Open Access Article
Open Access ArticleThe sulfation of biomimetic glycosaminoglycan substrates controls binding of growth factors and subsequent neural and glial cell growth†
Waddah
Malaeb‡
a,
Hisham F.
Bahmad‡
 b,
Wassim
Abou-Kheir
b,
Wassim
Abou-Kheir
 *b and
Rami
Mhanna
*b and
Rami
Mhanna
 *a
*a
aBiomedical Engineering Program, Maroun Semaan Faculty of Engineering and Architecture, American University of Beirut, Beirut 1107 2020, Lebanon. E-mail: rm136@aub.edu.lb
bDepartment of Anatomy, Cell Biology and Physiological Sciences, Faculty of Medicine, American University of Beirut, Beirut, Lebanon. E-mail: wa12@aub.edu.lb
First published on 6th August 2019
Abstract
Sulfated glycosaminoglycans (GAGs) are key structural and functional extracellular matrix (ECM) molecules involved in numerous signaling pathways mainly through their interaction with growth factors. Alginate sulfate mimics sulfated GAGs and binds growth factors such as basic fibroblast growth factor (FGF-2). Here, natural biomimetic substrates were engineered by immobilizing biotinylated alginate sulfates with varying degrees of sulfation (DS, from 0 to 2.7) on gold and polystyrene substrates using biotin–streptavidin binding. The build-up of films and the effect of the DS and biotinylation method on FGF-2 binding were assessed using quartz crystal microbalance with dissipation monitoring (QCM-D) and immunohistochemistry. The role of substrate sulfation and FGF-2 loading on the growth of A172 (human glioblastoma multiforme), SH-SY5Y (human neuroblastoma), and PC-12 (rat pheochromocytoma) cell lines was evaluated in vitro using proliferation and neurite outgrowth assessment. An increase in the DS of alginates resulted in augmented FGF-2 binding as evidenced by higher frequency and dissipation shifts measured with QCM-D and confirmed with immunostaining. All sulfated alginate substrates supported the attachment and growth of neural/glial cell lines better than controls with the highest increase in cell proliferation observed for the highest DS (p < 0.05 for all the cell lines). Moreover, FGF-2 loaded substrates with the highest DS induced the most significant increase in neurite-positive PC-12 cells and average neurite length. The developed biomimetic coatings can be used to functionalize substrates for biosensing applications (e.g. gold substrates) and to induce defined cellular responses via controlled growth factor delivery for basic and applied sciences.
1. Introduction
Glycosaminoglycans (GAGs) are key structural and functional extracellular matrix (ECM) polysaccharides that regulate numerous physiological and biological processes.1,2 GAGs undergo a series of posttranslational modifications in vivo by sulfotransferase and endosulfatase enzymes giving rise to a GAG sulfation code.3,4 The sulfation code has been found to be linked to developmental processes and several pathologies particularly in the brain.5,6 In this context, sulfation patterns of heparan sulfate (HS) and chondroitin sulfate (CS) exhibit various alterations during brain development7–9 and various HS motifs have been associated with the progression of neurodegenerative diseases such as Alzheimer's disease.10Molecular mechanisms that dictate the action of GAGs in biological events are starting to be elucidated and several studies have identified a correlation between GAGs sulfation and binding of growth factors as well as downstream cellular responses.11–14 The binding of fibroblast growth factor (FGF), a family comprising over 20 different proteins, and dimerization with its membrane receptors was shown to be dependent on HS sulfation.15 Moreover, different sulfation codes result in binding to different FGFs and FGF receptors (FGFRs). For instance, binding of FGF-2 to HS requires 2-O-sulfation and not 6-O-sulfation, whereas FGF-10 binding to HS demonstrates the opposite effect.16
Sulfation of HS has been also implicated in the repair of the central nervous system (CNS), particularly after injury and in neurodegenerative diseases.13 Similar to the action of HS is that of CS where neurite growth of hippocampal, dopaminergic and dorsal root ganglia (DRGs) neurons was shown to be promoted on highly sulfated CS-E tetrasaccharide-coated 2D substrates. CS-E stimulates neurite outgrowth by increased binding to brain derived neurotrophic factor (BDNF) and midkine which consequently activate cell surface receptors protein tyrosine phosphatase ζ (PTPζ) and tyrosine kinase B (TrkB).17–19 The highly sulfated CS-E also inhibits excitatory amino acid-induced neuronal cell death.20 Furthermore, sulfation regulates the proliferation and maturation of neural precursor cells in the developing mouse spinal cord21 and stimulates survival and proliferation of brain-derived neural stem cells.22,23
Natural GAGs exhibit a highly heterogeneous sulfation pattern that hampered their use for structure–activity studies and encouraged the synthesis of biomimetic GAGs.15,24,25 The fast degradation rate and high cost of some native GAGs such as HS further incentivize the development of biomimetic GAGs for biomedical applications. Mimetic polysaccharides that can be prepared in large amounts have been proposed as convenient and reliable models for systematic studies to assess the activity of GAGs and for potential use in therapeutic applications.14,26–29 In the current study, alginate, a natural biocompatible polysaccharide extracted from brown seaweed, was used as a starting material. Alginates are commercially available from various sources and are FDA-approved for wound dressing applications in humans.30–32 Alginate is commonly used as a gold standard for 3D cell encapsulation and has been extensively investigated in drug delivery and neural tissue engineering.33 Alginate sulfation can be achieved using sulfating agents including SO3 complexes, sulfuric acid-carbodiimide and chlorosulfonic acid-formamide.34 Most HN-binding growth factors have a high affinity to sulfated alginate; of particular importance is the binding of FGF-2 which is involved in a variety of neural processes.35–38 This suggests that alginate sulfate may have properties similar to HS and thus may be used to answer questions related to neuronal growth and development of nervous tissues. We therefore synthesized alginates with different degrees of sulfation (DSs) and studied the effect of the alginate sulfation degree on FGF-2 binding as its interactions with HN are the best studied so far. The sulfated molecules were then biotinylated and used to prepare thin films on two-dimensional (2D) gold substrates via streptavidin–biotin interactions. The films build-up and binding to FGF-2 were characterized in situ and then validated on polystyrene substrates. The effect of the sulfated nanofilms on the behavior of neuroblastoma, glioblastoma and pheochromocytoma cell lines was also studied.
