Open Access Article
Open Access ArticleDirected evolution of the optoelectronic properties of synthetic nanomaterials†
Benjamin
Lambert
 ,
Alice J.
Gillen
,
Alice J.
Gillen
 ,
Nils
Schuergers
,
Nils
Schuergers
 ,
Shang-Jung
Wu
,
Shang-Jung
Wu
 and
Ardemis A.
Boghossian
and
Ardemis A.
Boghossian
 *
*
Institute of Chemical Sciences and Engineering (ISIC), Ecole Polytechnique Fédérale de Lausanne (EPFL), 1015-Lausanne, Switzerland. E-mail: ardemis.boghossian@epfl.ch
First published on 27th February 2019
Abstract
Directed evolution is a powerful approach to tailor protein properties toward new or enhanced functions. Herein, we use directed evolution to engineer the optoelectronic properties of DNA-wrapped single-walled carbon nanotube sensors through DNA mutation. This approach leads to an improvement in the fluorescence intensity of 56% following two evolution cycles.
Synthetic nanomaterials are typically known to rely on a rational approach for tuning their physical and chemical properties. This approach involves modifying the morphology, composition, shape, size, or surface chemistry of the material in a predictive manner to achieve a desired outcome.1–3 For instance, the optoelectronic properties of quantum dots can be readily tuned by changing particle size.4 Nevertheless, this rational approach is limited to materials for which the relationship between material's structure and function is known.
Materials lacking a defined structure–function relationship are commonly encountered in the field of protein engineering. To engineer these biomaterials, directed evolution, which relies on the iterative screening and selection of mutants from a collection of mutated proteins, is used to enhance or change a desired protein function.5,6 Herein, we apply a directed evolution approach to engineer the optoelectronic properties of nanomaterials, in particular single-stranded DNA-wrapped single-walled carbon nanotubes (ssDNA-SWCNTs) complexes, which currently lack a defined structure–function relationship.
SWCNTs are cylindrical one-dimensional nanostructures that can be conceptualised as rolled-up sheets of graphene.7 The rolling angle of the graphene sheet determines the (n,m) chirality of the SWCNT, resulting in either metallic or semiconducting properties.7 Semiconducting SWCNTs have attracted particular interest in the field of optical biosensing owing to their near-infrared emission that presents many advantages over the emissions of conventional fluorophores. These advantages include fluorescence tunability and long-term photostability, as well as the ability to image in living tissue and blood, which absorb minimally in the near-infrared range.8–10 Furthermore, the surface of SWCNTs can be functionalised with biomolecules such as ssDNA. This functionalisation imparts both colloidal stability in aqueous solutions and analyte specificity for optical sensing applications.11–13 Previous studies have reported that the sequence of the DNA used to suspend the nanotube contributes to variations with respect to SWCNT chirality affinity, analyte selectivity, and the fluorescence intensity of the ssDNA-SWCNT complex.14–18 However, a general understanding of how the DNA sequence affects these properties is lacking.
An empirical approach has thus far been used to identify ssDNA sequences that yield complexes with a preferred fluorescence responses to specific analytes.13,16 In this approach, SWCNTs wrapped with different oligomers are incubated in the presence of various analytes and specificity is determined according to the observed fluorescence response. Sensors developed using this approach are currently used for in vitro and in vivo measurements.19–21 However, these sensors suffer from limited quantum yields (QYs), typically lower than surfactant-suspended SWCNTs which report maximum QYs of ∼1%.22–25 In addition, this current approach has been used to monitor the response of only 13 distinct DNA oligomers at a time. Given that a typical DNA sequence used in these studies is approximately 30 nucleotides long, these oligomers have a total of 430 different possible sequence combinations. As a result, the performance of the majority of these distinct ssDNA-SWCNT sensors is overlooked when using the current approach.
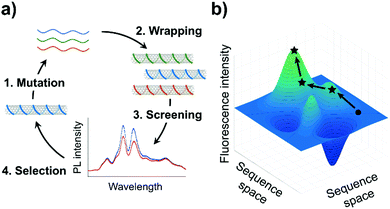
Directed evolution offers a guided search through this vast DNA sequence space. In this article, we use directed evolution to increase the fluorescence efficiency of a ssDNA-SWCNT sensor. The evolution of the ssDNA-SWCNT sensor was done through the cyclic procedure illustrated in Fig. 1a: (1) random mutation of a selected sensing oligomer; (2) wrapping of the mutated oligomers on SWCNTs; (3) screening of the ssDNA-SWCNT complexes for enhanced fluorescence; and (4) selection of the mutants showing the highest fluorescence intensity for the next round of mutation. This approach assumes a smooth fitness landscape (Fig. 1b) whereby the evolution of a particular ssDNA-SWCNT property, in this case the fluorescence intensity, is envisioned as a continuous path among peaks and valleys which represent the respective enhancement or attenuation of this property.
