A facile two-photon fluorescent probe: an endoplasmic reticulum tracker monitoring ER stress and vesicular transport to lysosomes†
Abstract
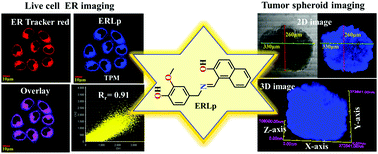
The morphological divergence of the Endoplasmic Reticulum (ER) during stress is a powerful indicator of several diseases. A new two-photon, non-cytotoxic, fluorescent probe (ERLp) was designed and synthesized in a facile manner for selective tracking of the Endoplasmic Reticulum (ER) with a high Pearson co-localization coefficient (0.91) in live cells and tumor spheroids. Further, ER stress during cell apoptosis and vesicular transport from the ER to the lysosomal compartment were also explored by employing ERLp. Therefore, ERLp can be used as a potent tool for examining vesicle transport or ER stress associated diseases in real time.


 Please wait while we load your content...
Please wait while we load your content...