2. Materials and methods
2.1. Materials
Dulbecco's Modified Eagle Media (DMEM) Ham's F-12, fetal bovine serum (FBS), penicillin/streptomycin, trypsin/EDTA, phosphate buffer saline (PBS), Alexa 488 goat anti-mouse conjugated IgG IgM (H + L), formaldehyde, heparin sodium salt (HN), sodium chloride (NaCl), DOWEX ion exchanger, tetrabutyl ammonium bromide, SO3/pyridine, cycloheximide, anhydrous dimethylformamide (DMF), hydrochloric acid (HCl), immunoglobin G (IgG), bovine serum albumin (BSA), sodium dodecyl chloride (SDS), and Triton X100 were purchased from Sigma Aldrich. A172 (human glioblastoma multiforme), SH-SY5Y (human neuroblastoma), and PC-12 (rat pheochromocytoma) cells were from ATCC, USA. Plasmocin™ prophylactic was obtained from InvivoGen, USA. Sodium alginate (Pronova UP LVG, low viscosity, 20–200 mPa s) in which at least 60% of the monomer units are α-L-guluronate was purchased from Novamatrix, Norway. Recombinant human FGF-2 (AF-100-18B), FGF-2 antibody (500-P18), and human FGF ELISA development kit (900-K08) were purchased from Peprotech. Cell strainers were from Becton Dickson. Oligo-ethylene glycol (OEG) disulfides with terminal hydroxyl (dS-OEG, structure: –[S–CH2–(CH2–O–CH2)7–CH2–OH]2, MW: 771.0 Da) and biotin groups (dS-OEG-b, structure: –[S–C2H4–CO–NH–(CH2–O–CH2)9–NH–CO–C4H8–biotin]2, MW: 1539.9 Da) were from Polypure, Norway. Biotinylated bovine serum albumin (b-BSA) was obtained from Thermo Fisher, USA.2.2. Preparation and characterization of sulfated alginate
Alginate sulfates were synthesized as previously described using SO3 as a sulfation agent.38–40 Briefly, alginate was dissolved at 5 mg mL−1 in water with a total volume of 400 mL. The DOWEX Marathon C ion exchanger was charged with an equal mass of tetrabutyl ammonium bromide and mixed with the alginate solution at a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 (w/v). The solution was stirred overnight, filtered and lyophilized. To create alginate sulfates with a DS of 0.8, the generated alginate tetrabutyl ammonium salt was suspended in DMF at a 1% (w/v) with 5-fold excess SO3/pyridine per disaccharide unit and stirred at room temperature for 1 h. For alginates with a DS of 2.0 and 2.7, the ratio of SO3/pyridine was increased to 9 and 15 respectively. The resulting solution was precipitated in acetone then brought to pH = 12 by titrating with ethanolic 0.1 M NaOH for 10 min and neutralized. The final precipitate was then filtered, dissolved in water, purified by dialysis, and lyophilized. The sulfur content was estimated using an automatic elemental analyzer (CHNS-932, Leco, Germany) to determine the DS of the products. The DS denotes the average number of sulfate groups per disaccharide repeating unit which ranges between 0 (native alginate) and 4.0 (all hydroxyl groups substituted by sulfates). The synthesized alginates had DSs = 0.8, 2.0 and 2.7 where non-modified alginate (DS = 0.0) was used as a control. The IR spectra were obtained using an FT-IR Spectrometer TS 175 (BIO RAD, Germany). The NMR spectra were determined in D2O at 343 K using a Bruker Advance 300 MHz spectrometer. Spectral data for the synthesized alginate sulfates were consistent with the literature for materials prepared by different procedures.28,34,37 IR (ATR): 3445, 2946, 1613, 1417, 1232, 1169, 1021, 951, 831, 796 cm−1. 13C-NMR (D2O): 178.1–177.6 (C-6), 102.3–101.8 (C-1), 80.9–69.9 (C-2, C-3, C-4), 66.3 ppm (C-5). Synthesis of alginate sulfate is schematically presented in Fig. S1.† End-on biotinilation of alginates and HN was performed via reductive amination where the reducing end of the polysaccharide was first reacted with biotin-LC-hydrazide, followed by a reduction with NaCNBH3 [synthesized molecules are denoted as b-AlgSulfn (were n represents the DS) or b-HN]. Side-on biotinylation of HN was achieved by 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC) reaction of biotin-LC-hydrazide in MES buffer at room temperature. The degree of substitution by biotin groups was assessed using biotin quantification kits, a fluorescent modified complex of avidin-HABA (40-hydroxyazobenzene-2-carboxylic acid). The biotin content of side-on b-HN used in the current study was 40.48 μg mg−1 (biotin/GAG) while for end-on functionalized samples the biotin content was 0.084 μg mg−1 for b-HN, 0.03 μg mg−1 for b-AlgSulf0.0, 0.013 μg mg−1 for b-AlgSulf0.8, 0.104 μg mg−1 for b-AlgSulf2.0 and 0.17 μg mg−1 for b-AlgSulf2.7.
10 (w/v). The solution was stirred overnight, filtered and lyophilized. To create alginate sulfates with a DS of 0.8, the generated alginate tetrabutyl ammonium salt was suspended in DMF at a 1% (w/v) with 5-fold excess SO3/pyridine per disaccharide unit and stirred at room temperature for 1 h. For alginates with a DS of 2.0 and 2.7, the ratio of SO3/pyridine was increased to 9 and 15 respectively. The resulting solution was precipitated in acetone then brought to pH = 12 by titrating with ethanolic 0.1 M NaOH for 10 min and neutralized. The final precipitate was then filtered, dissolved in water, purified by dialysis, and lyophilized. The sulfur content was estimated using an automatic elemental analyzer (CHNS-932, Leco, Germany) to determine the DS of the products. The DS denotes the average number of sulfate groups per disaccharide repeating unit which ranges between 0 (native alginate) and 4.0 (all hydroxyl groups substituted by sulfates). The synthesized alginates had DSs = 0.8, 2.0 and 2.7 where non-modified alginate (DS = 0.0) was used as a control. The IR spectra were obtained using an FT-IR Spectrometer TS 175 (BIO RAD, Germany). The NMR spectra were determined in D2O at 343 K using a Bruker Advance 300 MHz spectrometer. Spectral data for the synthesized alginate sulfates were consistent with the literature for materials prepared by different procedures.28,34,37 IR (ATR): 3445, 2946, 1613, 1417, 1232, 1169, 1021, 951, 831, 796 cm−1. 13C-NMR (D2O): 178.1–177.6 (C-6), 102.3–101.8 (C-1), 80.9–69.9 (C-2, C-3, C-4), 66.3 ppm (C-5). Synthesis of alginate sulfate is schematically presented in Fig. S1.† End-on biotinilation of alginates and HN was performed via reductive amination where the reducing end of the polysaccharide was first reacted with biotin-LC-hydrazide, followed by a reduction with NaCNBH3 [synthesized molecules are denoted as b-AlgSulfn (were n represents the DS) or b-HN]. Side-on biotinylation of HN was achieved by 1-ethyl-3-(3-dimethylaminopropyl) carbodiimide (EDC) reaction of biotin-LC-hydrazide in MES buffer at room temperature. The degree of substitution by biotin groups was assessed using biotin quantification kits, a fluorescent modified complex of avidin-HABA (40-hydroxyazobenzene-2-carboxylic acid). The biotin content of side-on b-HN used in the current study was 40.48 μg mg−1 (biotin/GAG) while for end-on functionalized samples the biotin content was 0.084 μg mg−1 for b-HN, 0.03 μg mg−1 for b-AlgSulf0.0, 0.013 μg mg−1 for b-AlgSulf0.8, 0.104 μg mg−1 for b-AlgSulf2.0 and 0.17 μg mg−1 for b-AlgSulf2.7.
2.3. Assessment of the build-up of biotinylated alginate sulfate films and effect of their DS on FGF-2 binding using QCM-D
The build-up of biotinylated alginate sulfate films and subsequent FGF-2 binding to the films was assessed using QCM-D (Q-Sense E4, Gothenburg, Sweden). Biotinilation of gold-coated QCM-D crystals was performed as previously described.41 Briefly, crystals were cleaned for 10 min in a 5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 solution of water, 25% ammonia and 30% hydrogen peroxide at 80 °C followed by thorough rinsing with water. The crystals were then dried with an N2 flow and incubated in a 0.5 mM ethanol solution of dS-OEG (99 mol%) and dS-OEG-biotin (1 mol%) overnight (>12 h) at room temperature to form a self-assembled monolayer (SAM). The crystals were then ultrasonicated for 3 min in ethanol to remove loosely bound molecules and washed in PBS 3× for 5 min each. The samples were mounted in the QCM-D flow cell, and after the stabilization of the signal, a 25 μg mL−1 streptavidin (SA) solution in PBS was injected for 10 min (100 μL min−1) with a peristaltic pump. The pump was stopped for 10 min to allow stabilization of the signal and films were rinsed with PBS for 5 min followed by a 5 min of no injection. After rinsing, solutions of b-AlgSulfn in PBS (1 mg mL−1) were injected using the same parameters as for SA. After the polysaccharide layer, a 10 μg mL−1 solution of FGF-2 in PBS was injected for 10 min (50 μL min−1) followed by no injection for 50 min. The films were finally rinsed with PBS 3× to ensure stability. Overtones 1, 3, 5, and 7 were monitored, and the 3rd overtone was used for the assessment of films’ build-up.