In this work, we applied a directed evolution approach to enhance the fluorescence intensity of ssDNA-SWCNT variants based on the (GT)15 sequence. The (GT)15 sequence was chosen because of its application in neurotransmitter sensing.16 Three random nucleotide substitutions were introduced computationally at randomized locations within the sequence (ESI†). These randomly generated (GT)15 derivative sequences were chemically synthesized (Microsynth AG, Switzerland) with a >99.25% coupling efficiency. For the first evolution cycle, 99 mutants of the (GT)15 sequence were screened for increased integrated fluorescence intensity and for increased intensity of individual SWCNT chiralities. A second round of mutagenesis was performed by introducing three random nucleotide substitutions to the mutants exhibiting increased integrated fluorescence intensity. For this round, a library size of only 10 additional sequences was needed to identify additional mutants that exhibited greater fluorescence enhancement. The absorbance and fluorescence of the SWCNT complexes were compared before and after wrapping to verify that the nanotubes were well suspended by the ssDNA (Fig. S1, S2 and S6, ESI†).
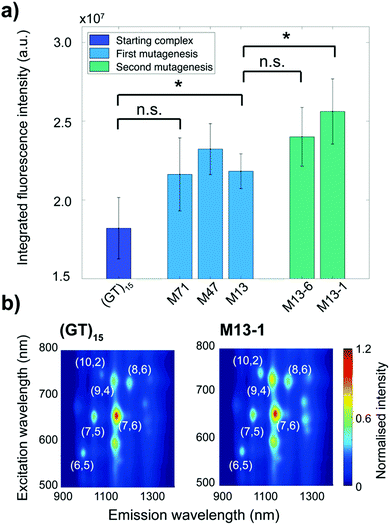
We identified several mutants in each cycle that exhibited higher fluorescence intensities compared to the starting (GT)15-SWCNT complex (Fig. 2a and Table 1). The sequences of the DNA mutants are provided in Table S1 (ESI†). In the first mutagenesis cycle, we identified two mutant complexes that demonstrated statistically increased integrated fluorescence intensity compared to the (GT)15-SWCNT complex: the M13- and M47-SWCNT mutants (Fig. 2a). The second mutagenesis cycle, that was performed on the M13 sequence, yielded two additional mutants, M13-1- and M13-6-SWCNT, that showed increased integrated fluorescence intensity compared to (GT)15-SWCNT complex. Furthermore, the M13-1-SWCNT mutant showed enhanced fluorescence compared to the M13-SWCNT mutant.
| Mutant | ALL | (6,5) | (7,5) | (7,6) | (9,4) | (10,2) | (8,6) |
|---|---|---|---|---|---|---|---|
| The error represents 1 standard deviation (n = 3 replicates). The * symbol indicates p < 0.05 for two-sample student t-tests between the complex and (GT)15-SWCNT. | |||||||
| M13 | 20 ± 14* | 1 ± 8 | 3 ± 9 | −3 ± 11 | 22 ± 17 | 35 ± 16* | 13 ± 18 |
| M47 | 28 ± 16* | 6 ± 10 | 7 ± 10 | −8 ± 11 | 21 ± 18 | 46 ± 17* | 40 ± 24* |
| M71 | 19 ± 18 | 3 ± 10 | 13 ± 13 | −2 ± 15 | 13 ± 20 | 38 ± 20* | 20 ± 24 |
| M13-1 | 41 ± 19* | 5 ± 11 | 1 ± 11 | 2 ± 13 | 43 ± 21* | 54 ± 19* | 56 ± 26* |
| M13-6 | 32 ± 17* | 2 ± 9 | −7 ± 10 | −5 ± 13 | 28 ± 20* | 44 ± 18* | 49 ± 25* |
In addition to increases in integrated fluorescence emissions, we noted a strong chirality dependence on fluorescence enhancement. As shown in the photoluminescence excitation maps (Fig. 2b), the increase in fluorescence intensity was larger for the (9,4), (10,2), and (8,6) chiralities. A quantitative comparison of the fluorescence changes for the different chiralities and the integrated intensity relative to the starting (GT)15-SWCNT complex is shown in Table 1. Overall, the M13-1-SWCNT showed the greatest fluorescence enhancement with an improvement in the integrated intensity of 41% compared to (GT)15-SWCNT. Moreover, the (9,4), (10,2), and (8,6) chiralities showed fluorescence intensity increases of up to 43%, 54%, and 56%, respectively. Although the increase in the integrated intensity of the M71-SWCNT was not statistically significant compared to (GT)15-SWCNT (Fig. 2a), significant increases (+38%) were observed for the (10,2) chirality (Table 1). Additional measurements involving surfactant replacement of the ssDNA18 verified that the observed fluorescence changes were neither due to changes in SWCNT concentration nor chirality distribution (Fig. S1 and S2, ESI†). The changes in fluorescence intensity were, thereby, assumed to be directly related to variations in the fluorescence QY of the complexes.
