Polyester-based ink platform with tunable bioactivity for 3D printing of tissue engineering scaffolds†
Abstract
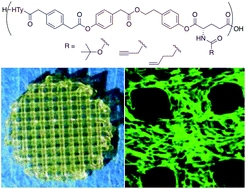
In this work, we synthesized a novel polymeric biomaterial platform with tunable functionalizability for extrusion-based 3D printing. Biodegradable polymers were synthesized using 4-hydroxyphenethyl 2-(4-hydroxyphenyl)acetate (HTy), which is derived from Tyrosol and 2-(4-hydroxyphenyl)acetic acid. p-Phenylenediacetic acid (PDA) was introduced to enhance crystallinity. To enable functionalizability without deteriorating printability, glutamic acid derivatives were introduced into the polymer design, forming copolymers including poly(HTy-co-45%PDA-co-5%Gluhexenamide ester) (HP5GH), poly(HTy-co-45%PDA-co-5%Glupentynamide ester) (HP5GP), and poly(HTy-co-45%PDA-co-5%BocGlu ester) (HP5BG). The resulting polymers have: two melting temperatures (125–131 °C and 141–147 °C), Young's moduli of 1.9–2.4 GPa, and print temperatures of 170–190 °C. The molecular weight (Mw) loss due to hydrolytic degradation was gradual with ∼30% Mw retained after 25 weeks for HP5BG, whereas it was much faster for HP5GP and HP5GH with only 18% Mw retained after 8 weeks. HP5GH and HP5GP were successfully functionalized in solution (bulk) or on the surface using click-based chemistry. Finally, the utilization of this novel platform was demonstrated by studying osteogenic differentiation of human mesenchymal stem cells (hMSCs) using 3D printed scaffolds from HP5GP. Scaffolds were functionalized with azide-Heparin (az-Heparin) to bind and deliver bone morphogenetic protein 2 (BMP-2). This sample group significantly enhanced osteogenic differentiation of hMSCs as compared to unfunctionalized scaffolds incubated directly with az-Heparin or BMP-2 prior to cell culture. This novel polymer platform with tunable functionalizability could be utilized for additive manufacturing of biodegradable devices and scaffolds with tailored mechanical and bioactive properties for a wide range of medical applications including bone fixation devices and scaffolds for bone regeneration.
- This article is part of the themed collection: Biomaterials Science Emerging Investigators


 Please wait while we load your content...
Please wait while we load your content...