1 solution of water, 25% ammonia and 30% hydrogen peroxide at 80 °C followed by thorough rinsing with water. The crystals were then dried with an N2 flow and incubated in a 0.5 mM ethanol solution of dS-OEG (99 mol%) and dS-OEG-biotin (1 mol%) overnight (>12 h) at room temperature to form a self-assembled monolayer (SAM). The crystals were then ultrasonicated for 3 min in ethanol to remove loosely bound molecules and washed in PBS 3× for 5 min each. The samples were mounted in the QCM-D flow cell, and after the stabilization of the signal, a 25 μg mL−1 streptavidin (SA) solution in PBS was injected for 10 min (100 μL min−1) with a peristaltic pump. The pump was stopped for 10 min to allow stabilization of the signal and films were rinsed with PBS for 5 min followed by a 5 min of no injection. After rinsing, solutions of b-AlgSulfn in PBS (1 mg mL−1) were injected using the same parameters as for SA. After the polysaccharide layer, a 10 μg mL−1 solution of FGF-2 in PBS was injected for 10 min (50 μL min−1) followed by no injection for 50 min. The films were finally rinsed with PBS 3× to ensure stability. Overtones 1, 3, 5, and 7 were monitored, and the 3rd overtone was used for the assessment of films’ build-up.
2.4. Studying the role of side-on compared to end-on biotinylation on FGF-2 loading
The effect of the biotinylation mode on FGF-2 loading was determined using HN molecules biotinylated in an end-on or side-on fashion and assessed using QCM-D. Films were prepared as described above where end-on or side-on b-HN molecules were adsorbed as a final layer followed by adsorption of FGF-2. Overtones 1, 3, 5, and 7 were monitored, and the 3rd overtone was used for the assessment of films’ build-up.2.5. Analysis of b-AlgSulfn films build-up and effect of sulfation on FGF-2 adsorption on polystyrene substrates using QCM-D
For cell culture applications, films build-up and FGF-2 adsorption on polystyrene-coated crystals was evaluated using polystyrene coated QCM-D crystals. For these studies, crystals were cleaned for 30 min in 2% SDS, rinsed with deionized water, then UV-treated for 30 min. The crystals were then incubated in a 2 mg mL−1 solution of b-BSA in PBS overnight (>12 h) at 4 °C. The crystals were then mounted in the flow cells and PBS was injected for 20 min (100 μL min−1) and the signal was allowed to stabilize. A 25 μg mL−1 SA solution in PBS was injected for 20 min (50 μL min−1) followed by no flow until the signal stabilized. The films were then rinsed with PBS followed by injection of solutions of b-AlgSulfn in PBS (1 mg mL−1) using the same parameters as for SA. After the polysaccharide layer, a 10 μg mL−1 solution of FGF-2 in PBS was injected for 10 min (50 μL min−1). The flow was stopped and the films were left in contact with the FGF-2 for an additional 50 min then rinsed with PBS 3× to ensure stability. Overtones 1, 3, 5, and 7 were monitored, and the 3rd overtone was used for the assessment of films’ build-up.2.6. FGF-2 immunostaining
Films of b-AlgSulfn and b-HN were prepared on gold-coated substrates and immersed in FGF-2 (2 μg mL−1) for 1 h. Controls without any molecules were also assayed. Substrates were then rinsed in PBS, blocked with BSA (30 min in 3% BSA in PBS), washed again and incubated with a primary antibody anti-FGF-2 (10 μg mL−1 in 1% BSA, PBS). After 1 h, the samples were washed and incubated with a rabbit anti-mouse Alexa Fluor 488 secondary antibody (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 in 1% w/v BSA/PBS) for 1 h. Samples without the primary antibody incubation were assayed to assess non-specific binding of the fluorescent staining. Films were finally washed 3 times with PBS and imaged with an LSM710 laser scanning confocal microscope (Zeiss). The fluorescent intensity of the samples was quantified using the image analysis tools with the Zeiss ZEN 2012 image-analysis software.
500 in 1% w/v BSA/PBS) for 1 h. Samples without the primary antibody incubation were assayed to assess non-specific binding of the fluorescent staining. Films were finally washed 3 times with PBS and imaged with an LSM710 laser scanning confocal microscope (Zeiss). The fluorescent intensity of the samples was quantified using the image analysis tools with the Zeiss ZEN 2012 image-analysis software.
2.7. Cell culture
A172, SH-SY5Y and PC-12 cells were cultured and maintained in DMEM Ham's F-12 cell culture medium supplemented with 10% heat inactivated FBS, 1% penicillin/streptomycin and 5 μg mL−1 Plasmocin™ prophylactic. Cells were incubated at 37 °C in a humidified incubator containing 5% CO2.2.8. Trypan blue cell viability assay
Cells were seeded in duplicates in 24-well plates at a density of 10 × 103 cells per well, in 6 different conditions, on different substrates: (1) b-AlgSulf0.0 + FGF-2 (control), (2) b-AlgSulf0.8 + FGF-2, (3) b-AlgSulf2.0 + FGF-2, (4) end-on b-HN + FGF-2 as positive control, and (5) b-AlgSulf0.0 w/o FGF-2 and (6) b-AlgSulf2.0 w/o FGF-2 as negative controls. Cells were cultured with the predetermined conditions up to 72 h. Viable cells were then collected and counted using trypan blue dye exclusion method. Cell viability was expressed as the ratio of viable cells at 72 h representing cell growth (comparisons were made as fold changes relative to control wells). The data are derived from the mean of duplicate wells of three independent experiments.2.9. Neurite outgrowth assessment and quantification
PC-12 cells were chosen to assess the effect of sulfation on neurite outgrowth. Cells were cultured with the 6 predetermined conditions for 72 h, and then washed once gently with PBS. Photos were taken for assessment and quantification of neurite outgrowth. Images were visualized by an Axiovert inverted microscope at 20× magnification and analyzed by Carl Zeiss Zen 2012 image software. Ten different fields were chosen from three independent experiments, as described by Bahrini et al. (2015),42 and percentage of PC-12 cells with sprouting neurites (referred to as neurite (+) cells) as well as the average length of neurites were quantified by analyzing 100 cells selected from the 10 selected fields (Fig. S4†).2.10. Statistical analysis