The QY of ssDNA-SWCNTs is believed to depend on a variety of factors. One such factor is DNA coverage on the SWCNT surface. The DNA wrapping reduces the amount of exposed SWCNT surface, shielding it from oxygen.26,27 Since oxygen induces fluorescence quenching, denser coverage can, therefore, yield increased fluorescence emission.26–28 The dependence of fluorescence intensity on SWCNT surface coverage also partially explains the chirality- and sequence-dependence of the fluorescence changes observed in this work. In agreement with these observations, previous studies have shown that the binding energy and wrapping angle of the DNA on the surface of the SWCNT can depend on the DNA sequence and the SWCNT chirality.29
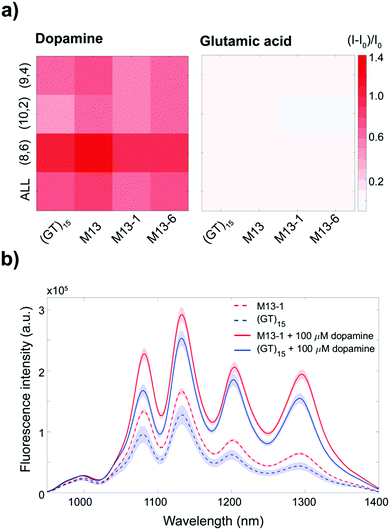
The starting (GT)15-SWCNT complex is responsive towards certain neurotransmitters, including dopamine, which on addition results in a fluorescence increase.16 Dopamine detection assays were performed on the (GT)15-SWCNTs and the mutant complexes in order to test whether the sensing capability was compromised post-mutation. A summary of these sensing responses is provided in Table S2 (ESI†). Dopamine was found to increase fluorescence in a chirality-specific manner (Fig. 3a), with more pronounced responses for the (8,6) chirality observed for both the (GT)15- and M13-SWCNTs (p-value < 0.05). No significant differences in dopamine response were observed between the (GT)15 and mutant complexes for neither the (9,4) chirality nor the integrated intensity. However, for the (10,2) chirality, the M13-, M13-1-, and M13-6-SWCNTs showed overall stronger responses towards dopamine compared to the starting (GT)15-SWCNT complex (p-value < 0.05). No response was observed for the (GT)15- or mutant complexes towards other neurotransmitters, including glutamic acid, γ-aminobutyric acid (GABA), acetylcholine, and glycine (Fig. 3a and Fig. S4, ESI†), demonstrating that selectivity towards dopamine is retained post-mutation. A comparison of the calibration curves for the (GT)15- and M13-SWCNT complexes (Fig. S5, ESI†) confirmed that the mutant and original sensors are similarly able to detect 100 nM of dopamine.
These results show that the mutant complexes demonstrate increased fluorescence intensities while simultaneously retaining the dopamine responsivity of the original sensor. In addition, the fluorescence enhancement of the mutant complexes relative to the (GT)15-SWCNT complex is sustained even following the addition of dopamine (Fig. 3b and Fig. S3, ESI†). This improvement in the fluorescence intensity of these sensors, both before and after dopamine addition, can enable in vivo imaging at penetration depths up to 193 μm deeper30 (ESI†), without compromising dopamine detection.
