for real RNA samples. Both WO-Seq and TAWO-Seq could potentially solve the false positive issue of bisulfite sequencing since they directly detect modified cytosine without affecting unmodified cytosine. Further improvement of both methods to increase the conversion rate and apply to mRNA samples using next-generation sequencing technology are underway in the lab. We believe this method could be highly useful for the identification of unexplored m5C/hm5C distribution and function in the transcriptome.
We would like to acknowledge P. Spingardi, G. Berridge and B. Kessler for helping with the HPLC-MS/MS; C. He for the mTet1; F. Howe for editing the manuscript. This work was supported by the Ludwig Institute for Cancer Research. Work in the C.-X. Song lab is also supported by Cancer Research UK (C63763/A26394 and C63763/A27122), NIHR Oxford Biomedical Research Centre, and Conrad N. Hilton Foundation. The views expressed are those of the authors and not necessarily those of the NHS, the NIHR or the Department of Health. F. Yuan and Y. Bi are supported by China Scholarship Council.
Conflicts of interest
There are no conflicts to declare.Notes and references
- Y. Saletore, K. Meyer, J. Korlach, I. D. Vilfan, S. Jaffrey and C. E. Mason, Genome Biol., 2012, 13, 175 CrossRef CAS PubMed.
- M. A. Machnicka, K. Milanowska, O. Osman Oglou, E. Purta, M. Kurkowska, A. Olchowik, W. Januszewski, S. Kalinowski, S. Dunin-Horkawicz, K. M. Rother, M. Helm, J. M. Bujnicki and H. Grosjean, Nucleic Acids Res., 2013, 41, D262–D267 CrossRef CAS PubMed.
- M. Frye, S. R. Jaffrey, T. Pan, G. Rechavi and T. Suzuki, Nat. Rev. Genet., 2016, 17, 365 CrossRef CAS PubMed.
- J. E. S. a. B. S. Cooperman, Biochemistry, 1992, 31, 10825–10834 CrossRef.
- H. S.-G. Y. Chen, R. Guenther, K. Everett and P. F. Agris, Biochemistry, 1993, 32, 10249–10253 CrossRef PubMed.
- Y. Motorin and M. Helm, Biochemistry, 2010, 49, 4934–4944 CrossRef CAS PubMed.
- M. Schaefer, T. Pollex, K. Hanna and F. Lyko, Nucleic Acids Res., 2009, 37, e12 CrossRef PubMed.
- J. E. Squires, H. R. Patel, M. Nousch, T. Sibbritt, D. T. Humphreys, B. J. Parker, C. M. Suter and T. Preiss, Nucleic Acids Res., 2012, 40, 5023–5033 CrossRef CAS PubMed.
- X. Yang, Y. Yang, B.-F. Sun, Y.-S. Chen, J.-W. Xu, W.-Y. Lai, A. Li, X. Wang, D. P. Bhattarai, W. Xiao, H.-Y. Sun, Q. Zhu, H.-L. Ma, S. Adhikari, M. Sun, Y.-J. Hao, B. Zhang, C.-M. Huang, N. Huang, G.-B. Jiang, Y.-L. Zhao, H.-L. Wang, Y.-P. Sun and Y.-G. Yang, Cell Res., 2017, 27, 606 CrossRef CAS PubMed.
- F. W. Benjamin Delatte, L. V. Ngoc, E. Collignon, E. Bonvin, R. Deplus, E. Calonne, B. Hassabi, P. Putmans, S. Awe, C. Wetzel, J. Kreher, R. Soin, C. Creppe, P. A. Limbach, C. Gueydan, V. Kruys, A. Brehm, S. Minakhina, M. Defrance, R. Steward and F. Fuks, Science, 2016, 351, 282–285 CrossRef PubMed.
- H. Y. Zhang, J. Xiong, B. L. Qi, Y. Q. Feng and B. F. Yuan, Chem. Commun., 2016, 52, 737–740 RSC.
- L. Fu, C. R. Guerrero, N. Zhong, N. J. Amato, Y. Liu, S. Liu, Q. Cai, D. Ji, S. G. Jin, L. J. Niedernhofer, G. P. Pfeifer, G. L. Xu and Y. Wang, J. Am. Chem. Soc., 2014, 136, 11582–11585 CrossRef CAS PubMed.
- S. Hussain, J. Aleksic, S. Blanco, S. Dietmann and M. Frye, Genome Biol., 2013, 14, 215 CrossRef PubMed.
- W. V. Gilbert, T. A. Bell and C. Schaening, Science, 2016, 352, 1408–1412 CrossRef CAS PubMed.
- A. Shafik, U. Schumann, M. Evers, T. Sibbritt and T. Preiss, Biochim. Biophys. Acta, Gene Regul. Mech., 2016, 1859, 59–70 CrossRef CAS PubMed.
- S. Edelheit, S. Schwartz, M. R. Mumbach, O. Wurtzel and R. Sorek, PLoS Genet., 2013, 9, e1003602 CrossRef CAS PubMed.
- V. Khoddami and B. R. Cairns, Nat. Biotechnol., 2013, 31, 458–464 CrossRef CAS PubMed.
- S. Hussain, A. A. Sajini, S. Blanco, S. Dietmann, P. Lombard, Y. Sugimoto, M. Paramor, J. G. Gleeson, D. T. Odom, J. Ule and M. Frye, Cell Rep., 2013, 4, 255–261 CrossRef CAS PubMed.
- A. Okamoto, K. Sugizaki, A. Nakamura, H. Yanagisawa and S. Ikeda, Chem. Commun., 2011, 47, 11231–11233 RSC.
- G. Hayashi, K. Koyama, H. Shiota, A. Kamio, T. Umeda, G. Nagae, H. Aburatani and A. Okamoto, J. Am. Chem. Soc., 2016, 138, 14178–14181 CrossRef CAS PubMed.
- S. Mohr, E. Ghanem, W. Smith, D. Sheeter, Y. Qin, O. King, D. Polioudakis, V. R. Iyer, S. Hunicke-Smith, S. Swamy, S. Kuersten and A. M. Lambowitz, RNA, 2013, 19, 958–970 CrossRef CAS PubMed.
- S. T. Carrell, Z. Tang, S. Mohr, A. M. Lambowitz and C. A. Thornton, Nucleic Acids Res., 2018, 46, e1–e1 CrossRef PubMed.
- J. E. Pais, N. Dai, E. Tamanaha, R. Vaisvila, A. I. Fomenkov, J. Bitinaite, Z. Sun, S. Guan, I. R. Corrêa, C. J. Noren, X. Cheng, R. J. Roberts, Y. Zheng and L. Saleh, Proc. Natl. Acad. Sci. U. S. A., 2015, 112, 4316–4321 CrossRef CAS PubMed.
- M. G. Goll, F. Kirpekar, K. A. Maggert, J. A. Yoder, C.-L. Hsieh, X. Zhang, K. G. Golic, S. E. Jacobsen and T. H. Bestor, Science, 2006, 311, 395–398 CrossRef CAS PubMed.
- M. Schaefer, T. Pollex, K. Hanna and F. Lyko, Nucleic Acids Res., 2009, 37, e12–e12 CrossRef PubMed.
- W. A. Cantara, P. F. Crain, J. Rozenski, J. A. McCloskey, K. A. Harris, X. Zhang, F. A. P. Vendeix, D. Fabris and P. F. Agris, Nucleic Acids Res., 2011, 39, D195–D201 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9cc00274j |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2019 |