Quantitative data were obtained from at least 3 independent experiments and expressed as the mean ± standard error of the mean. The significance of the data was analyzed using the one-way ANOVA followed by Bonferroni's multiple comparison test, or two-way ANOVA followed by post-hoc Tukey's multiple comparison test. P-Values of p < 0.05 (*), p < 0.01 (**) and p < 0.001 (***) were considered significant. Statistical analysis was performed using GraphPad Prism 6 analysis software.3. Results
3.1. Build-up of b-AlgSulfn films on gold substrates and effect of DS on FGF-2 loading
The build-up of b-AlgSulfn films was first investigated on gold substrates and showed that an increase in the DS of the mimetic GAGs increased FGF-2 binding. A schematic of the build-up of the films and FGF-2 adsorption is depicted in Scheme 1. The adsorption of SA caused the same frequency and dissipation shifts for all substrates whereas adsorption of b-AlgSulfn resulted in increased frequency shifts with increasing DS as follows: b-AlgSulf2.7 > b-AlgSulf2.0 > b-AlgSulf0.8 > b-AlgSulf0.0 (Fig. 1a and b, and ESI Table S1†). The dissipation shifts for b-AlgSulf2.7 and b-AlgSulf2.0 were similar and were both higher than those of the lower DS molecules. The adsorption of FGF-2 onto the above films resulted in bigger frequency and dissipation shifts for all the sulfated materials when compared to the non-sulfated alginate (one-way ANOVA, p < 0.001). Frequency shifts of b-AlgSulf2.7, b-AlgSulf2.0 and b-AlgSulf0.8 were higher than b-AlgSulf0.0 control by 5.4-fold (p < 0.001), 5.8-fold (p < 0.001) and 2.4-fold (p < 0.01) respectively. Dissipation shifts showed a similar trend as that of frequency; however, only AlgSulf2.7 and b-AlgSulf2.0 were significantly higher than b-AlgSulf0.0 control (p < 0.001) by ∼12 fold (Fig. 1a and b, and ESI Table S1†).3.2. Assessment of the effect of DS on FGF-2 loading using immunostaining
Immunofluorescent analysis of FGF-2 binding to the constructed b-AlgSulfn films confirmed the QCM-D results where sulfated substrates had a higher fluorescent intensity compared to non-sulfated substrates (Fig. 2a). Quantification of the relative fluorescent intensity (% fluorescence relative to samples w/o 1° antibody) showed a highly significant increase in fluorescence for b-AlgSulf2.0 (p < 0.001) and a significant increase (p < 0.01) for b-AlgSulf2.7 but not for b-AlgSulf0.8 compared to b-AlgSulf0.0 control (Fig. 2b).3.3. Characterization of the build-up of b-AlgSulfn films on polystyrene substrates and effect of DS on FGF-2 loading using QCM-D
The build-up of b-AlgSulfn films on polystyrene substrates was in agreement with those on gold where increased DS resulted in increased adsorption of FGF-2. The frequency and dissipation shifts observed on polystyrene substrates were however less than those on gold (Fig. 3). Before the build-up of the films, we assessed adsorption of b-BSA using QCM-D and immunostaining with avidin-FITC which clearly showed an increase in b-BSA adsorption with increased concentration (ESI Fig. S2 and Table S2†). The fluorescent intensity of avidin-FITC on adsorbed b-BSA was more than 2-fold higher than controls where a bare polystyrene substrate or a substrate to which non-biotinylated BSA was adsorbed (Fig. S2b and S2c†). The adsorption of SA to the b-BSA caused the same frequency and dissipation shifts for the studied substrates. The frequency shift of adsorbed b-AlgSulf2.0 was higher than b-AlgSulf0.0 (control) by 2.5-fold (p < 0.001) (Fig. 3a and b, and ESI Table S3†). The adsorption of FGF-2 onto b-AlgSulf2.0 films was 3.5-fold (p < 0.001) higher compared to the non-sulfated b-AlgSulf0.0 control (Fig. 3a and b, and ESI Table S3†). Finally, adsorption of FGF-2 to the bare polystyrene sensor or to a b-BSA coated sensor was similar to the adsorption to b-AlgSulf0.0 (ESI Fig. S3†).3.4. Effect of side-on vs. end-on biotinylation on FGF-2 loading
The adsorption of FGF-2 to end-on b-HN was significantly higher than side-on b-HN as determined via QCM-D and immunostaining of FGF-2 (Fig. 4). The frequency shift following adsorption of FGF-2 to end-on b-HN was 2-fold higher (p < 0.01) than that of side-on b-HN (Fig. 4a and b). The dissipation shift for end-on b-HN was slightly positive indicating that FGF-2 binds on top of the film while the dissipation shift after FGF-2 binding to end-on b-HN was negative and larger than that of side-on b-HN indicating that FGF-2 penetrates the film and makes it more compact. Immunohistochemical assessment of FGF-2 bound to the side-on and end-on HN films showed higher fluorescence signals for end-on films (p < 0.001) which is in agreement with the QCM-D results (Fig. 4c and d).3.5. SH-SY5Y, A172 and PC-12 cell viability was increased with DS increase from 0.0 to 2.0
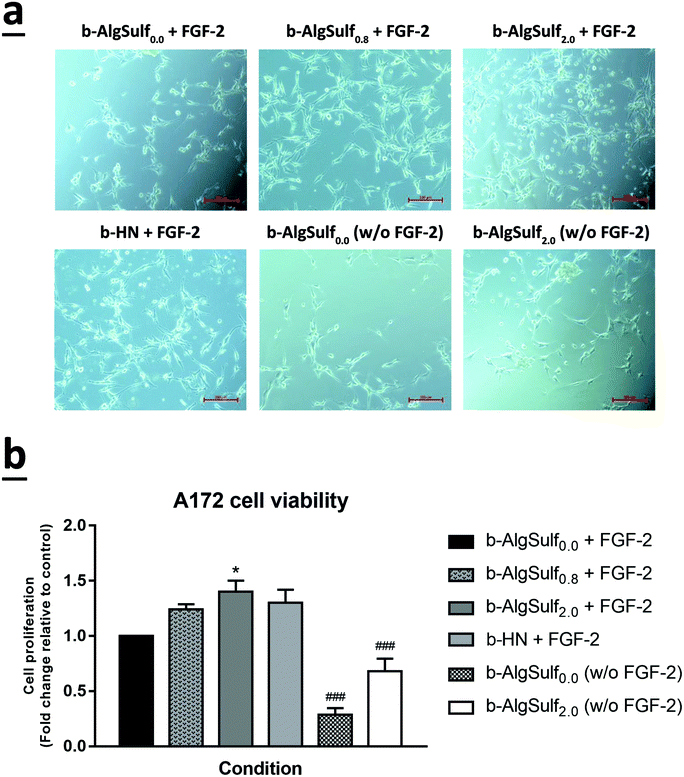
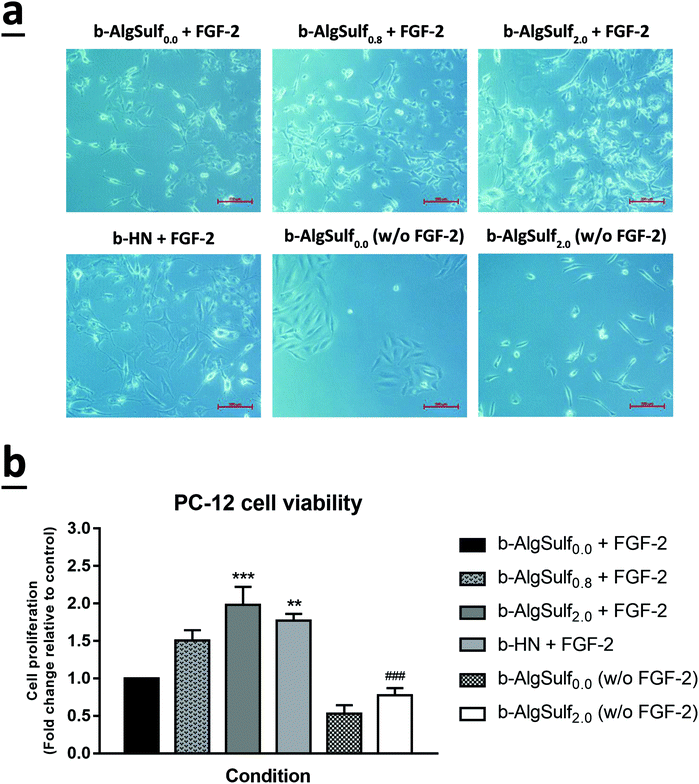
In consistence with the QCM-D results demonstrating that binding of FGF-2 to the sulfated substrates increases with higher DSs, with an optimal DS being 2.0, an increase in cell viability among the three cell lines was observed, at 72 h after plating, with the increase in DSs from 0.0 to 2.0. Accordingly, cells grown on b-AlgSulf2.0 + FGF-2 demonstrated the highest significant increase in cell viability by 2.02 folds for SH-SY5Y (Fig. 5a and b; p < 0.001), 1.40 folds for A172 (Fig. 6a and b; p < 0.05), and 1.98 folds for PC-12 cells (Fig. 7a and b; p < 0.001), followed by b-AlgSulf0.8 + FGF-2 substrates by 1.49 folds for SH-SY5Y (Fig. 5a and b; p < 0.05), 1.24 folds for A172 (Fig. 6a and b), and 1.51 folds for PC-12 cells (Fig. 7a and b), compared to b-AlgSulf0.0 + FGF-2 (control).It is noteworthy mentioning that we also observed a comparable increase in the cell viability in b-HN + FGF-2 substrate (used as positive control) among A172 (Fig. 6a and b) and PC-12 cells (Fig. 7a and b), by 1.30 and 1.77 folds respectively. Interestingly, this increase in cell viability was higher than that observed on b-AlgSulf0.8 + FGF-2 substrates but lower than that of b-AlgSulf2.0 + FGF-2. As for SH-SY5Y cells, cell viability on b-HN + FGF-2 substrates was increased by 1.19 folds, which is higher than that in b-AlgSulf0.0 + FGF-2, but lower than both b-AlgSulf0.8 + FGF-2 and b-AlgSulf2.0 + FGF-2 substrates (Fig. 5a and b).