In this article, we evolved brighter ssDNA-SWCNT sensors through directed evolution of the DNA wrapping. While the reasons for the fluorescence intensity enhancement remain unclear, we demonstrate a guided approach that can be used to tune ssDNA-SWCNT properties in the absence of a defined structure–function relationship. Whereas previous work has focused on monitoring the response of a relatively small collection of random DNA sequences against a variety of analytes, we have instead focused on screening the performances of a larger library of mutants for a single characteristic, in this case fluorescence intensity. Although the observed SWCNT fluorescence increase is lower than other reported methods,26,31–35 the enhancement described herein was achieved without the addition of any exogenous compounds. This achievement is of particular importance for in vivo sensing applications since additives, such as reducing agents or nanoparticles, can lead to increased cytotoxicity and diminished sensitivity and/or selectivity of the sensor.26,31,32,36
While further enhancement may be achieved by refinements, such as screening even larger DNA libraries over additional evolution cycles, the results of this study aim to demonstrate the applicability of a new technique in navigating the fluorescence intensity landscape of ssDNA-SWCNTs. More generally, this work establishes a new, guided technique that can be applied not only to improve properties of existing optical SWCNT sensors, such as sensitivity and selectivity, but also to engineer new sensors with capabilities that are yet to be discovered.
We are thankful for financial support from the Swiss National Science Foundation Assistant Professor (AP) Energy Grant.
Conflicts of interest
There are no conflicts to declare.Notes and references
- Y. Wang, S. De and N. Yan, Chem. Commun., 2016, 52, 6210–6224 RSC.
- A. Albanese, P. S. Tang and W. C. W. Chan, Annu. Rev. Biomed. Eng., 2012, 14, 1–16 CrossRef CAS PubMed.
- C. Kinnear, T. L. Moore, L. Rodriguez-Lorenzo, B. Rothen-Rutishauser and A. Petri-Fink, Chem. Rev., 2017, 117, 11476–11521 CrossRef CAS PubMed.
- S. Baskoutas and A. F. Terzis, J. Appl. Phys., 2006, 99, 013708 CrossRef.
- J. C. Moore and F. H. Arnold, Nat. Biotechnol., 1996, 14, 303–308 CrossRef PubMed.
- F. H. Arnold, Acc. Chem. Res., 1998, 31, 125–131 CrossRef CAS.
- M. S. Dresselhaus, G. Dresselhaus and P. C. Eklund, Science of Fullerenes and Carbon Nanotubes, 1996 Search PubMed.
- A. A. Boghossian, J. Zhang, P. W. Barone, N. F. Reuel, J. H. Kim, D. A. Heller, J. H. Ahn, A. J. Hilmer, A. Rwei, J. R. Arkalgud, C. T. Zhang and M. S. Strano, ChemSusChem, 2011, 4, 848–863 CrossRef CAS PubMed.
- S. Diao, J. L. Blackburn, G. Hong, A. L. Antaris, J. Chang, J. Z. Wu, B. Zhang, K. Cheng, C. J. Kuo and H. Dai, Angew. Chem., Int. Ed., 2015, 54, 14758–14762 CrossRef CAS PubMed.
- P. W. Barone and M. S. Strano, J. Diabetes Sci. Technol., 2009, 3, 242–252 CrossRef PubMed.
- M. Zheng, A. Jagota, E. D. Semke, B. A. Diner, R. S. McLean, S. R. Lustig, R. E. Richardson and N. G. Tassi, Nat. Mater., 2003, 2, 338–342 CrossRef CAS PubMed.
- K. Umemura, Nanomaterials, 2015, 5, 321–350 CrossRef CAS PubMed.
- J. Zhang, A. A. Boghossian, P. W. Barone, A. Rwei, J. H. Kim, D. Lin, D. A. Heller, A. J. Hilmer, N. Nair, N. F. Reuel and M. S. Strano, J. Am. Chem. Soc., 2011, 133, 567–581 CrossRef CAS PubMed.
- A. Shankar, J. Mittal and A. Jagota, Langmuir, 2014, 30, 3176–3183 CrossRef CAS PubMed.
- G. Ao, J. K. Streit, J. A. Fagan and M. Zheng, J. Am. Chem. Soc., 2016, 138, 16677–16685 CrossRef CAS PubMed.
- S. Kruss, M. P. Landry, E. Vander Ende, B. M. A. Lima, N. F. Reuel, J. Zhang, J. Nelson, B. Mu, A. Hilmer and M. Strano, J. Am. Chem. Soc., 2014, 136, 713–724 CrossRef CAS PubMed.
- R. Haggenmueller, S. S. Rahatekar, J. A. Fagan, J. Chun, M. L. Becker, R. R. Naik, T. Krauss, L. Carlson, J. F. Kadla, P. C. Trulove, D. F. Fox, H. C. DeLong, Z. Fang, S. O. Kelley and J. W. Gilrnan, Langmuir, 2008, 24, 5070–5078 CrossRef CAS PubMed.