The three cell lines were also grown on b-AlgSulf0.0 and b-AlgSulf2.0 without FGF-2 to be used as negative controls. Interestingly, our results showed that in both conditions, cell viability was always significantly lower than that observed on b-AlgSulfn + FGF-2 substrates. Additionally, upon plating SH-SY5Y, A172, and PC-12 cells directly on the plates without substrate (with media alone), no significant change in cell viability was noted compared to b-AlgSulf0.0 + FGF-2 (data not shown). Yet, upon addition of FGF-2 directly to the media (media + FGF-2 w/o substrate), there was an increase in cell viability compared to b-AlgSulf0.0 + FGF-2 in all 3 cell lines, which is an expected effect of FGF-2.
3.6. Neurite outgrowth of PC-12 cells is increased with the increase in DS from 0.0 to 2.0
Since PC-12 cell lines represent a well-appreciated model to study development of neurites and synapses,43 we sought to determine the effect of the DS on neurite outgrowth on those cells using the method described by Bahrini et al. (2015).42 Accordingly, the number of neurites longer than one cell body diameter were counted, and their lengths were measured. Cells retaining more than one neurite were referred to as neurite (+) cells, and quantification of the percentage of neurite (+) cells was determined. Average neurite length was also estimated by quantifying the sum of the total neurite length and dividing it by the number of neurites from a single cell.42 In the absence of FGF-2, PC-12 cells either failed to develop neurites or neurites sprouting from some PC-12 cells were degenerated within 48 h after plating (Fig. 8 and ESI Fig. S4†).Our results revealed that the percentage of neurite (+) PC-12 cells increased with the increase in the DS from 73% on b-AlgSulf0.0 + FGF-2 to 78% on b-AlgSulf0.8 + FGF-2 (p = 0.18146) and reaching 87% on b-AlgSulf2.0 + FGF-2 (p < 0.05) but decreased to 8% on b-HN + FGF-2 (p < 0.001) (Fig. 8b). Similarly, the average length of neurites also increased from 58.3 μm on b-AlgSulf0.0 + FGF-2 to 62.3 μm on b-AlgSulf0.8 + FGF-2, and 66.2 μm on b-AlgSulf2.0 + FGF-2, but slightly decreased to 56.2 μm on b-HN + FGF-2 (Fig. 8c).
4. Discussion
Recently, we developed alginate sulfates as sulfated GAG mimics, and showed a direct correlation between their DS and the binding of FGF-2 in solution. We then used the anionic alginate sulfates to construct layer-by-layer (LbL) films by coupling to cationic type I collagen at acidic pH showing that these films can increase FGF-2 loading with increased DS of the anionic pair.38 However, these natural LbL films are known to have an inherent low stability as they rely on electrostatic binding which is further compromised in neutral pH (e.g. cell culture medium). The instability of these LbL films necessitates their chemical crosslinking using toxic molecules which makes the films less appealing for cell-based applications, especially in vivo.44To overcome the film stability and toxicity associated with crosslinked LbL films, we biotinylated sulfated alginates at different DSs and used them to construct biomimetic sulfated GAG films. Unlike the ionic LbL interactions, the biotin–streptavidin bond is rapid, unaffected by extremes of pH, temperature, and denaturing agents and is the strongest non-covalent interaction between a ligand and a receptor, with a Ka ∼1015 M−1.45–47 In addition to increased stability, the biotin–streptavidin films also increase FGF-2 loading compared to LbL films as evident by an increased frequency shift of 20 Hz for similar alginate sulfate DS. The higher adsorption is possibly because LbL films have a stiffer structure similar to films made with side-on biotinylated GAGs which limit growth factor loading. Moreover, in LbL assemblies, electronegative sulfate groups are occupied with binding to positive groups of the polycation which reduces the FGF-2 binding sites. Although LbL films are highly versatile, the need for type I collagen or a cationic polymer, in general, adds complexity to the system and makes the biotin–streptavidin sulfated GAG mimetic films more feasible for biomedical applications and an industrial setting.
We used SO3/pyridine for the alginate sulfation process as it has been shown to allow excellent control of the DS of GAGs.48,49 Alginate sulfate has high affinity to most heparin binding growth factors and therefore the developed films can be used for the loading of a variety of growth factors depending on the target applications.37 The current films are also advantageous over previously developed substrates functionalized with –SO3H/–OH groups by self-assembly which show three times lower frequency shifts.50 While such substrates are simple to prepare, have a well-defined surface chemistry, and are reproducible, they are far from mimicking the native sulfated GAG-FGF-2 binding.
The films were first prepared on gold crystals given the well-established chemistry for stable gold biotinylation and the common use of gold in biosensing applications.41,51,52 The films can therefore be used for biosensing of growth factors which are highly relevant in health and disease. Growth factors are particularly important in cancer diagnostics where increased growth factor production is typically associated with progression and metastasis of breast, colon and prostate cancers among others.53–55 For cell culture applications, it was necessary to assess the build-up of the films using a polystyrene substrate and to change the initial biotinylated layer. We opted to use b-BSA to provide biotin molecules on polystyrene as it has been successfully utilized for similar applications previously and it provides a natural passivation against non-specific binding.56–58 However, to be confident of the polystyrene surface coverage with b-BSA, we evaluated adsorption in situ using QCM-D and confirmed the adsorption by immunostaining with avidin-FITC. Moreover, assessment of the full build-up is necessary as the underlying substrate (gold compared to polystyrene) not only influences initial film adsorption but may influence subsequent layers including FGF-2.44,59
We demonstrated that the higher DS biotinylated alginates (DS = 2.0 and 2.7), bind more FGF-2 than lower (DS = 0.8) or non-sulfated controls which is in agreement with our previous study and other previous reports.14,60,61 The current results indicate that the DS of mimetic GAGs can be used to control growth factor binding and induce predefined cellular responses. Noteworthy was the observation that films with a DS = 2.0 exhibited the highest FGF-2 binding, even higher than DS = 2.7 which is in agreement with previous studies showing the highest mitogenic activity in average sulfation mimetic GAGs.14 We believe that this ideal DS exists because brushes resulting from a film with DS = 2.7 have lower flexibility than those with DS = 2.0 which limits FGF-2 loading. Interestingly, FGF-2 binding to biotinylated alginate at a DS similar to HN (i.e. DS ≥ 2.0) was higher than that of the native GAG. These results are in agreement with our previous work and the study of Freeman et al.37 which showed higher FGF-2 binding to sulfated alginate when compared to native HN pointing out to the potential role of other factors (e.g. steric hindering, molecular weight, and regioselectivity) on growth factor affinity.62 Moreover, the biotinylation method (side-on vs. end-on) affects GAGs biofunctionality and recognition by enzymes due to changes in the primary and secondary structural arrangements of the GAG.63 The role of the biotinylation method on films build-up and growth factor binding was assessed using b-HN prepared in an end-on or side-on fashion. Adsorption of end-on b-HN led to stronger dissipation shifts compared to side-on b-HN indicative of highly hydrated films as shown in previous reports.64 FGF-2 binding to end-on b-HN was higher than binding to side-on b-HN possibly due to the more accessible growth factor binding sites on end-on b-HN films. This finding is in agreement with previous studies showing that structural arrangements of GAGs were preserved in end-on biotinylation unlike in side-on, where major enzymatic recognitions were blocked.65
The sulfation patterns of native GAGs like HS, CS and dermatan sulfate have a known role in promoting neural survival and neurite outgrowth.5,20,23,66–68 However, sulfation of GAGs can also inhibit neurite outgrowth and have been linked to CNS repair and the progression of neuronal diseases such as Alzheimer's and multiple sclerosis.10,13,69,70 The engineered b-AlgSulfn materials and substrates provide a systematic means to study structure–activity relationship and can be used to promote (high DS) or inhibit (low DS) cell and neurite growth. While other approaches proposed the use of small tetrasaccharide or octasaccharide GAG mimetics with success in identifying structure-relation activity,18,19 alginate sulfates have a number of advantages including ease of synthesis, low cost, native ability to form 3D hydrogels, and resistance to enzymatic degradation, hence rendering them highly biocompatible. The ability to form 3D hydrogels using divalent cations (e.g. Ca++, Ba++, Sr++) without the need for further chemical modifications is of high relevance for tissue engineering and regenerative medicine applications and we have been able to grow a variety of primary cells in sulfated alginates including chondrocytes, adipose derived stem cells cortical and hippocampal neurons.
Here, we showed that the effects of the sulfation degree on cell behavior were most evident upon FGF-2 supplementation, where samples that were not loaded with FGF-2 had no significant differences in cell proliferation or neurite regulation. This is in agreement with the hypothesis that sulfation induces its effects through binding to growth factors.11–16,71 The QCM-D results also showed that the dissipation shifts for b-AlgSulf2.7 and b-AlgSulf2.0 were higher than those of the lower DS molecules. The high DS alginate sulfates are expected to adsorb in a brush-like configuration compared to a more mushroom-like configuration for the low DS alginates resulting in higher dissipation for the higher DS alginates which is in agreement with previous studies.64 The stiffness of natural films has been shown earlier to modulate cell growth, where relatively stiff chemically crosslinked layer-by-layer films induced better cell attachment and growth than native films.72 While previously reported dissipation differences between soft and stiff substrates were 10-fold higher than differences between our samples (b-AlgSulf0.0 and b-AlgSulf2.0), the softer b-AlgSulf2.0 films induced better cell attachment than b-AlgSulf0.0 indicating that the chemical cues were more important than mechanical cues. Differences in dissipation were not anymore evident upon FGF-2 supplementation thus ruling out the potential role of substrate stiffness on further cell attachment. Film crosslinking upon supplementation of cell culture media (cell culture media contains 1 mM CaCl2) might also influence surface stiffness. However, the very low concentration of CaCl2 is not likely to cause a significant effect. While FGF-2 is a key neuronal growth factor, other growth factors relevant in neural physiology such as FGF-1, FGF-9, BDNF, transforming growth factor (TGF)-α, TGF-β, insulin-like growth factor and nerve growth factor (NGF) also have a strong affinity to HN and are thus expected to bind to our alginate sulfate substrate.62,73–76 Substrates can be therefore designed to induce more controlled responses or inhibit unwanted processes in the case of disease by binding to growth factors.
We have shown previously that alginate sulfate can be applied in combination with FGF-2 to maintain stemness of stem cells and for regeneration of GAG-rich tissues such as cartilage by three-dimensional encapsulation of chondrocytes.40 Alginate sulfate also induces neurite extension of encapsulated mice primary cortical neurons and supports the proliferation and viability of mesenchymal stem cells.29,40,77 Herein, we showed that the DS of engineered biotinylated alginate sulfate films indeed controls FGF-2 binding. The current results indicate that biotinylated alginate sulfates can be confidently used as sulfated GAG mimics to control growth factor binding and subsequent cell responses including adhesion, differentiation and proliferation. Moreover, defined cellular niches can be constructed to precisely present stabilized growth factors with spatial resolution that dictate required pathways.
5. Conclusion
In conclusion, we engineered biomimetic sulfated GAG substrates using biotinylated sulfated polysaccharides and showed that their DS can be used to control FGF-2 binding. The films were constructed on gold substrates which could be suitable for biosensing and possibly controlled growth factor delivery by coupling to gold particles. We further validated the formation of the films and DS-dependent FGF-2 binding on polystyrene substrates which makes the engineered films suitable for cell culture applications. Substrates engineered using the biomimetic biotinylated alginate sulfate polysaccharides showed an ability to control FGF-2 loading, maintain the growth of several neural/glial cell lines and regulate neurite outgrowth. The versatility of the biotinylated alginate sulfate substrates stems from the multiple structural and functional roles that sulfated GAGs play in health and disease physiology mostly through growth factor and cytokine binding. The developed substrates can thus be used to answer fundamental questions on sulfated GAG affinities to growth factors and cytokines and decipher their underlying effects on cell function. The stability of the biotin–streptavidin bond used herein allows the sulfated substrates to be applied for a wide range of biomedical applications particularly in tissue engineering and neural repair. The ability of the DS to control growth factor binding enables the developed films to be used to increase proliferation, biosynthesis and neurite growth as well as inhibiting cell growth in diseases like cancer.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We acknowledge the Lebanese National Council for Scientific Research (CNRS), the University Research Board and the Collaborative Research Stimulus at the American University of Beirut for funding. We would like to thank all members in the Biomimetics Engineering Laboratory (BEL) and the Abou-Kheir's Laboratory (The WAK Lab) for their help in this work. In addition, we would like to thank all members of the core facilities in the DTS Building at the American University of Beirut (AUB) for their help and support.References
- T. Mikami and H. Kitagawa, Glycoconjugate J., 2017, 34, 725–735 CrossRef CAS PubMed.