- P. V. Jena, M. M. Safaee, D. A. Heller and D. Roxbury, ACS Appl. Mater. Interfaces, 2017, 9, 21397–21405 CrossRef CAS PubMed.
- S. Kruss, D. P. Salem, L. Vukovic, B. Lima, E. Vander Ende, E. S. Boyden and M. S. Strano, PNAS, 2017, 114, 1789–1794 CrossRef CAS PubMed.
- J. P. Giraldo, M. P. Landry, S. Y. Kwak, R. M. Jain, M. H. Wong, N. M. Iverson, M. Ben-Naim and M. S. Strano, Small, 2015, 11, 3973–3984 CrossRef CAS PubMed.
- J. T. D. Bonis-O’Donnell, R. H. Page, A. G. Beyene, E. G. Tindall, I. R. McFarlane and M. P. Landry, Adv. Funct. Mater., 2017, 27, 1–10 Search PubMed.
- M. J. O’Connell, S. M. Bachilo, C. B. Huffman, V. C. Moore, M. S. Strano, E. H. Haroz, K. L. Rialon, P. J. Boul, W. H. Noon, C. Kittrell, J. Ma, R. H. Hauge, R. B. Weisman and R. E. Smalley, Science, 2002, 297, 593–596 CrossRef PubMed.
- S. Lebedkin, K. Arnold, F. Hennrich, R. Krupke, B. Renker and M. M. Kappes, New J. Phys., 2003, 5, 140 CrossRef.
- M. Jones, C. Engtrakul, W. K. Metzger, R. J. Ellingson, A. J. Nozik, M. J. Heben and G. Rumbles, Phys. Rev. B: Condens. Matter Mater. Phys., 2005, 71, 1–9 Search PubMed.
- J. Crochet, M. Clemens and T. Hertel, J. Am. Chem. Soc., 2007, 129, 8058–8059 CrossRef CAS PubMed.
- D. P. Salem, X. Gong, A. T. Liu, V. B. Koman, J. Dong and M. S. Strano, J. Am. Chem. Soc., 2017, 139, 16791–16802 CrossRef CAS PubMed.
- A. J. Gillen, J. Kupis-Rozmysłowicz, C. Gigli, N. Schuergers and A. A. Boghossian, J. Phys. Chem. Lett., 2018, 9, 4336–4343 CrossRef CAS PubMed.
- Y. Zheng, S. M. Bachilo and R. B. Weisman, J. Phys. Chem. Lett., 2017, 8, 1952–1955 CrossRef CAS PubMed.
- M. L. Mayo, Z. Q. Chen and S. V. Kilina, J. Phys. Chem. Lett., 2012, 3, 2790–2797 CrossRef CAS.
- M. A. Lee, F. T. Nguyen, K. Scott, N. Y. L. Chan, N. A. Bakh, K. K. Jones, C. Pham, P. Garcia-Salinas, D. Garcia-Parraga, A. Fahlman, V. Marco, V. B. Koman, R. J. Oliver, L. W. Hopkins, C. Rubio, R. P. Wilson, M. G. Meekan, C. M. Duarte and M. S. Strano, ACS Sens., 2019, 4, 32–43 CrossRef CAS PubMed.
- F. Sen, A. A. Boghossian, S. Sen, Z. W. Ulissi, J. Zhang and M. S. Strano, ACS Nano, 2012, 6, 10632–10645 CrossRef CAS PubMed.
- A. J. Lee, X. Wang, L. J. Carlson, J. a. Smyder, B. Loesch, X. Tu, M. Zheng and T. D. Krauss, Nano Lett., 2011, 11, 1636–1640 CrossRef CAS PubMed.
- E. Polo and S. Kruss, J. Phys. Chem. C, 2016, 120, 3061–3070 CrossRef CAS.
- J. Yang, Q. Zhao, M. Lyu, Z. Zhang, X. Wang, M. Wang, Z. Gao and Y. Li, Small, 2016, 12, 3164–3171 CrossRef CAS PubMed.
- T. Shiraki, T. Shiraishi, G. Juhász and N. Nakashima, Sci. Rep., 2016, 6, 1–9 CrossRef PubMed.
- K. D. Held and D. C. Melder, Radiat. Res., 1987, 112, 544–554 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c8cc08670b |
| This journal is © The Royal Society of Chemistry 2019 |