- I. Pashkuleva and R. L. Reis, J. Mater. Chem., 2010, 20, 8803–8818 RSC.
- S. Hemmerich, Drug Discovery Today, 2001, 6, 27–35 CrossRef CAS PubMed.
- M. Kusche-Gullberg and L. Kjellén, Curr. Opin. Struct. Biol., 2003, 13, 605–611 CrossRef CAS PubMed.
- A. J. Hayes and J. Melrose, Biochem. J., 2018, 475, 2511–2545 CrossRef CAS PubMed.
- D. S. d. Costa, R. L. Reis and I. Pashkuleva, Annu. Rev. Biomed. Eng., 2017, 19, 1–26 CrossRef PubMed.
- Y. G. Brickman, M. D. Ford, J. T. Gallagher, V. Nurcombe, P. F. Bartlett and J. E. Turnbull, J. Biol. Chem., 1998, 273, 4350–4359 CrossRef CAS PubMed.
- H. Kitagawa, K. Tsutsumi, Y. Tone and K. Sugahara, J. Biol. Chem., 1997, 272, 31377–31381 CrossRef CAS PubMed.
- C. M. Galtrey and J. W. Fawcett, Brain Res. Rev., 2007, 54, 1–18 CrossRef CAS PubMed.
- J.-P. Li, M. L. E. Galvis, F. Gong, X. Zhang, E. Zcharia, S. Metzger, I. Vlodavsky, R. Kisilevsky and U. Lindahl, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 6473–6477 CrossRef CAS PubMed.
- C. Rider, Biochem. Soc. Trans., 2006, 34, 458 CrossRef CAS PubMed.
- M. Lyon, G. Rushton and J. T. Gallagher, J. Biol. Chem., 1997, 272, 18000–18006 CrossRef CAS PubMed.
- J. R. Higginson, S. M. Thompson, A. Santos-Silva, S. E. Guimond, J. E. Turnbull and S. C. Barnett, J. Neurosci., 2012, 32, 15902–15912 CrossRef CAS PubMed.
- A. Weltrowski, M.-L. da Silva Almeida, D. Peschel, K. Zhang, S. Fischer and T. Groth, Macromol. Biosci., 2012, 12, 740–750 CrossRef CAS PubMed.
- C. I. Gama and L. C. Hsieh-Wilson, Curr. Opin. Chem. Biol., 2005, 9, 609–619 CrossRef CAS PubMed.
- S. Ashikari-Hada, H. Habuchi, Y. Kariya, N. Itoh, A. H. Reddi and K. Kimata, J. Biol. Chem., 2004, 279, 12346–12354 CrossRef CAS PubMed.
- C. I. Gama, S. E. Tully, N. Sotogaku, P. M. Clark, M. Rawat, N. Vaidehi, W. A. Goddard, A. Nishi and L. C. Hsieh-Wilson, Nat. Chem. Biol., 2006, 2, 467–473 CrossRef CAS PubMed.
- S. Nadanaka, A. Clement, K. Masayama, A. Faissner and K. Sugahara, J. Biol. Chem., 1998, 273, 3296–3307 CrossRef CAS PubMed.
- S. E. Tully, R. Mabon, C. I. Gama, S. M. Tsai, X. Liu and L. C. Hsieh-Wilson, J. Am. Chem. Soc., 2004, 126, 7736–7737 CrossRef CAS PubMed.
- K. Nakanishi, M. Ito, Y. Sato and A. Oohira, Neurosci. Res., 2012, 74, 223–229 CrossRef CAS PubMed.
- M. Karus, S. Samtleben, C. Busse, T. Tsai, I. D. Dietzel, A. Faissner and S. Wiese, Neural Dev., 2012, 7, 20 CrossRef CAS PubMed.
- M. Ida, T. Shuo, K. Hirano, Y. Tokita, K. Nakanishi, F. Matsui, S. Aono, H. Fujita, Y. Fujiwara, T. Kaji and A. Oohira, J. Biol. Chem., 2006, 281, 5982–5991 CrossRef CAS PubMed.
- M. Tham, S. Ramasamy, H. T. Gan, A. Ramachandran, A. Poonepalli, Y. H. Yu and S. Ahmed, PLoS One, 2010, 5, e15341 CrossRef CAS PubMed.
- L. L. Kiessling and J. C. Grim, Chem. Soc. Rev., 2013, 42, 4476–4491 RSC.
- R. A. Scott and A. Panitch, WIREs Nanomed. Nanobiotechnol., 2013, 5, 388–398 CrossRef CAS PubMed.
- K. T. Adjou, S. Simoneau, N. Salès, F. Lamoury, D. Dormont, D. Papy-Garcia, D. Barritault, J.-P. Deslys and C. I. Lasmézas, J. Gen. Virol., 2003, 84, 2595–2603 CrossRef CAS PubMed.
- D. Papy-Garcia, V. Barbier-Chassefière, V. Rouet, M.-E. Kerros, C. Klochendler, M.-C. Tournaire, D. Barritault, J.-P. Caruelle and E. Petit, Macromolecules, 2005, 38, 4647–4654 CrossRef CAS.
- H. Ronghua, D. Yumin and Y. Jianhong, Carbohydr. Polym., 2003, 52, 19–24 CrossRef.
- M. Askari, S. Bonakdar, M. H. Anbouhi, H. Shahsavarani, S. Kargozar, V. Khalaj and M. A. Shokrgozar, J. Mater. Sci.: Mater. Med., 2019, 30, 7 CrossRef PubMed.
- Y. Suzuki, Y. Nishimura, M. Tanihara, K. Suzuki, T. Nakamura, Y. Shimizu, Y. Yamawaki and Y. Kakimaru, J. Biomed. Mater. Res., 1998, 39, 317–322 CrossRef CAS PubMed.
- B.-S. Kim, C. E. Baez and A. Atala, World J. Urol., 2000, 18, 2–9 CrossRef CAS PubMed.
- N. L. Francis, M. S. Shanbhag, I. Fischer and M. A. Wheatley, J. Microencapsulation, 2011, 28, 353–362 CrossRef CAS PubMed.
- Y.-C. Kuo and Y.-H. Chang, Colloids Surf., B, 2013, 102, 405–411 CrossRef CAS PubMed.
- L. H. Fan, L. Jiang, Y. M. Xu, Y. Zhou, Y. A. Shen, W. G. Xie, Z. H. Long and J. P. Zhou, Carbohydr. Polym., 2011, 83, 1797–1803 CrossRef CAS.
- X. Qian, A. A. Davis, S. K. Goderie and S. Temple, Neuron, 1997, 18, 81–93 CrossRef CAS PubMed.
- T. D. Palmer, J. Ray and F. H. Gage, Mol. Cell. Neurosci., 1995, 6, 474–486 CrossRef CAS PubMed.
- I. Freeman, A. Kedem and S. Cohen, Biomaterials, 2008, 29, 3260–3268 CrossRef CAS PubMed.
- R. Mhanna, J. Becher, M. Schnabelrauch, R. L. Reis and I. Pashkuleva, Adv. Biosyst., 2017, 1, 1700043 CrossRef.
- R. Mhanna, A. Kashyap, G. Palazzolo, Q. Vallmajo-Martin, J. Becher, S. Möller, M. Schnabelrauch and M. Zenobi-Wong, Tissue Eng., Part A, 2014, 20, 1454–1464 CrossRef CAS PubMed.
- M. Zenobi-Wong, G. Palazzolo, R. Mhanna, J. Becher, S. Moller and M. Schnabelrauch, US Pat9937257, 2018 Search PubMed.
- E. Nilebäck, L. Feuz, H. Uddenberg, R. Valiokas and S. Svedhem, Biosens. Bioelectron., 2011, 28, 407–413 CrossRef PubMed.
- I. Bahrini, J.-h. Song, D. Diez and R. Hanayama, Sci. Rep., 2015, 5, 7989 CrossRef CAS PubMed.
- C.-Y. Jeon, J.-K. Jin, Y.-H. Koh, W. Chun, I.-G. Choi, H.-J. Kown, Y.-S. Kim and J.-B. Park, Synapse, 2010, 64, 765–772 CAS.
- R. F. Mhanna, J. Vörös and M. Zenobi-Wong, Biomacromolecules, 2011, 12, 609–616 CrossRef CAS PubMed.
- C. E. Chivers, A. L. Koner, E. D. Lowe and M. Howarth, Biochem. J., 2011, 435, 55–63 CrossRef CAS PubMed.
- M. González, L. A. Bagatolli, I. Echabe, J. L. R. Arrondo, C. E. Argaraña, C. R. Cantor and G. D. Fidelio, J. Biol. Chem., 1997, 272, 11288–11294 CrossRef PubMed.
- P. Weber, D. Ohlendorf, J. Wendoloski and F. Salemme, Science, 1989, 243, 85–88 CrossRef CAS PubMed.
- V. Hintze, S. Moeller, M. Schnabelrauch, S. Bierbaum, M. Viola, H. Worch and D. Scharnweber, Biomacromolecules, 2009, 10, 3290–3297 CrossRef CAS PubMed.
- J. Becher, S. Möller, T. Riemer, J. Schiller, V. Hintze, S. Bierbaum, D. Scharnweber, H. Worch and M. Schnabelrauch, in Functional Materials from Renewable Sources, American Chemical Society, 2012, vol. 1107, ch. 17, pp. 315–328 Search PubMed.
- S. Amorim, R. A. Pires, D. S. d. Costa, R. L. Reis and I. Pashkuleva, Langmuir, 2013, 29, 7983–7992 CrossRef CAS PubMed.
- M. Frasconi, F. Mazzei and T. Ferri, Anal. Bioanal. Chem., 2010, 398, 1545–1564 CrossRef CAS PubMed.
- I. H. El-Sayed, X. Huang and M. A. El-Sayed, Nano Lett., 2005, 5, 829–834 CrossRef CAS PubMed.
- J. M. Chan, M. J. Stampfer, E. Giovannucci, P. H. Gann, J. Ma, P. Wilkinson, C. H. Hennekens and M. Pollak, Science, 1998, 279, 563–566 CrossRef CAS PubMed.
- E. Giovannucci, J. Nutr., 2001, 131, 3109S–3120S CrossRef CAS PubMed.
- M. Zajkowska, E. K. Głażewska, G. E. Będkowska, P. Chorąży, M. Szmitkowski and S. Ławicki, Mediators Inflammation, 2016, 2016, 5962946 Search PubMed.
- F.-B. Wu, S.-Q. Han, T. Xu and Y.-F. He, Anal. Biochem., 2003, 314, 87–96 CrossRef CAS PubMed.
- P. Vincent and D. Samuel, J. Immunol. Methods, 1993, 165, 177–182 CrossRef CAS PubMed.
- E. A. Bayer, H. Ben-Hur and M. Wilchek, Anal. Biochem., 1986, 154, 367–370 CrossRef CAS PubMed.
- O. Guillaume-Gentil, R. Zahn, S. Lindhoud, N. Graf, J. Vörös and T. Zambelli, Soft Matter, 2011, 7, 3861–3871 RSC.
- D. Peschel, K. Zhang, N. Aggarwal, E. Brendler, S. Fischer and T. Groth, Acta Biomater., 2010, 6, 2116–2125 CrossRef CAS PubMed.
- D. Peschel, K. Zhang, S. Fischer and T. Groth, Acta Biomater., 2012, 8, 183–193 CrossRef CAS PubMed.
- J. Angulo, R. Ojeda, J.-L. de Paz, R. Lucas, P. M. Nieto, R. M. Lozano, M. Redondo-Horcajo, G. Giménez-Gallego and M. Martín-Lomas, ChemBioChem, 2004, 5, 55–61 CrossRef CAS PubMed.
- R. I. W. Osmond, W. C. Kett, S. E. Skett and D. R. Coombe, Anal. Biochem., 2002, 310, 199–207 CrossRef CAS PubMed.
- R. P. Richter, K. K. Hock, J. Burkhartsmeyer, H. Boehm, P. Bingen, G. Wang, N. F. Steinmetz, D. J. Evans and J. P. Spatz, J. Am. Chem. Soc., 2007, 129, 5306–5307 CrossRef CAS PubMed.
- N. Altgärde, E. Nilebäck, L. de Battice, I. Pashkuleva, R. L. Reis, J. Becher, S. Möller, M. Schnabelrauch and S. Svedhem, Acta Biomater., 2013, 9, 8158–8166 CrossRef PubMed.
- M. Hikino, T. Mikami, A. Faissner, A. C. E. Vilela-Silva, M. S. Pavão and K. Sugahara, J. Biol. Chem., 2003, 278(44), 43744–43754 CrossRef CAS PubMed.
- K. Sugahara and T. Mikami, Curr. Opin. Struct. Biol., 2007, 17, 536–545 CrossRef CAS PubMed.
- T. Kinnunen, E. Raulo, R. Nolo, M. Maccarana, U. Lindahl and H. Rauvala, J. Biol. Chem., 1996, 271, 2243–2248 CrossRef CAS PubMed.
- T. Laabs, D. Carulli, H. M. Geller and J. W. Fawcett, Curr. Opin. Neurobiol., 2005, 15, 116–120 CrossRef CAS PubMed.
- R. A. Sobel and A. S. Ahmed, J. Neuropathol. Exp. Neurol., 2001, 60, 1198–1207 CrossRef CAS PubMed.
- O. Ostrovsky, B. Berman, J. Gallagher, B. Mulloy, D. G. Fernig, M. Delehedde and D. Ron, J. Biol. Chem., 2002, 277, 2444–2453 CrossRef CAS PubMed.
- L. Richert, F. Boulmedais, P. Lavalle, J. Mutterer, E. Ferreux, G. Decher, P. Schaaf, J.-C. Voegel and C. Picart, Biomacromolecules, 2004, 5, 284–294 CrossRef CAS PubMed.
- S. E. Sakiyama-Elbert and J. A. Hubbell, J. Controlled Release, 2000, 69, 149–158 CrossRef CAS PubMed.
- S. Guimond, M. Maccarana, B. B. Olwin, U. Lindahl and A. C. Rapraeger, J. Biol. Chem., 1993, 268, 23906–23914 CAS.
- S. Nakamura, T. Todo, S. Haga, T. Aizawa, Y. Motoi, A. Ueki, T. Kurokawa and K. Ikeda, Neurosci. Lett., 1997, 221, 181–184 CrossRef CAS PubMed.
- H. A. Cameron, T. G. Hazel and R. D. G. McKay, J. Neurobiol., 1998, 36, 287–306 CrossRef CAS PubMed.
- E. Öztürk, Ø. Arlov, S. Aksel, L. Li, D. M. Ornitz, G. Skjåk-Bræk and M. Zenobi-Wong, Adv. Funct. Mater., 2016, 26, 3649–3662 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/c9bm00964g |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2019 |